Abstract
Methicillin resistant Staphylococcus aureus causing bovine mastitis has been very well investigated worldwide. However, there are only limited reports on the characterization of methicillin resistant and sensitive coagulase negative staphylococci (CoNS) across the globe. Hence, in the present study, we aim to determine the phenotypic traits based on antimicrobial susceptibility profile and genotypic characterization by verifying the presence of resistance determinants, virulence and toxin genes present in the CoNS causing clinical mastitis. We obtained 62 CoNS isolates from 167 mastitic milk samples collected from three different states of India. The 62 isolates comprises of 10 different CoNS species S. sciuri, S. haemolyticus, S. chromogenes, S. saprophyticus, S. xylosus, S. simulans, S. agnetis, S. epidermidis, S. gallinarum, and S. cohinii. Susceptibility screening against 11 antibiotics determined 45.16% isolates as multidrug resistant (resistant to more than two class of antibiotic), 46.74% resistant (one or two antibiotic class) and 8.06% isolates were pan-sensitive (sensitive to all drugs). High resistance was observed against oxacillin and cefoxitin, whereas all isolates were susceptible toward vancomycin and linezolid. Fifty three isolates were methicillin resistant and 9 isolates were sensitive as determined by oxacillin susceptibility assay. The methicillin resistance gene, mecA was found in 95.16% isolates and staphylococcal cassette chromosome mec (SCCmec) typing predominantly revealed Type III (n = 34) and Type V (n = 18). Interestingly, 11.9% of mecA positive isolates were oxacillin susceptible and referred as oxacillin susceptible mecA positive staphylococci (OS-MRS). Additionally, genes encoding for enterotoxin, (sea, seb, seh, see) toxic shock syndrome (tsst), exfoliatin (eta, etb, etd) and virulence (pvl, Y-hlg) were also screened. Of all the genes examined, 67.74% of isolate were positive for the Y-hlg gene, followed by the sea gene in 25.8% whereas in none of the isolates the eta and the etb gene was amplified. The study also highlights the incidence of clinical isolates of CoNS, which are harboring the toxin and the virulence genes rendering them as a more potential threat. This is the first report of animal origin OS-MRS from India, which emphasizes on the inclusion of both the genetic and phenotypic test for proper characterization of CoNS and preventing resistant strain misidentification.
Keywords: coagulase negative staphylococci, bovine mastitis, antibiotic susceptibility, OS-MRS, pvl gene
Introduction
Coagulase negative staphylococci (CoNS) belong to the group of opportunistic pathogen causing infection in humans and animals. In humans, they are associated with endocarditis, septicemia and blood stream infection, etc., whereas in dairy cattle they mainly cause inflammation of the udder resulting in bovine mastitis (Becker et al., 2014). There are two major forms of mastitis in dairy cattle, clinical and sub-clinical mastitis. In clinical mastitis, the symptoms like swelling, redness or hardness of udder, decrease in milk yield with appearance of clots, flakes, pus or change in milk color, etc., are clearly observed, which makes the diagnosis easy. While in sub-clinical mastitis cases, the symptoms are invisible and hence, difficult to diagnose and results in major cost implications. Mastitis greatly affects the animal health negatively impacting the economy of countries like India, which is one of the largest producers of milk in the world (Preethirani et al., 2015). Bovine mastitis is mainly caused by the group of bacterial species belonging to Streptococcus sp. Escherichia coli, and Staphylococci sp., etc. (Krishnamoorthy et al., 2017). CoNS were earlier considered as the minor group of bacteria with less pathogenicity, as they were only reported in sub-clinical mastitis cases and hence, less attention was paid toward them (Pyörälä and Taponen, 2009). However, reports of clinical mastitis infection caused by various CoNS species have now surfaced largely and they have emerged as an important pathogen (Pyörälä and Taponen, 2009; Preethirani et al., 2015; Srednik et al., 2015).
Infections caused by methicillin resistant staphylococci (MRS) are more harmful due to prolonged treatment duration and limited drug options. Recent reports from different parts of the world revealed MRS as an emerging cause of infection and potent threat to the dairy industry and public health due to its zoonotic potential (Luthje and Schwarz, 2006; Walther and Perreten, 2007; Moodley and Guardabassi, 2009; Weiβ et al., 2013). However, methicillin resistant Staphylococcus aureus (MRSA) have been more widely studied and well characterized in comparison to MRS especially in India (Monecke et al., 2007; Fessler et al., 2010; Türkyilmaz et al., 2010). Increased drug exposure and use of antibiotics in animal diseases also poses a hazard to human health whose impact and effect is still not well characterized but may result in emergence of antibiotic resistant strains making the surveillance of antibiotic susceptibility very crucial (Pyörälä and Taponen, 2009; Fessler et al., 2010; Preethirani et al., 2015; Srednik et al., 2015). Another, concern associated with CoNS is that they may also act as reservoirs of antimicrobial resistance genes (Becker et al., 2014), and can transfer the resistance gene into the S. aureus genome leading to the development of new and multidrug resistant strains (Otto, 2012; Vitali et al., 2014).
