Abstract Abstract
DNA barcoding and morphological characters were used to identify adult snails belonging to the genus Biomphalaria from 17 municipalities in the state of São Paulo, Brazil. The DNA barcode analysis also included twenty-nine sequences retrieved from GenBank. The final data set of 104 sequences of the mitochondrial cytochrome oxidase I (COI) gene was analyzed for K2P intraspecific and interspecific divergences, through tree-reconstruction methods (Neighbor-Joining, Maximum Likelihood and Bayesian inference), and by applying different models (ABGD, bPTP, GMYC) to partition the sequences according to the pattern of genetic variation. Twenty-seven morphological parameters of internal organs were used to identify specimens. The molecular taxonomy of Biomphalaria agreed with the morphological identification of specimens from the same collection locality. DNA barcoding may therefore be a useful supporting tool for identifying Biomphalaria snails in areas at risk for schistosomiasis.
Keywords: Biomphalaria, COI, DNA barcoding, morphological taxonomy, schistosomiasis, species identification
Introduction
Brazil contains one of the richest faunas of freshwater snails of the genus Biomphalaria (Agostinho et al. 2005, Scholte et al. 2012). The state of São Paulo, in southeast Brazil, is of enormous epidemiological importance, as all the three Neotropical intermediate hosts of Schistosoma mansoni (Sambon, 1907), Biomphalaria glabrata (Say, 1818), B. straminea (Dunker, 1848), B. tenagophila (d’Orbigny, 1835), are distributed in streams, ponds, dams and reservoirs in this municipality. Biomphalaria occidentalis (Paraense, 1981), B. peregrina (d’Orbigny, 1835), B. intermedia (Paraense & Deslandes, 1962), B. oligoza (Paraense, 1975) and B. schrammi (Crosse, 1864) are also distributed in São Paulo state (Vaz 1989; Teles 2005; Ohlweiler et al. 2010).
Identification of Biomphalaria specimens to the species level and analysis of infection by S. mansoni are key elements of surveillance strategies for schistosomiasis control and elimination (PAHO 1968, WHO 2013). Shell morphology is of limited use for identifying different species of snails in this genus (Paraense 1966; Jarne and Théron 2001), and therefore the anatomical characters described by Paraense (1961, 1975, 2001) are used instead. However, identification of Biomphalaria solely based on morphological characters is constrained by phenotypic plasticity, the limited descriptions of cryptic species, and the difficulty in applying species-diagnostic characters to juvenile specimens (Carvalho et al. 2008; Teodoro et al. 2010). The issue of how useful molecular tools may be in the identification of Biomphalaria snails has become particularly important in recent years as there is consensus among malacologists that morphological identification using internal anatomical parameters is susceptible to error, especially when the snails being analyzed belong to complexes of morphologically similar species (Paraense 1972, 1974, 1988; Spatz et al. 1999; Vidigal et al. 2000). To overcome these limitations and difficulties associated with traditional taxonomy, various methodologies based on molecular markers have been developed.
PCR-RFLP analysis of mitochondrial and nuclear genes (Spatz et al. 1999; Vidigal et al. 1998, 2000; Caldeira et al. 2000, 2009), fingerprinting techniques using nonspecific primers (Abdel-Hamid et al. 1999; Al-Quraishy et al. 2014) and sequence analysis of COI and r16RNA genes and ITS-1 and ITS-2 sequences (Woolhouse and Chandiwana 1989; Langand et al. 1998; Vidigal et al. 2000; Campbell et al. 2000; DeJong et al. 2003; Wethington et al. 2007; Tuan and Santos 2007; Tuan et al. 2012) have all produced results that allowed significant genetic differences in species and populations to be identified.
When used in conjunction with bioinformatics tools and sequence databases, DNA barcoding routinely facilitates the identification of biological species (Ratnasingham and Hebert 2007; Casiraghi et al. 2010). This technique is based on the polymorphism of a short region (approximately 600 bp long) of the mitochondrial cytochrome c oxidase 1 (COI) gene (Hebert et al. 2003). DNA barcode includes a series of strategies for delimiting species into molecular operational taxonomic units (MOTUs) using a combination of laboratory and bioinformatics methods (Fontaneto et al. 2013). The most important strategies for identifying MOTUs include analysis of intraspecific and interspecific genetic distances, and analyses based on population and phylogenetic models. These approaches include (ABGD) (Puillandre et al. 2012) and the barcode index number (BIN) system (Ratnasingham and Hebert 2013), which use algorithms based on the partition of molecular data according distance methods, and the generalized mixed Yule coalescent (GMYC) method (Fujisawa and Barraclough 2013) and Bayesian Poisson Tree Processes (bPTP) method (Zhang et al. 2013).
DNA barcoding has been used to augment morphological identification of Bulinus in Africa (Kane at al. 2008; Stothard et al. 2013; Standley et al. 2014), and yielded better results than identifications based on shell characters. Although there are over 500 COI sequences in GenBank from snails of the genus Biomphalaria found in African and Neotropical regions, most DNA barcoding studies use African species. There is therefore a dearth of knowledge about the effectiveness of DNA barcoding in taxonomic identification of Neotropical species of Biomphalaria (Standley et al. 2011; Tuan et al. 2012).
Here, we investigate the utility of analysis of distributions of intraspecific and interspecific COI divergences based on genetic distances, tree reconstruction methods based on Bayesian inference, Maximum Likelihood (ML), and K2P-Neighbor-Joining (NJ) grouping of sequences, and the ABGD, GMYC and bPTP methods for delimitation of Biomphalaria species in conjunction with schistosomiasis field surveys.