Therefore, susceptibility profiling of the CoNS clinical isolates will provide us with valuable information regarding the efficacy of the antibiotics used in the field and warn us about the emergence of antibiotic resistant strain. Overall, the knowledge will be helpful to veterinarians for designing an effective treatment regime and to policy makers, which can formulate a better strategy to control bovine mastitis. Another aspect of CoNS biology, which is understudied, is the evaluation of the toxin genes in the clinical isolates. Recently, the presence of toxin genes in CoNS strains has been reported, which may be responsible for food poisoning, especially the enterotoxin genes which are heat stable and may not get destroyed after boiling of milk or proper cooking of dairy products and meat (Veras et al., 2008).
A very little information is available on the CoNS isolates in the context of phenotypic and genotypic traits correlation of MRS, their response against other antimicrobials and the prevalence of virulence or toxin genes. Therefore, the present study aims to molecularly and phenotypically characterize CoNS causing bovine mastitis in India.
Materials and Methods
Isolation and Characterization of CoNS Isolates
Sixty-two CoNS isolates have been isolated from 167 milk samples of cows suffering from clinical bovine mastitis belonging to different farms (organized and unorganized) in three different states of India, Telangana (n = 78), Andhra Pradesh (n = 50) and Tamil Nadu (n = 39). Before milk collection, oral consent was obtained from the farm owners and the professional veterinarian collected samples. Milk samples were cultured in trypticase soy broth (TSB, Himedia, Mumbai, India) and incubated overnight at 37°C, further, the broth was streaked on mannitol salt agar plate. Pinkish-white colonies on the mannitol salt agar plate (Himedia, Mumbai, India) were presumed to be CoNS. Further, confirmation of the CoNS colonies was done by biochemical test [Gram staining kit (Sigma, St. Louis, MO, United States), and catalase test (BD, New Delhi, India)] followed by sequence analysis of the amplified universal 16S rRNA PCR product (>1.2 Kb) for species confirmation. Briefly, in order to perform PCR, genomic DNA was extracted from each isolate using Wizard genomic DNA kit (Promega, Madison, WI, United States). Briefly, 2 mL of overnight grown culture in TSB were pelleted at 10,000 × g for 3 min. The pellet was then washed thrice with 1 × PBS at 8000 × g, 3 min and resuspended in 500 μl of 50 mM EDTA, lysostaphin (100 μg/ml; Sigma, St. Louis, MO, United States) and lysozyme (100 μg/ml; Sigma, St. Louis, MO, United States) for 1 hr at 37°C followed by manufacturer’s instructions. The 16S rRNA gene PCR was carried out according to the conditions described elsewhere (Srinivasan et al., 2015; Mistry et al., 2016).
PCR Assay and SCC mec Typing
All strains were checked for the presence of mecA, mecC, and van A using PCR as described previously (Stegger et al., 2012). Further, SCC mec cassette element classification was done for all mecA positive isolates, as described earlier (Zhang et al., 2005).
Toxin Genes Profiling
Genomic DNA isolated from the CoNS isolates were subjected to amplification of five enterotoxin genes (sea, seb, sed, see, and seh), three exfoliatin gene (eta, etb, and etd), toxic shock syndrome gene (tsst) and two virulence gene (Panton-Valentine leukocidin gene, pvl and Ẏ-hemolysin, Ý-hlg gene). All primers and PCR condition used for toxin genes amplification was performed as described previously (Moore and Lindsay, 2001; Xie et al., 2011; Okolie et al., 2015). Additionally, sequencing of few amplified PCR products was carried in order to confirm the toxin genes sequences. List of primers and their sequences are mentioned in Table 1.
Table 1.
Details of primers used in the study.
| S. No. | Gene name | Sequence | Amplicon Size | Reference |
|---|---|---|---|---|
| 1 | 16S rRNA | F: AGAGTTTGATCCTGGCTCAG | >1.2 Kb | Srinivasan et al., 2015 |
| R:TAC GYT ACC TTG TTA CGA CTT | ||||
| 2 | mecA | F:TCCAGATTACAACTTCACCAGG | 162 bp | Stegger et al., 2012 |
| R:CCACTTCATATCTTGTAACG | ||||
| 3 | mecC | F:GAAAAAAAGGCTTAGAACGCCTC | 138 bp | Stegger et al., 2012 |
| R:GAAGATCTTTTCCGTTTTCAGC | ||||
| 4 | sea | F: ATTAACCGAAGGTTCTGTAGA | 552 bp | Xie et al., 2011 |
| R:TTGCGTAAATCTGAA TT | ||||
| 5 | seb | F:TGTATGTATGGAGGTGTAAC | 270 bp | Xie et al., 2011 |
| R: ATAGTGACGAGTTAGGTA | ||||
| 6 | sed | F:CTAGTTTGGTAATATCTCCT | 317 bp | Xie et al., 2011 |
| R: TAATGCTATATCTTATAGGG | ||||
| 7 | see | F:TAGATAAAGTTAAAACAAGC | 170 bp | Xie et al., 2011 |
| R:TAACTTACCGTGGACCCTTC | ||||
| 8 | seh | F:CACATCATATGCGAAAGCAGA | 617 bp | Xie et al., 2011 |
| R:CCTTTTAAATCATAAATGTCGAATGA | ||||
| 9 | eta | F:ACTGTAGGAGCTAGTGCATTTGT | 190 bp | Xie et al., 2011 |
| R:TGGATACTTTTGTCTATCTTTTTC ATCAAC | ||||
| 10 | etb | F:CAGATAAAGAGCTTTATA CAC ACATTAC | 612 bp | Xie et al., 2011 |
| R:AGTGAACTTATCTTTCTATTG AAAAACACTC | ||||
| 11 | etd | F:CGCAAATACATATGAAGAATCTGA | 452 bp | Xie et al., 2011 |
| R:TGTCACCTTGTTGCAAATCTATAG | ||||
| 12 | tsst | F:TGCAAAAGCATCTACAAACGA | 499 bp | Xie et al., 2011 |
| R:TGTGGATCCGTCATTCATTG | ||||
| 13 | Y-hlg | F:CCAATCCGTTATTAGAAAATGC | 937 bp | Moore and Lindsay, 2001 |
| R:CCATAGACGTAGCAACGGAT | ||||
| 14 | pvl | F:TTACACAGTTAAATATGAAGTGAACTGGA | 118 bp | Okolie et al., 2015 |
| R:AGCAAAAGCAATGCAATTGATG | ||||
Antimicrobial Susceptibility
Disk diffusion and micro-broth dilution assays were used to determine antimicrobial susceptibility of the 62 clinical isolates as per CLSI guidelines. All the cultures were inoculated into Mueller–Hinton Broth (Himedia, Mumbai, India) and incubated overnight at 37°C. The turbidity of the cultures was adjusted to 0.5 McFarland standard (Himedia, Mumbai, India) and was streaked onto Mueller–Hinton Agar (Himedia, Mumbai, India) plates for disk diffusion while Mueller–Hinton broth was used for micro-broth dilution assay. The disk diffusion assay was done for 8 antibiotics; clindamycin (2 μg), erythromycin (15 μg), gentamicin (10 μg), ciprofloxacin (5 μg), tetracycline (30 μg), rifampicin (5 μg), cefoxitin (30 μg), and teicoplanin (30 μg). All the antibiotic disks were procured from Himedia, Mumbai, India. The MICs against oxacillin (Sigma, St. Louis, MO, United States), vancomycin (Sigma, St. Louis, MO, United States) and linezolid (Sigma, St. Louis, MO, United States) were evaluated by micro-broth dilution method using resazurin dye as described previously (Sarker et al., 2007; Mistry et al., 2016). ATCC 29213 and ATCC 25923 were used as a recommended quality control strains for antibiotic susceptibility assays as per CLSI guidelines (Clinical and Laboratory Standards Institute, 2014).
Results
CoNS Species Causing Bovine Mastitis
The 62 clinical isolates obtained belonged to 10 different CoNS species identified were S. sciuri (n = 20), S. haemolyticus (n = 13), and S. chromogenes (n = 10). Few infections were also caused by, S. saprophyticus (n = 6), S. xylosus (n = 5), S. simulans (n = 2), S. agnetis (n = 2), S. epidermidis (n = 2), S. gallinarum (n = 1), and S. cohinii (n = 1).
Antibiotic Resistance Genes PCR and SCCmec Classification
PCR of resistance genes mecA, mecC, and vanA were done for all isolates. In none of the isolates, mecC and vanA were amplified. The mecA gene was detected in 95.16% isolates. SCCmec classification of those 59 isolates revealed type III (n = 34) and type V (n = 18) with seven non-typeable strains.
Toxin Gene Profiling
Toxin and virulence genes were profiled in 62 CoNS isolates. The sea gene was amplified in 9.16% isolates belonging to five different species, S. sciuri (n = 6), S. haemolyticus (n = 7), S. xylosus (n = 1), S. saprophyticus (n = 1), and S. chromogenes (n = 1). The amplification of the seh gene was observed in three S. chromogenes, two S. haemolyticus followed by one isolate belonging to S. saprophyticus, S. epidermidis. Amplification for the seb gene was observed in single isolate of S. sciuri while in one S. chromogenes isolate the see gene was positive.