Materials and methods
Experimental design
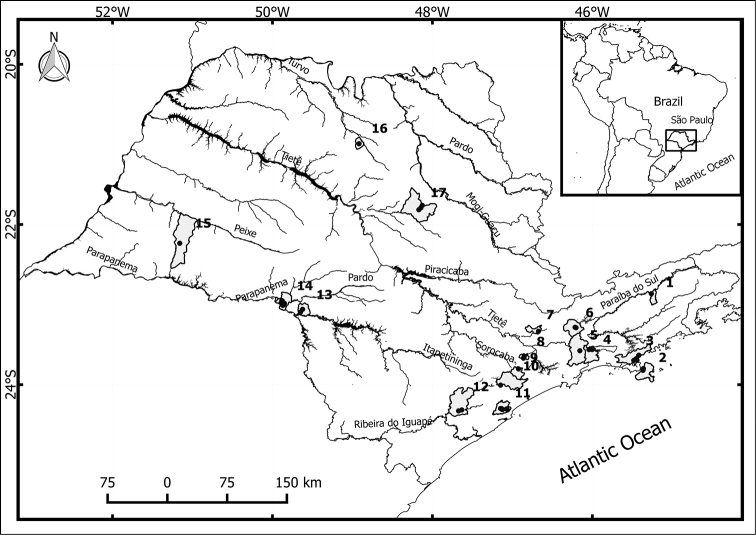
Planorbids were collected in 17 municipalities in the state of São Paulo, Brazil between May 2012 and January 2013 (Fig. 1). The collection points were georeferenced with a Garmin ETrex Summit® GPS (Table 1).
Figure 1.
Locations of the 17 municipalities in São Paulo (Brazil) where the snails were collected. 1 Aparecida 2 Ilhabela 3 Caraguatatuba 4 Biritiba Mirim 5 Mogi das Cruzes 6 Santa Isabel 7 Franco da Rocha 8 Embu das Artes 9 São Lourenço da Serra 10 Juquitiba 11 Itariri 12 Juquiá 13 Ipaussu 14 Ourinhos 15 Martinópolis 16 Novais 17 Araraquara (coordinates are detailed in Table 1).
Table 1.
Collection localities, sample information, and GenBank accession numbers for COI sequences used in this study.
| Sample Sites/ Country | Map locality | Municipality | Latitude; Longitude | COI sequence | GenBank accession number |
|---|---|---|---|---|---|
| São Paulo, Brazil | 1 | Aparecida | 22°51'52.0"S; 45°15'46.0"W | 589, 588, 591 | KF926184, KF926196, KF926186 |
| 2 | Ilhabela |
23°49'17.5"S; 45°22'01.4"W | 564,555 | KF926191, KF926187 | |
| 23°47'56.4"S; 45°21'44.0"W | 593,554 | KF926213, KF926212 | |||
| 3 | Caraguatatuba | 23°37'55.7"S; 45°25'08.7"W | 563 | KF926218 | |
| 23°37'59.6"S; 45°25'11.4"W | 517 | KF926105 | |||
| 23°38'04.2"S; 45°25'14.7"W | 579, 580 | KF926221, KF926217 | |||
| 23°38'3.25"S; 45°25'14.3"W | 516 | KF926106 | |||
| 23°40'26.1"S; 45°26'54.3"W | 592 | KF926215 | |||
| 23°40'42.2"S; 45°27'18.5"W | 568 | KF926214 | |||
| 23°41'34.8"S; 45°26'58.1"W | 569 | KF926216 | |||
| 23°41'46.4"S; 45°28'57.9"W | 565, 571 | KF926219, KF926220 | |||
| 23°41'49.5"S; 45°26'30.8"W | 523 | KF926222 | |||
| 4 | São Paulo | 23°33'43.0"S; 45°59'66.0"W | 551 | KF926204 | |
| 23°33'44.0"S; 46°02'35.0"W | 548, 549 | KF926203, KF926205 | |||
| 5 | 23°33'95.0"S; 46°09'24.0"W | 547 | KF926202 | ||
| 6 | Santa Isabel | 23°17'16.8"S; 46°12'16.1"W | 544 | KF926174 | |
| 23°17'00.2"S; 46°12'59.1"W | 545, 546, 550, 552 | KF926177, KF926189, KF926195, KF926190 | |||
| 7 | Franco da Rocha | 23°20'02.0"S; 46°40'28.0"W | - | ||
| 8 | Embu das Artes | 23°38'50.5"S; 46°51'11.3"W | 524 | KF926197 | |
| 23°40'08.5"S; 46°51'41.7"W | 640 | KF926198 | |||
| 9 | Embu-Guaçu | 23°48'11.0"S; 46°55'27.0"W | 630 | KF926201 | |
| 10 | Juquitiba | 24°00'21.0"S; 47°08'52.0"W | - | ||
| 11 | Itariri | 24°17'53.6"S; 47°08'55.0"W | 537 | KF926188 | |
| 24°17'55.0"S; 47°08'06.8"W | 536 | KF926211 | |||
| 24°18'26.3"S; 47°03'58.9"W | 618 | KF926207 | |||
| 24°18'39.9"S; 47°07'31.4"W | 503 | KF926206 | |||
| 24°18'11.8"S; 47°04'04.1"W | 532, 627, 534 | KF926209, KF926185, KF926208 | |||
| 24°18'13.5"S; 47°04'31.7"W | 535 | KF926210 | |||
| São Paulo, Brazil | 12 | Juquiá | 24°18'55.1"S; 47°37'58.6"W | 650, 651, 653 | KT225577, KT225578, KT225579 |
| 24°19'39.5"S; 47°40'25.0"W | 655 | KT225580 | |||
| 13 | Ipaussu | 23°05'39.6"S; 49°39'01.5"W | 756, 761, 755 | KX354441-KX354442, KX354440 | |
| 14 | Ourinhos | 22°57'00.2"S; 49°52'33.1"W | 764 | KX354435 | |
| 22°58'02.5"S; 49°52'27.1"W | 572, 543, 573 | KF926181, KF926182, KF926183 | |||
| 22°58'03.4"S; 49°52'28.9"W | 735, 733, 766 | KX354437-KX354438, KX354433 | |||
| 22°59'08.0"S; 49°50'59.9"W | 577 | KF926192 | |||
| 22°58'29.5"S; 49°53'29.4"W | 538, 578 | KF926165, KF926193 | |||
| 23°00'24.8"S; 49°51'48.7"W | 739 | KX354444 | |||
| 23°00'32.2"S; 49°52'21.9"W | 763, 765 | KX354436, KX354434 | |||
| 23°00'11.5"S; 49°51'41.4"W | 747 | KX354443 | |||
| 22°57'11.6"S; 49°52'41.9"W | 636, 540 | KF926194, KF926166 | |||
| 22°57'11.6"S; 49°52'41.9"W | 575, 542 | KF926168, KF926167 | |||
| 22°59'42.4"S; 49°52'27.6"W | 541 | KF926178 | |||
| 15 | Martinópolis | 22°14'04.4"S; 51°09'36.4"W | 581, 582 | KX354445, KF926180 | |
| 16 | Novais | 20°59'30.0"S; 48°55'05.0"W | 570, 586, 587 | KF926179, KF926169, KF926171 | |
| 17 | Araraquara | 21°45'37.9"S; 48°07'40.1"W | 595, 599, 601 | KF926170, KF926173, KF926172 | |
| 21°47'30.3"S; 48°08'41.1"W | 594, 596 | KF926199, KF926200 | |||
| 21°48'57.1"S; 48°10'13.1"W | 602 | KF926175 | |||
| Argentina | ARG_1, ARG_2, ARG_3, ARG_4 |
JN621901, JN621902 JN621903, GU168593 |
|||
| Brazil | BRA_1 | AF199090 | |||
| RS_BRA_2, RS_BRA_3, RS_BRA_4, MG_BRA_5, BRA_6, BRA_7, BRA_8, BRA_9 |
KF926107, KF926108 KF926109 AF199094 AF199091 , AF199092, AF199095 , AF199096 |
||||
| RS_BRA_10 | KX354439 | ||||
| BRA_11 | AF199084 | ||||
| Brazil | RS_BRA_12, RS_BRA_13, RS_BRA_14, RS_BRA_15, RS_BRA_16 |
KF926155-KF926156 KX354446-KX354447 KX354448 |
|||
|
BRA_17 RS_BRA_18, RS_BRA_19 |
AF199089 EF433576, NC010220 |
||||
| Egypt | EGY_2, EGY_1 |
DQ084823 AF199111 |
|||
| Hong Kong | HKG | AF199085 | |||
| M-Line * | AY380567 | ||||
| Puerto Rico | PUR | DQ084824 | |||
| Venezuela | VEN | AF199093 |
*M-Line refers to a laboratory strain of Biomphalaria glabrata derived from a Puerto Rico pigmented snail and an albino Brazilian snail (Mulvey and Bandoni 1994).
Samples were collected from freshwater habitats in the Paranapanema, Tietê, Ribeira do Iguape and Paraíba do Sul River basins and the northern coast of São Paulo that had been previously surveyed and classified according to the risk for schistosomiasis transmission as part of a program to monitor snails that are intermediate hosts of S. mansoni (Biomphalaria).
In accordance with the methods described in the Brazilian Ministry of Health Schistosomiasis Surveillance and Control Program (Ministry of Health 2008), snails were collected at sampling stations in each freshwater body and grouped into batches according to their origin. Most of the snails in each batch were then exposed to artificial light in the laboratory to determine whether they were infected with cercariae. At least two specimens from each batch were used for morphological analysis and at least two for the DNA barcode analysis.
DNA barcoding was applied to 75 adult snails taken from samples collected in the field. Only snails that did not have any parasite larvae in their digestive gland and ovotestis were used for molecular identification. Shells were removed by compressing each snail between two slides. After removing the shell fragments, each crushed snail was transferred to a clean Petri dish. The portion of the cephalopodal mass corresponding to the foot was excised under a stereo microscope with forceps and scissors and used as starting material for isolation of total DNA. To maximize the efficiency of genomic DNA purification we used fresh material that had not been fixed. Each specimen was then dissected and identified to the species level based on the presence or absence of the renal ridge and the most informative characters of the male and female copulatory organs. DNA barcoding was carried out in a blind fashion, i.e., without prior knowledge of the general morphological characteristics identified in the animal.
An additional 118 adult specimens were taken from the same field samples (at least two per batch) and scored for 27 morphological characters used by Paraense (1975, 1981, 1984, 2001) in his descriptions of Neotropical species of the genus Biomphalaria. The soft parts were removed from the shell after placing the snails in 70°C for 40 seconds and then fixing them in Railliet-Henry’s solution (distilled water 930 mL, sodium chloride 6 g, formalin 50 mL and glacial acetic acid 20 mL). After at least 24 hours of fixation, the specimens were dissected following Deslandes’ (1951) protocols to examined the renal tube and reproductive system. Specimens were not anesthetized prior to fixation to ensure that the procedure followed was the same as that used in our malacology laboratories.
The longitudinal renal ridge is considered the gold-standard character for differentiating B. glabrata (Paraense and Deslandes 1959) from other species in the genus, in which the ridge is absent. The anterior and posterior regions of the vagina were examined. The proportions for the diameter and length of the oviduct were based on the nidamental gland; for the diameter of the uterus, the cephalic portion of the nidamental gland was used, for the length of the uterus, the posterior region of the vagina; for the length of the spermathecal duct, the body of the spermatheca; and for the length of the anterior region of the vagina, the posterior region of the vagina. The relative proportions of the organs or structures were used for comparisons together with the shell and mantle pigmentation pattern.
DNA extraction, amplification and sequencing
DNA isolation was carried out with the DNeasy Tissue Kit (Qiagen®). A fragment of the COI gene (~600 bp) was amplified with the LCO/HCO primers (Folmer et al. 1994). Polymerase chain reaction (PCR) was carried out in a total volume of 50 µL and the following reaction mixture: 10-100 ng of DNA, 0.2 mM of each dNTP, 0.10 μM of each primer and 1 U of Taq DNA polymerase in the supplied reaction buffer. The cycling conditions consisted of an initial 3 min step at 95°C for denaturation; 25 cycles of 1 min at 95°C, 1 min at 47°C and 1 min 30 s at 72°C and a final extension step of 7 min at 72°C (Tuan et al. 2012). PCR products were purified with a Qiagen purification kit and then sequenced in the Biotechnology Center at the Butantan Institute in an ABI3100 automated sequencer (Applied Biosystems®).
Molecular data analysis
The electropherograms obtained from forward and reverse sequencing of each specimen were corrected using CHROMAS (Technelysium Pty Ltd.) and then aligned with CLUSTALX version 1.8 (Thompson et al. 1997). The aligned sequences were edited with BIOEDIT version 7.0 (Hall 1999), and the general polymorphism of the sequences was calculated in DNAsp version 5 (Librado and Rozas 2009).