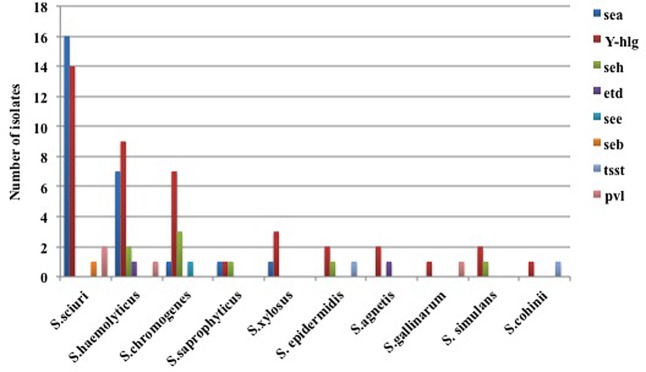
Out of the three exfoliatin genes, the etd gene was amplified in two isolates (S. agnetis and S. haemolyticus) while the eta and the etb were not amplified. The tsst gene was amplified in only two isolates belonging to S. agnetis and S. haemolyticus species. The virulence genes; Ý-hlg were positive in 42 clinical isolates belonging to all species with predominance of S. sciuri (n = 14) and S. haemolyticus (n = 9) whereas only four isolates, two of S. sciuri and one each of S. haemolyticus and S. gallinarum were positive for the pvl gene (Figure 1).
FIGURE 1.
Presence of toxin genes in the clinical isolates of coagulase negative staphylococci (CoNS). The toxin genes sea, seb, see, seh, etd, tsst, Ý-hlg, and pvl were screened using PCR. The bar indicates the different species of CoNS showing amplification of respective toxin genes.
Antimicrobial Susceptibility Profile
A high resistance was observed toward oxacillin 85.5% (53/62, 83.9%) and cefoxitin (53/62, 83.9%) and moderate resistance were seen against rifampicin (23/62, 37.1%), clindamycin (20/62, 32.3%), erythromycin (16/62, 25.8%), and tetracycline (13/62, 20.9%) (Figure 2). Resistance against ciprofloxacin (7/62, 11.3%) and gentamycin (6/62, 9.7%) were low, while all strains were susceptible to vancomycin, teicoplanin and linezolid (Table 2). Overall, susceptibility profile revealed 45.16% isolates as multidrug resistant (resistant to three or more class of antibiotics), 46.74% as resistant and only 8.06% as pan sensitive (susceptible to all drugs).
FIGURE 2.
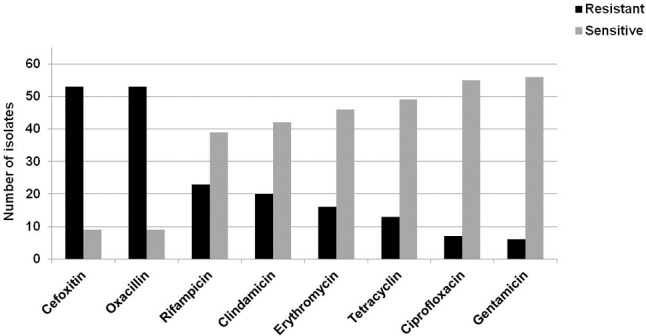
Antibiotic susceptibility profiling of the clinical isolates of CoNS causing bovine mastitis. Antibiotic susceptibility profiling of all the 62 strains was done against oxacillin, clindamycin, erythromycin, gentamicin, ciprofloxacin, tetracycline, and rifampicin. The bars represent the number of isolates sensitive or resistant strain against the respective antibiotic.
Table 2.
Characterization of the coagulase negative staphylococci clinical isolates causing bovine mastitis.
| Name of the Species | Regional Distribution | Number of Isolates | Multidrug Resistant | Resistant | Sensitive | Toxin genes | Virulence genes |
|---|---|---|---|---|---|---|---|
| S. sciuri | Telangana-14 | 20 | 10 | 10 | 0 | sea, seb | Ý-hlg, pvl |
| Andhra Pradesh-6 | |||||||
| S. haemolyticus | Telangana-2 | 13 | 3 | 9 | 1 | sea, seh, etd | Ý-hlg, pvl |
| Andhra Pradesh-7 | |||||||
| Tamil Nadu-4 | |||||||
| S. chromogenes | Telangana-4 | 10 | 7 | 1 | 2 | sea, seh, see | Ý-hlg |
| Tamil Nadu-6 | |||||||
| S. saprophyticus | Telangana-6 | 6 | 1 | 5 | 0 | sea, seh | Ý-hlg |
| S. xylosus | Telangana-5 | 5 | 5 | 0 | 0 | sea | Ý-hlg |
| S. simulans | Tamil Nadu-2 | 2 | 2 | 0 | seh | Ý-hlg | |
| S. agnetis | Telangana-1 | 2 | 1 | 1 | 0 | Etd | Ý-hlg |
| Tamil Nadu-1 | |||||||
| S. epidermidis | Andhra Pradesh-1 | 2 | 1 | 1 | seh, tsst | Ý-hlg | |
| Tamil Nadu-1 | |||||||
| S. gallinarum | Telangana-1 | 1 | 1 | 0 | 0 | – | Ý-hlg, pvl |
| S. cohnii | Telangana-1 | 1 | 0 | 1 | 0 | Tsst | Ý-hlg |
Multidrug resistant refer to CoNS exhibiting resistance against three or more classes of drugs. Resistant refer to CoNS exhibiting resistance against one or two class of drugs. Sensitive refer to CoNS susceptible against all class of drugs.