The final alignment consisted of a matrix of 75 COI sequences from the collected specimens (36 B. tenagophila, 12 B. occidentalis, 10 B. glabrata, 9 B. straminea, 1 B. intermedia, 7 B. peregrina) and 29 COI sequences of Biomphalaria from other Neotropical areas that were retrieved from GenBank (Table 1).
Intraspecific and interspecific genetic distances (Kimura 1980) were calculated by pairwise comparison of the sequences of all the individuals using the Kimura 2-parameter (K2P) method with the MEGA 6 (Molecular Evolutionary Genetics Analysis) package (Tamura et al. 2013). Three tree-based methods were performed for phylogenetic reconstructions. The K2P distance matrix was used to reconstruct a Neighbor-Joining (NJ) tree. MEGA 6 was also used to perform Maximum Likelihood analysis. In the ML analysis, the GTR+I+G model of sequence evolution was chosen using the Akaike information criterion as implemented in MODELTEST 2.3 (Nylander 2004). The reliability of NJ and ML topologies was evaluated using bootstrap support with 1000 replicates. The parameters estimated by MODELTEST were also used in a Bayesian Markov-Chain Monte Carlo (MCMC) analysis in MRBAYES 3.1 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). Two simultaneous independent searches were run for 1.5 x 106 generations, with trees saved every 100 generations, and the first 1.500 sampled trees of each search discarded as “burn-in”.
The barcode gap analysis was performed with the ABGD (Puillandre et al. 2012), bPTP (Zhang et al. 2013) and GMYC methods (Fujisawa and Barraclough 2013). ABGD, bPTP and GMYC were run on the http://www.abi.snv.jussieu.fr/ public/abgd/, http://species.h-its.org/ and http://species.h-its.org/gmyc/ web servers, respectively, using default parameters.
All the molecular analysis was performed on the 104 sequences (39 B. tenagophila, 23 B. glabrata, 13 B. occidentalis, 11 B. straminea, 12 B. peregrina, 1 B. intermedia, and 5 sequences from B. tenagophila guaibensis) (Table 1). Biomphalaria oligoza was excluded from the analysis because we were unable to amplify its DNA.
Results
Morphological analysis
The morphological identifications of the 118 adult snails that were studied are presented in Table 2. The results of morphological analysis revealed the following: Shell: the presence of a carina, the shape of the whorls and the shape of the shell aperture distinguished B. tenagophila and B. occidentalis from the other species in the group. Renal tube: The presence of renal ridge was observed in all the B. glabrata specimens studied. Pigmentation of the mantle: adult specimens of B. tenagophila, B. glabrata and B. occidentalis had more uniform pigmentation than the four other species studied, which had blotchy pigmentation. Reproductive system: the presence of a vaginal pouch in B. tenagophila and its absence in B. occidentalis differentiates these two species. Biomphalaria straminea and B. intermedia had marked variation in the posterior region of the vagina; in the former, the corrugation in this region was markedly wavy while in the latter it was swollen.
Table 2.
Morphological characters used to identify 118 Biomphalaria specimens from the state of São Paulo.
| Morphological character |
B.
glabrata
(n= 9) |
B.
tenagophila
(n= 56) |
B.
occidentalis
(n= 18) |
B.
oligoza
(n= 10) |
B.
peregrina
(n= 9) |
B.
intermedia
(n=10) |
B.
straminea
(n= 6) |
|---|---|---|---|---|---|---|---|
| Carinate shell | Absent | Present | Present | Absent | Absent | Absent | Absent |
| Shape of the whorls on the shell | Rounded | Angular | Angular | Rounded | Rounded | Rounded | Rounded |
| Shell aperture | Rounded | Transverse, low or deltoid | Transverse, low or deltoid | Rounded | Rounded slightly to the right | Rounded | Rounded |
| Mantle pigmentation | Tends to be homogeneous | Tends to be homogeneous | Tends to be homogeneous | Spotted or blotchy | Spotted or blotchy | Spotted or blotchy | Spotted or blotchy |
| Longitudinal renal ridge | Present | Absent | Absent | Absent | Absent | Absent | Absent |
| Number of ovotestis diverticula | More than 100 | More than 100 | More than 100 | 18 to 37 | More than 100 | Around 60 | More than 100 |
| Shape of the ovotestis diverticula | Elongate, simple or subdivided | Elongate, simple or subdivided | Elongate, simple or subdivided | Bulging and simple | Elongate, simple or subdivided | Elongate, simple or subdivided | Elongate, simple or subdivided |
| Differentiation of the ovotestis diverticula | Weakly differentiated | Weakly differentiated | Weakly differentiated | Well differentiated | Well differentiated | Well differentiated | Well differentiated |
| Diameter of the oviduct | Narrow | Narrow | Narrow | Wide | Narrow | Wide | Wide |
| Length of the oviduct | Long | Long | Long | Short | Long | Long | Short |
| Appearance of the oviduct pouch | Clearly defined | Clearly defined | Clearly defined | Bulky | Clearly defined | Clearly defined | Bulky |
| Diameter of the uterus | Narrow | Narrow | Narrow | Wide | Wide | Narrow | Wide |
| Length of the uterus | Long | Long | Long | Short | Short | Long | Long |
| Length of the anterior region of the vagina | Long | Long | Long | Short | Short | Long | Long |
| Corrugation on the dorsal wall of the posterior region of the vagina | Absent | Absent | Absent | Absent | Absent | Present | Present |
| Type of vaginal Corrugation | - | - | - | - | - | Swollen | Strongly wavy |
| Vaginal pouch on the ventral wall of the posterior region of the vagina | Present | Present | Absent | Present | Present | Present | Absent |
| Shape of the vaginal pouch | Elongate | Bulging | _ | Elongate | Elongate | Elongate | _ |
| Appearance of the vaginal pouch | Clearly defined | Clearly defined | _ | Discrete | Clearly defined | Discrete | _ |
| Length of the spermathecal duct | Long | Long | Long | Short | Short | Long | Long |
| Shape of the prostate diverticula | Tree-like | Tree-like | Tree-like | Simple or subdivided | Tree-like | Tree-like | Tree-like |
| Number of prostate diverticula | Around 30 | Around 30 | Around 20 | 1 to 4 | Around 13 | Around 13 | Around 20 |
| Length of the penial sheath | Approx. the same length as the prepuce | Approx. the same length as the prepuce | Shorter than the prepuce | Approx. the same length as the prepuce | Longer than the prepuce | Approx. the same length as the prepuce | Longer than the prepuce |
| Diameter of the penial sheath | Narrow | Narrow | Narrow | Wide | Wide | Wide | Wide |
| Shape of the prepuce | Free end is wider | Free end is wider | Same diameter along its whole length | Same diameter along its whole length | Free end is wider | Free end is wider | Free end is wider |
| Seminal vesicle extensions | Finger-like | Finger-like | Finger-like | Nodular | Finger-like | Finger-like | Finger-like |
| Appearance of the seminal vesicle | Developed | Developed | Poorly developed | Poorly developed | Developed | Developed | Developed |
Biomphalaria peregrina differed from the species in the B. straminea complex (B. straminea and B. intermedia) in the width of the oviduct, the length of the uterus, the length of the spermathecal duct and the length of the anterior region of the vagina. Biomphalaria intermedia differed from B. straminea in the number of ovotestis diverticula, the length of the oviduct, the presence of an oviduct pouch, the number of prostate diverticula and the width of the uterus.
Biomphalaria oligoza, B. peregrina, B. intermedia and B. straminea are differentiated by the number and shape of the ovotestis diverticula, appearence and size of seminal vesicle, the number and shape of the prostate diverticula, and the shape of the prepuce. The 27 morphological characters used to identify Biomphalaria are detailed in Table 2.
All these findings are in agreement with Paraense and Deslandes (1959), Paraense (1961, 1974, 1975, 1981, 1984).
Molecular analysis
The final alignment matrix for the 104 sequences consisted of 549 characters including 25% polymorphic, 21% parsimony-informative and 12 unique sites (Table 3).
Table 3.
Sample size (N), number of haplotypes (H), haplotype diversity (Hd), nucleotide diversity (π, Nei 1987, equation 10.5) and average number of nucleotide differences (K, Tajima 1983, equation A3) calculated in DNAsp v.5 (Librado and Rozas 2009) for a 549 bp region of the COI gene in the six Biomphalaria species and one Biomphalaria subspecies.
| Species | N | H | Hd | π | K |
|---|---|---|---|---|---|
| Biomphalaria | 104 | 36 | 0.946 | 0.06805 | |
| B. straminea | 11 | 6 | 0.836 | 0.01199 | 6.582 |
| B. occidentalis | 13 | 1 | 0.000 | 0.00000 | 0.000 |
| B. peregrina | 12 | 6 | 0.848 | 0.01954 | 10.727 |
| B. glabrata | 23 | 10 | 0.862 | 0.01914 | 10.506 |
| B. tenagophila | 39 | 11 | 0.803 | 0.01222 | 6.707 |
| B. t. guaibensis | 5 | 1 | 0.000 | 0.00000 | 0.000 |
| B. intermedia | 1 | 1 | - | - | - |
The K2P sequence divergence for intraspecific comparisons ranged from 0.0% to 4.0%, while for interspecific comparisons the corresponding figure varied from 4.0% to 12% (Table 4). The greatest intraspecific genetic distances were observed between specimens of B. peregrina from SP and Rio Grande do Sul (southern Brazil) (4.0%) and specimens of B. glabrata from Rio Grande do Sul and Puerto Rico (3.9%).
Table 4.
Intraspecific and interspecific genetic distances (COI) generated using the Kimura 2-parameter model (K2P, Kimura 1980) in MEGA6 (Tamura et al. 2013).
| Species 1 | Species 2 | Minimum distance | Mean distance | Maximum distance |
|---|---|---|---|---|
| Intraspecific | ||||
| B. glabrata | 0.00 | 0.03 | 0.04 | |
| B. tenagophila | 0.00 | 0.02 | 0.03 | |
| B. straminea | 0.00 | 0.01 | 0.03 | |
| B. occidentalis | 0.00 | 0.00 | 0.00 | |
| B. peregrina | 0.00 | 0.02 | 0.04 | |
| B. intermedia | 0.00 | 0.00 | 0.00 | |
| B. t. guaibensis | 0.00 | 0.00 | 0.00 | |
| Interspecific | ||||
| B. glabrata | B. tenagophila | 0.07 | 0.09 | 0.10 |
| B. straminea | 0.07 | 0.09 | 0.10 | |