Further, multidrug resistance phenotype was observed in species of S. chromogenes (n = 7), S. haemolyticus (n = 3), S. sciuri (n = 10), S. xylosus (n = 5), S. saprophyticus (n = 1), S. gallinarum (n = 1), and S. epidermisis (n = 1). Resistance against one or two class of antibiotics was observed in S. sciuri (n = 10), S. haemolyticus (n = 9), S. saprophyticus (n = 5), S. agnetis (n = 1), S. chromogenes (n = 1), S. cohnii (n = 1), and S. simulans (n = 2). Pan sensitive phenotype was observed for the four species, S. chromogenes (n = 2), S. haemolyticus (n = 1), S. agnetis (n = 1), and S. epidermidis (n = 1).
Classification of Oxacillin Susceptible and mecA Positive Strain (OS-MRS)
Oxacillin susceptibility determined 53 oxacillin resistant and 9 oxacillin sensitive isolates. Seven out of the nine susceptible isolates contain the mecA gene representing (OS-MRS) staphylococci. This characteristic was observed among four different species of CoNS, oxacillin susceptible mecA positive S. chromogenes (OS-MRSC, n = 3), oxacillin susceptible mecA positive S. agnetis (OS-MRSAg, n = 2), oxacillin susceptible mecA positive S. sciuri (OS-MRSS, n = 1), and oxacillin susceptible mecA positive S. epidermidis (OS-MRSE, n = 1) (Table 3).
Table 3.
Characteristics of the seven oxacillin susceptible mecA positive Coagulase negative Staphylococci (OS-MRS).
| Isolate ID | Species | Region | Oxacillin MIC (μg/ml) | mecA gene | SCC mec type | Multidrug resistant (MDR) / Resistant (R) / Sensitive (S) | Toxin genes | Virulence genes |
|---|---|---|---|---|---|---|---|---|
| TG-25 | S. chromogenes | Telangana | 0.31 | + | 3 | R | Nil | Nil |
| TG-31 | S. agnetis | Telangana | 0.31 | + | 3 | S | etd | Nil |
| TG-58 | S. sciuri | Telangana | 0.31 | + | 3 | R | sea, seb | Ý-hlg |
| TG-68 | S. chromogenes | Telangana | 0.31 | + | 3 | S | Nil | Ý-hlg |
| TN-29 | S. agnetis | Tamil Nadu | 0.31 | + | 3 | S | Nil | Ý-hlg |
| TN-30 | S. epidermidis | Tamil Nadu | 0.31 | + | 3 | S | seh, tsst | Ý-hlg |
| TN-37 | S. chromogenes | Tamil Nadu | 0.31 | + | 3 | S | seh | Ý-hlg |
Multidrug Resistant refer to CoNS exhibiting resistance against three or more classes of drugs. Resistant refer to CoNS exhibiting resistance against one or two class of drugs. Sensitive refer to CoNS susceptible against all class of drugs.
Overall, susceptibility profiling of the OS-MRS isolates revealed three resistant and four pan-sensitive isolates. OS-MRS belonged to Telangana (n = 4) and Tamil Nadu (n = 3). All OS-MRS isolates belonged to same SCCmec type III. Toxin genes found in OS-MRS isolates were Ý-hlg (n = 7), seh (n = 3), sea (n = 1), etd (n = 1), tsst (n = 1), and pvl (n = 1) genes.
However, we also found an isolate belonging to S. chromogenes species from Tamil Nadu, which was oxacillin resistant and negative for mecA gene.