| B. occidentalis | 0.09 | 0.09 | 0.09 | |
| B. peregrina | 0.10 | 0.12 | 0.15 | |
| B. intermedia | 0.06 | 0.08 | 0.09 | |
| B. t. guaibensis | 0.07 | 0.09 | 0.09 | |
| B. tenagophila | B. straminea | 0.08 | 0.09 | 0.10 |
| B. occidentalis | 0.04 | 0.05 | 0.06 | |
| B. peregrina | 0.10 | 0.12 | 0.15 | |
| B. intermedia | 0.05 | 0.08 | 0.09 | |
| B. t. guaibensis | 0.04 | 0.04 | 0.05 | |
| B. straminea | B. occidentalis | 0.09 | 0.09 | 0.10 |
| B. peregrina | 0.09 | 0.01 | 0.10 | |
| B. intermedia | 0.05 | 0.05 | 0.06 | |
| B. t. guaibensis | 0.08 | 0.08 | 0.09 | |
| B. occidentalis | B. peregrina | 0.10 | 0.11 | 0.13 |
| B. intermedia | 0.08 | 0.08 | 0.08 | |
| B. t. guaibensis | 0.03 | 0.03 | 0.03 | |
| B. peregrina | B. intermedia | 0.09 | 0.09 | 0.10 |
| B. t. guaibensis | 0.10 | 0.12 | 0.13 | |
| B. intermedia | B. t. guaibensis | 0.08 | 0.08 | 0.08 |
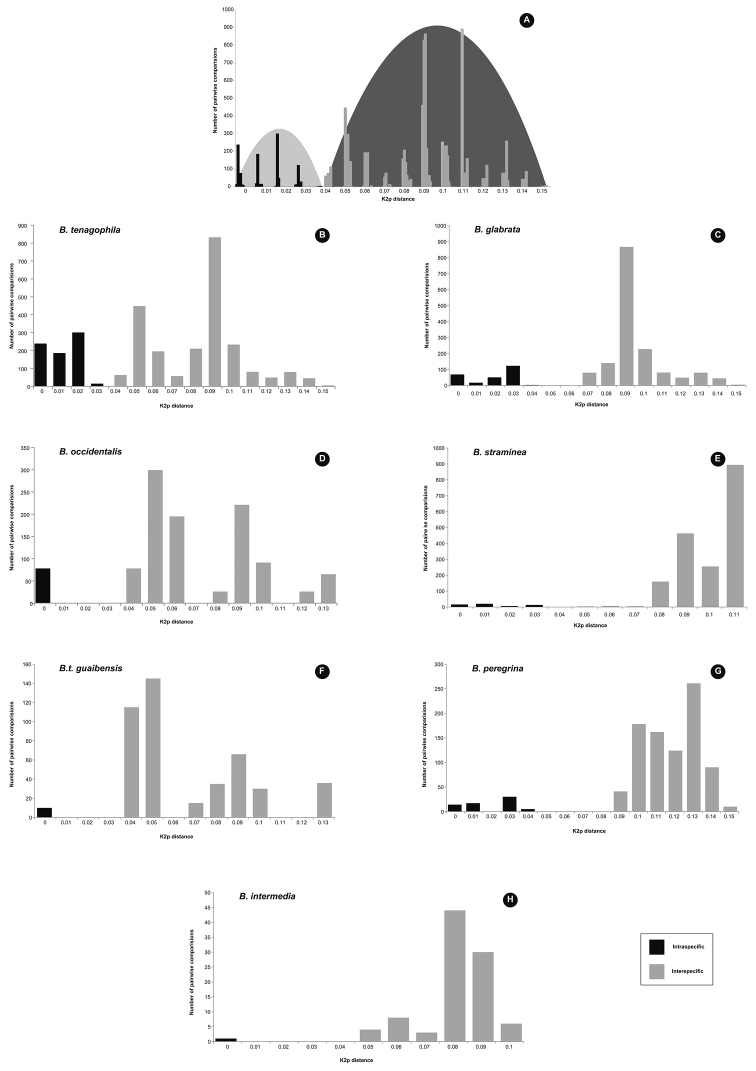
The frequency distribution of the 104 analyzed sequences indicates that although there were some extreme pairwise distances (>3%) in B. glabrata, B. tenagophila, B. peregrina and B. straminea; intraspecific and interspecific divergences did not overlap (Fig. 2A). Nevertheless, a typical barcode gap was not observed in this dataset. A closer inspection of the distances for each taxonomic group shows that there is a clear barcode gap between B. glabrata, B. straminea, B. peregrina and B. intermedia. There was no clear barcode gap between closely related B. tenagophila, B. t. guaibensis and B. occidentalis (interspecific distance 3-4%) (Fig. 2 C, D, E, F).
Figure 2.
A histogram showing pairwise Kimura 2-parameter intraspecific and interspecific distances for 104 Biomphalaria cytochrome oxidase I sequences B–H pairwise distances between each species and the other taxa analyzed.
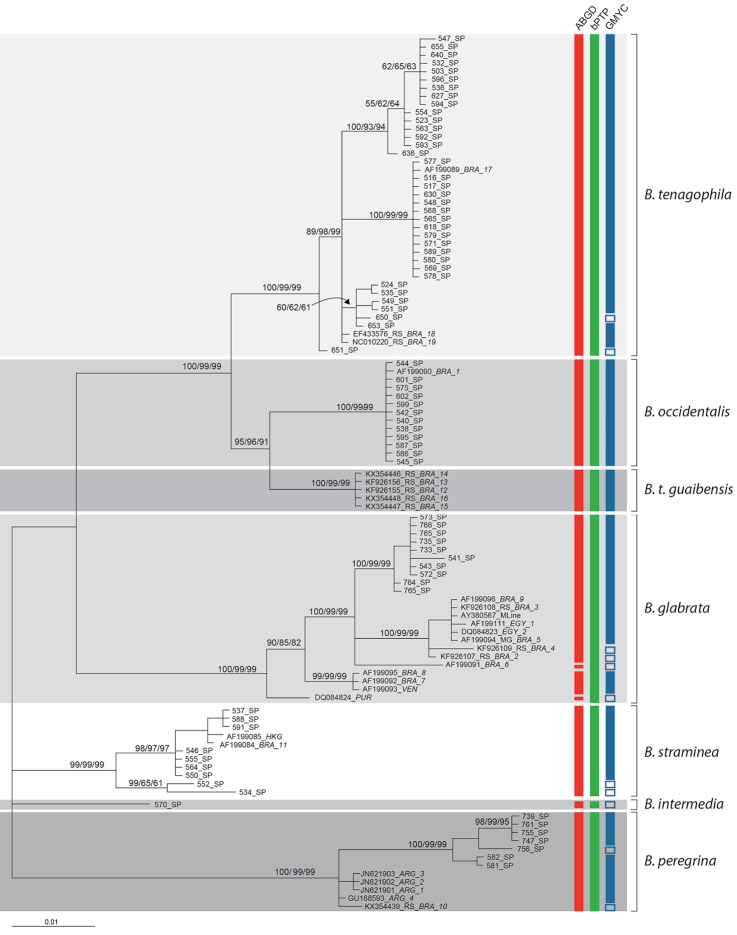
The total number of MOTUs within the same taxon (Fig. 3) varied depending on the model used to partition the COI data (GMYC, bPTP or ABGD). Only bPTP recovered all seven groups identified by traditional morphology. GMYC revealed various sequences that were not consistent with morphological identifications: B. peregrina sequences from Rio Grande do Sul (BRA_10/KX354439) and São Paulo (756/KX354441), B. straminea sequences from Santa Isabel (SP) and Itariri (SP) (552/KF926190, 534/KF926185), one B. intermedia sequence (570/KF926179), two B. tenagophila sequences from Juquiá (SP) and four B. glabrata sequences from GenBank (RS_BRA_2/KF926107, RS_BRA_4/KF926109, BRA_6/AF199091 and PUR/DQ084824).
Figure 3.