Discussion
Bovine mastitis is an economically most dangerous disease impacting the livestock industry with a pooled prevalence rate of 41% (sub-clinical mastitis) and 27% (clinical mastitis) from India (Krishnamoorthy et al., 2017). A total loss of 7165.51 crore rupees was estimated due to mastitis in India (Bansal and Gupta, 2009). A recently published meta-analysis report from India for the period of 2005–2016 indicates Staphylococcus sp (45%) to be most predominant mastitis pathogen followed by E. coli (14%) and Streptococcus sp (13%) (Krishnamoorthy et al., 2017). MRSA have been widely studied in India whereas reports of CoNS are limited, however, they have emerged as predominant pathogen now across the world (Pyörälä and Taponen, 2009). In the current study, we have characterized 62 CoNS isolates belonging to 10 different species causing bovine mastitis in India. S. sciuri (32.26%), S. haemolyticus (20.97%), and S. chromogenes (16.13%) were seen as a dominant species causing mastitis which was also observed in a previous finding from India (Preethirani et al., 2015). The oxacillin susceptibility revealed 53 isolates as MRS and 9 isolates as methicillin sensitive staphylococci (MSS). Further, susceptibility profiling against other antibiotics revealed maximum isolates as multidrug resistant or resistant. In the present study, S. sciuri and S. haemolyticus appeared as the most predominant species in which maximum isolates were methicillin resistant. However, a report from Germany revealed S. haemolyticus and S. epidermidis as frequently observed MRS species (Fessler et al., 2010).
Methicillin resistance determinant, mecA was found in 59 isolates out of which 7 were oxacillin susceptible and are referred as OS-MRS. The OS-MRS belonged to four species: OS-MRSC, OS-MRSAg, OS-MRSS, and OS-MRSE. Recently, the first report of oxacillin susceptible mecA positive S. haemolyticus isolate has surfaced from China (Li et al., 2015) causing bovine mastitis while from India no incidence of OS-MRS has been reported till date. These OS-MRS isolates were observed from different farms belonging to Telangana and Tamil Nadu. Overall, CoNS with variable traits make it difficult to characterize it as true MRS on the basis of a single test. It demands the need to include both the phenotypic and the genetic test in order to properly screen MRS isolates. Further, treatment of such isolates with β-lactam antibiotics may result in treatment failure leading to the death of the animal and can develop into more resistant population on exposure to antibiotics. In the present study, we also found an isolate, which was mecA negative and was oxacillin resistant with a MIC value of 10 μg/ml. A similar finding with nine mecA negative and oxacillin resistant CoNS were reported, in which the oxacillin MICs were 0.5 or 1 μg/ml and were determined sensitive using oxacillin or cefoxitin disk diffusion assay (Fessler et al., 2010).
Additionally, we have also screened for toxin genes using PCR in CoNS, which is an important contributing factor in food poisoning. However, to the best of the available knowledge, they are no reports of the toxin genes profiling from CoNS causing bovine mastitis from India. We found enterotoxin genes mainly, the sea and the seh present in 25.8% and 12.9% of CoNS, respectively, which was in line with the previous finding from Brazil where the prevalence of the seh (25.2%) and the sea (18.2%) genes was predominant in CoNS isolates from mastitis cases (Mello et al., 2016). Further, studies on CoNS isolated from food products have also revealed the sea gene as the most frequent enterotoxin gene present and responsible for outbreaks of food poisoning (Argudín et al., 2010; Rall et al., 2010). In the present study, the sea gene was found majorly in clinical isolates of S. haemolyticus and S. sciuri from all the three states, however, the seh gene was found only in isolates of Telangana and Tamil Nadu.
Virulence or the cytotoxin gene; Ý-hlg was found in the higher percentage of isolates amounting to 67.7% as compared to the pvl gene which was present only in 6.5% of the total population. Presence of Ý-hlg gene in the isolates may impart them with increased cytolytic activity. Interestingly, Ý-hlg was present in all the 10 different species dominantly in S. sciuri and S. haemolyticus. The presence of the pvl gene in the CoNS isolates indicates a worrisome situation, as these genes impart enhanced virulence capacity and are mostly reported in CoNS isolates of human origin (Baba et al., 2002; Mistry et al., 2016). The data on the prevalence of the pvl gene in animal origin CoNS isolates is limited, we found 4/62 isolates positive for the pvl gene which is similar to a recent study in which they found 3/81 cows harboring the pvl gene, although all isolates were methicillin sensitive which was not observed in our study (Unal and Cinar, 2012). We also found two isolates to be positive for the tsst gene, which is not reported earlier in CoNS of animal origin. Therefore, it is crucial to screen for these genes, which make them more potent threat and are only well studied in S. aureus causing human and animal infection in comparison to CoNS.
Conclusion
The present report features the emergence of CoNS isolates with variable phenotypes and genotypes, especially the OS-MRS causing bovine mastitis in India. The findings also stressed the need to include both the susceptibility and genetic tests in order to properly characterize and differentiate among MRS and OS-MRS isolates for correct treatment regime. Further, the majority of multidrug resistant CoNS isolates contain virulence genes, which may result in more serious infection state and a worrisome condition for the veterinarian in the field with limited treatment options. Therefore, studies involving screening of the prevailing population is very crucial, as it will provide us with the updated status regarding the emergence of any genotypic or phenotypic variable antibiotic resistant strain. In future, we plan to investigate the resistance mechanism of such emerging variable strains, which will enhance our understanding and provide with useful insight to design new treatment or diagnostic approaches.