Bayesian phylogram. Support values for individual branches are given as Bayesian credibility/ML bootstrap/NJ bootstrap and are depicted above each node. The different shades of gray identify morphological species. The red, green and blue bars indicate species delimitations based on the distance-based (ABGD) and tree-based (bPTP and GMYC) models, respectively.
When run using the default settings, ABGD recovered five different subunits of B. glabrata. This result may be explained by the pronounced genetic variation in this species, but the possibility that these subgroups represent cryptic taxa cannot be ruled out.
The trees generated by the Bayesian, ML and NJ methods (Fig. 3) delineated six well supported groups (posterior probabilities and bootstrap values ≥90) congruent with the current classification of Biomphalaria. The only B. intermedia sequence appeared in a distinct branch supported by low Bayesian and bootstrap values.
Discussion
This study sought to determine the utility of DNA barcoding in delimiting species in freshwater snails of the genus Biomphalaria. The Bayesian, ML and NJ analyses (Fig. 3, Suppl. material 1) yielded trees with well-supported internal branches (≥90), resolving six out of the seven taxa as monophyletic groups.
The assessment of the potential of DNA barcode for species differentiation in Biomphalaria essentially revolves around the comparison of results of the morphological and molecular analysis of closely similar or taxonomically ambiguous species. In the case of the three taxa in the B. tenagophila complex, one character that is normally effective for specific identification is the vaginal pouch, which is present in B. tenagophila and B. t. guaibensis but not in B. occidentalis. (The anatomical features of these three taxa were illustrated by Tuan et al. 2012). Although we did not observe this in our material, in some specimens of B. occidentalis there is a slight projection of the ventral wall of the vagina (Paraense 1981), which raises questions regarding the distinctness of this taxon.
The intraspecific genetic distance within B. tenagophila showed values with a range from 0 to 3% (Table 4, Figs 2, 3). A high level of genetic divergence within this species was obtained for sequences associated with specimens collected in Juquiá (650,651,653), Itariri (535), Embu das Artes (524,535) and São Paulo (549, 551). Due to these values we could not assign a clear barcode gap between B. tenagophila and B. occidentalis and B. t. guaibensis (Fig. 2 b, d, f). However, the Bayesian tree inferred from COI data (Fig. 3), as well as the ABGD and both bPTP and GMYC analyses recovered these close related taxa as distinct groups. We suggest that in geographical areas where B. tenagophila species complex have the same geographical distribution.
The application of the 3-4% cutoff value for maximum intraspecific divergence may be appropriate for our dataset as 36% of the intraspecific comparisons reached this value (Table 4). The highest values for intraspecific divergence (>3%) do not appear to be a consequence of geographic distance given that the greatest divergence in B. tenagophila was between closely proximal localities in São Paulo state (Fig. 3).
Biomphalaria glabrata and B. tenagophila, are differentiated by the renal ridge, which is present in the former and absent in the latter. Paraense and Deslandes (1959) described a false ridge that runs obliquely to the renal tube and is attached to the pneumostome, in specimens of B. tenagophila from Macaé, RJ. The presence of this false ridge in B. tenagophila may lead to incorrectly identify this species, particularly in juvenile specimens or specimens that have not been properly fixed. Five specimens of B. tenagophila in our study (three from São Lourenço da Serra and two from São Paulo) had a membrane on the renal tubes similar to that described by Paraense and Deslandes. The genetic distance of 9% between B. glabrata and B. tenagophila observed with both genetic distance and tree-based approaches show that DNA barcoding is an important tool for identifying these closely similar taxa.
The ABGD analysis partitioned B. glabrata into five distinct groups, while the GMYC analysis yielded a more cohesive group. Despite the pronounced COI divergence within B. glabrata, in all the specimens analyzed here the renal ridge has been considered a robust and consistent taxonomical character, suggesting that morphology is more effective than DNA barcode in this case. However, bBTP analysis and phylogenetic reconstruction supported B. glabrata as single and well supported MOTU, a result congruent with the morphological identification.
Another group of morphologically similar and frequently misidentified congeners includes B. intermedia and B. straminea; the latter a natural intermediate host of S. mansoni. Of the seventeen diagnostic characters common to B. straminea and B. intermedia, the degree of corrugation in the dorsal wall of the vagina has been used to these taxa as a species complex (Paraense 1975) . The vaginal corrugation, which is markedly wavy in B. straminea appears as swollen in B. intermedia. The large genetic divergence between B. straminea and B. intermedia, which was 9% greater than the intraspecific values in both species, indicates that these two species can be identified by DNA barcode. Note, however, that our study only included two of the three species in the B. straminea complex, as B. kuhniana does not occur in São Paulo state (Paraense 1988, Teodoro et al. 2010). In addition, we were unable to collect many specimens of B. intermedia owing to its rareness in São Paulo state.
Our findings show Biomphalaria species delimitation by phylogenetic approaches and bPTP yielded the same groups identified by traditional taxonomy. The use of DNA barcode to identify species in conjunction with Biomphalaria surveys requires the application of both evolutionary and bioinformatics criteria, making it a time-consuming approach that is dependent on specialist knowledge. Morphological identification also requires specialist knowledge. However, as shown in this study DNA barcoding can identify subtle (genetic) differences between intraspecific populations that are not detectable by traditional morphological study.
Furthermore, morphological identification of Biomphalaria species depends on subjective interpretation of anatomical variations, as these are measured in terms of relative rather than absolute sizes. We therefore agree with Hebert and Gregory (2005, p. 853), who stated that by reversing the logic of standard taxonomic approaches that “operate in an a priori fashion—seeking…morphological discontinuities”, DNA barcoding may, as “a posteriori approach”, direct the study of morphological variation in genetically divergent groups of Biomphalaria.
Acknowledgments
This work was supported by a grant from the Superintendency for the Control of Endemic Diseases (SUCEN) (ref. no. 0375/2012) to R. Tuan and M. C. A. Guimarães. The acquisition of equipment for the molecular analysis was funded by FAPESP grants to R. Tuan.
Citation
Palasio RGS, Guimarães MCA, Ohlweiler FP, Tuan R (2017) Molecular and morphological identification of Biomphalaria species from the state of São Paulo, Brazil. ZooKeys 668: 11–32. https://doi.org/10.3897/zookeys.668.10562
Supplementary materials
Figure S1
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Author: Roseli Tuan
Data type: molecular data
Explanation note: ML and NJ trees generated with MEGA 6 software from the alignment of the 104 Biomphalaria COI sequences.