Author Contributions
VB conceived and designed the experiment. SM, HM, and VB performed experiment. VB and PS data analysis. VB, PS, and RS provided reagents and sample. VB and PS drafted the manuscript. SC and VB manuscript editing and final preparation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge Veterinary Biological and Research Institute (VBRI), Hyderabad, India and the farm owners from Tamil Nadu, Telangana, and Andhra Pradesh for providing mastitic milk samples.
Footnotes
Funding. The work was funded by DST/INSPIRE Faculty Award/2016/DST/INSPIRE/04/2015/000242 and intramural funds by National Institute of Animal Biotechnology-DBT, Hyderabad.
References
- Argudín M. A., Mendoza M. C., Rodicio M. R. (2010). Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2 1751–1773. 10.3390/toxins2071751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T., Takeuchi F., Kuroda M., Yuzawa H., Aoki K., Oguchi A., et al. (2002). Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359 1819–1827. [DOI] [PubMed] [Google Scholar]
- Bansal B. K., Gupta D. K. (2009). Economic analysis of bovine mastitis in India and Punjab. Ind. J. Dairy Sci. 62 337–345. [Google Scholar]
- Becker K., Heilmann C., Peters G. (2014). Coagulase-negative staphylococci. Clin. Microbiol. Rev. 27 870–926. 10.1128/CMR.00109-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2014). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fourth Informational Supplement M100-S24. Wayne, PA: CLSI. [Google Scholar]
- Fessler A. T., Billerbeck C., Kadlec K., Schwarz S. (2010). Identification and characterization of methicillin-resistant coagulase-negative staphylococci from bovine mastitis. J. Antimicrob. Chemother. 65 1576–1582. 10.1093/jac/dkq172 [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy P., Suresh P. K., Saha S., Govindaraj G., Shome B. R., Roy P. (2017). Meta-analysis of prevalence of subclinical and clinical mastitis, major mastitis pathogens in dairy cattle in India. Int. J. Curr. Microbiol. Appl. Sci. 6 1214–1234. 10.20546/ijcmas.2017.603.141 [DOI] [Google Scholar]
- Li L., Zhou L., Wang L., Xue H., Zhao X. (2015). Characterization of methicillin-resistant and susceptible staphylococcal isolates from bovine milk in northwestern china. PLoS ONE 10:e0116699 10.1371/journal.pone.0116699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthje P., Schwarz S. (2006). Antimicrobial resistance of coagulase-negative staphylococci from bovine subclinical mastitis with particular reference to macrolide–lincosamide resistance phenotypes and genotypes. J. Antimicrob. Chemother. 57 966–969. 10.1093/jac/dkl061 [DOI] [PubMed] [Google Scholar]
- Mello P. L., Moraes Riboli D. F., Pinheiro L., de Almeida Martins L., Vasconcelos Paiva Brito M. A., Ribeiro de Souza da Cunha Mde L. (2016). Detection of enterotoxigenic potential and determination of clonal profile in Staphylococcus aureus and coagulase-negative staphylococci isolated from bovine subclinical mastitis in different Brazilian states. Toxins 8:104 10.3390/toxins8040104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry H., Sharma P., Mahato S., Saravanan R., Kumar P. A., Bhandari V. (2016). Prevalence and characterization of oxacillin susceptible mecA-positive clinical isolates of Staphylococcus aureus causing bovine mastitis in India. PLoS ONE 9:e0162256 10.1371/journal.pone.0162256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke S., Kuhnert P., Hotzel H., Slickers P., Ehricht R. (2007). Microarray based study on virulence-associated genes and resistance determinants of Staphylococcus aureus isolates from cattle. Vet. Microbiol. 125 128–140. 10.1016/j.vetmic.2007.05.016 [DOI] [PubMed] [Google Scholar]
- Moodley A., Guardabassi L. (2009). Clonal spread of methicillin-resistant coagulase negative staphylococci among horses, personnel and environmental sites at equine facilities. Vet. Microbiol. 137 397–401. 10.1016/j.vetmic.2009.01.034 [DOI] [PubMed] [Google Scholar]
- Moore P. C. L., Lindsay J. A. (2001). Genetic variation among hospital isolates of methicillin-sensitive Staphylococcus aureus: evidence for horizontal transfer of virulence genes. J. Clin. Microbiol. 39 2760–2767. 10.1128/JCM.39.8.2760-2767.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okolie C. E., Wooldridge K. G., Turner D. P., Cockayne A., James R. (2015). Development of a heptaplex PCR assay for identification of Staphylococcus aureus and CoNS with simultaneous detection of virulence and antibiotic resistance genes. BMC Microbiol. 15 157 10.1186/s12866-015-0490-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2012). Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection. Bioessays 35 4–11. 10.1002/bies.201200112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preethirani P. L., Isloor S., Sundareshan S., Nuthanalakshmi V., Deepthikiran K., Sinha A. Y., et al. (2015). Isolation, biochemical and molecular identification, and in-vitro antimicrobial resistance patterns of bacteria isolated from bubaline subclinical mastitis in south India. PLoS ONE 10:e0142717 10.1371/journal.pone.0142717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyörälä S., Taponen S. (2009). Coagulase-negative staphylococci-emerging mastitis pathogens. Vet. Microbiol. 134 3–8. 10.1016/j.vetmic.2008.09.015 [DOI] [PubMed] [Google Scholar]
- Rall V. L., Sforcin J. M., de Deus M. F., de Sousa D. C., Camargo C. H., Godinho N. C., et al. (2010). Polymerase chain reaction detection of enterotoxins genes in coagulase-negative staphylococci isolated from Brazilian Minas Cheese. Foodborne Pathog. Dis. 7 1121–1123. 10.1089/fpd.2009.0478 [DOI] [PubMed] [Google Scholar]
- Sarker S. D., Nahar L., Kumarasamy Y. (2007). Microtitre plate-based antibacterial assay incorporating resazurin as an indicator of cell growth, and its application in the in vitro antibacterial screening of phytochemicals. Methods 42 321–324. 10.1016/j.ymeth.2007.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srednik M. E., Grieben M. A., Bentancor A., Gentilini E. R. (2015). Molecular identification of coagulase-negative staphylococci isolated from bovine mastitis and detection of β-lactam resistance. J. Infect. Dev. Ctries. 9 1022–1027. 10.3855/jidc.5871 [DOI] [PubMed] [Google Scholar]
- Srinivasan R., Karaoz U., Volegova M., MacKichan J., Kato-Maeda M., Miller S., et al. (2015). Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS ONE 10:e0117617 10.1371/journal.pone.0117617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegger M., Andersen P. S., Kearns A., Pichon B., Holmes M. A., Edwards G., et al. (2012). Rapid detection differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA (LGA251). Clin. Microbiol. Infect. 18 395–400. 10.1111/j.1469-0691.2011.03715.x [DOI] [PubMed] [Google Scholar]
- Türkyilmaz S., Tekbiyik S., Oryasin E., Bozdogan B. (2010). Molecular epidemiology and antimicrobial resistance mechanisms of methicillin-resistant Staphylococcus aureus isolated from bovine milk. Zoonoses Public Health 57 197–203. 10.1111/j.1863-2378.2009.01257.x [DOI] [PubMed] [Google Scholar]
- Unal N., Cinar O. D. (2012). Detection of stapylococcal enterotoxin, methicillin-resistant and Panton-Valentine leukocidin genes in coagulase-negative staphylococci isolated from cows and ewes with subclinical mastitis. Trop. Anim. Health Prod. 44 369–375. 10.1007/s11250-011-0032-x [DOI] [PubMed] [Google Scholar]
- Veras J. F., do Carmo L. S., Tong L. C., Shupp J. W., Cummings C., Dos Santos D. A., et al. (2008). A study of the enterotoxigenicity of coagulase-negative and coagulase-positive staphylococcal isolates from food poisoning outbreaks in Minas Gerais, Brazil. Int. J. Infect. Dis. 12 410–415. 10.1016/j.ijid.2007.09.018 [DOI] [PubMed] [Google Scholar]
- Vitali L. A., Petrelli D., Lamikanra A., Prenna M., Akinkunmi E. O. (2014). Diversity of antibiotic resistance genes and staphylococcal cassette chromosome mec elements in faecal isolates of coagulase negative staphylococci from Nigeria. BMC Microbiol. 14:106 10.1186/1471-2180-14-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther C., Perreten V. (2007). Methicillin-resistant Staphylococcus epidermidis in organic milk production. J. Dairy Sci. 90 5351 10.3168/jds.2007-0547 [DOI] [PubMed] [Google Scholar]
- Weiβ S., Kadlec K., Fessler A. T., Schwarz S. (2013). Identification and characterization of methicillin-resistant Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus haemolyticus and Staphylococcus pettenkoferi from a small animal clinic. Vet. Microbiol. 167 680–685. 10.1016/j.vetmic.2013.07.036 [DOI] [PubMed] [Google Scholar]
- Xie Y., He Y., Gehring A., Hu Y., Li Q., Tu S. I., et al. (2011). Genotypes and toxin gene profiles of Staphylococcus aureus clinical isolates from China. PLoS ONE 6:e28276 10.1371/journal.pone.0028276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., McClure J. A., Elsayed S., Louie T., Conly J. M. (2005). Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43 5025–5033. 10.1128/JCM.43.10.5026-5033.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]