References
- Abdel-Hamid AHZ, Molfetta JBD, Fernandez V, Rodrigues V. (1999) Genetic variation between susceptible and non-susceptible snails to Schistosoma infection using random amplified polymorphic DNA analysis (RAPDs). Revista do Instituto de Medicina Tropical de São Paulo 41(5): 291–295. http://doi.org/10.1590/S0036-46651999000500005 [DOI] [PubMed] [Google Scholar]
- Agostinho AA, Thomaz SM, Gomes LC. (2005) Conservação da biodiversidade em águas continentais do Brasil. Megadiversidade 1(1): 70–78. [Google Scholar]
- Al-Quraishy SA, Dajem SMB, Mostafa OM, Ibrahim EH, Al-Qahtani A. (2014) Development of species-specific primers for identification of Biomphalaria arabica, the intermediate host of Schistosoma mansoni in Saudi Arabia. Saudi Journal of Biological Sciences 21(1): 65–70. http://doi.org/10.1016/j.sjbs.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldeira RL, Vidigal THDA, Martinela L, Simpson AJG, Carvalho OS. (2000) Identification of planorbids from Venezuela by Polymerase Chain Reaction Amplification and Restriction Fragment Length Polymorphism of ITS of the RNA ribosomal gene. Memórias do Instituto Oswaldo Cruz 95: 171–177. http://doi.org/10.1590/S0074-02762000000200007 [DOI] [PubMed] [Google Scholar]
- Caldeira RL, Jannotti-Passos LK, Carvalho OS. (2009) Molecular epidemiology of Brazilian Biomphalaria: A review of the identification of species and the detection of infected snails. Acta Tropica 111: 1–6. http://doi.org/10.1016/j.actatropica.2009.02.004 [DOI] [PubMed] [Google Scholar]
- Campbell G, Jones CS, Lockyer AE, Hughes S, Brown D, Noble LR, Rollinson D. (2000) Molecular evidence supports an African affinity of the Neotropical freshwater gastropod, Biomphalaria glabrata, Say 1818, an intermediate host for Schistosoma mansoni. Proceedings of the Royal Society of London B Biological Sciences 7(267): 2351–2358. http://doi.org/10.1098/rspb.2000.1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho OS, Jannotti-Passos LK, Caldeira RL. (2008) Importância Epidemiológica e Biologia Molecular Aplicada ao Estudo dos moluscos do gênero Biomphalaria. In: Carvalho OS, Zech Coelho PM, Lenzi HL. (Eds) Schistosoma mansoni e esquistossomose: uma visão multidisciplinar. Rio de Janeiro, Brasil. Editora Fiocruz, 311–346. https://doi.org/10.7476/9788575413708
- Casiraghi M, Labra M, Ferri E, Galimberti A, De Mattia F. (2010) DNA barcoding: a six-question tour to improve users’ awareness about the method. Briefings in Bioinformatics 11(4): 440–53. https://doi.org/10.1093/bib/bbq003 [DOI] [PubMed] [Google Scholar]
- DeJong RJ, Morgan JA, Wilson WD, Al-Jaser MH, Appleton CC, Coulibaly G. (2003) Phylogeography of Biomphalaria glabrata and Biomphalaria pfeifferi, important intermediate hosts of Schistosoma mansoni in the New and Old World tropics. Molecular Ecology 12: 3041–3056. https://doi.org/10.1046/j.1365-294X.2003.01977.x [DOI] [PubMed] [Google Scholar]
- Deslandes N. (1951) Técnica de dissecção e exame de planorbídeos. Revista Serviço Especial Saúde Pública 4(2): 371–382. [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3(5): 294–299. [PubMed] [Google Scholar]
- Fontaneto D, Hortal J. (2013) At least some protist species are not ubiquitous. Molecular Ecology 22(20): 5053–5055. https://doi.org/10.1111/mec.12507 [DOI] [PubMed] [Google Scholar]
- Fujisawa T, Barraclough TG. (2013) Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent (GMYC) approach: a revised method and evaluation on simulated datasets. Systematic Biology syt033. https://doi.org/10.1093/sysbio/syt033 [DOI] [PMC free article] [PubMed]
- Guimarães MCDA, Menezes RMTD, Tuan R. (2016) Experimental study on reproduction of the freshwater snail Biomphalaria tenagophila (d’Orbigny, 1835). Invertebrate Reproduction & Development 60(2): 145–151. https://doi.org/10.1080/07924259.2016.1174156 [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 9. Nucleic Acids Symposium Series 41(41): 95–98. [Google Scholar]
- Hebert PD, Ratnasingham S, Waard JR. (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society of London B: Biological Sciences 270(Suppl 1): S96–S99. https://doi.org/10.1098/rsbl.2003.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PD, Gregory TR. (2005) The promise of DNA barcoding for taxonomy. Systematic biology 54(5): 852–859. https://doi.org/10.1080/10635150500354886 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8): 754–755. https://doi.org/10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jarne P, Théron A. (2001) Genetic structure in natural populations of flukes and snails: a practical approach and review. Parasitology 123(07): 27–40. https://doi.org/10.1017/S0031182001007715 [DOI] [PubMed] [Google Scholar]
- Kane RA, Stothard JR, Emery AM, Rollinson D. (2008) Molecular characterization of freshwater snails in the genus Bulinus: a role for barcodes?. Parasites & Vectors 1(1): 15 https://doi.org/10.1186/1756-3305-1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16(2): 111–120. https://doi.org/10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Langand J, Jourdane J, Coustau C, Delay B, Morand S. (1998) Cost of resistance, expressed as a delayed maturity, detected in the host–parasite system Biomphalaria glabrata/Echinostoma caproni. Heredity 80(3): 320–325. https://doi.org/10.1046/j.1365-2540.1998.00291.x [Google Scholar]
- Librado P, Rozas J. (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11): 1451–1452. https://doi.org/10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- Ministry of Health (2008) Vigilância e controle de moluscos de importância epidemiológica: diretrizes técnicas: Programa de Vigilância e Controle da Esquistossomose (PCE). 2. ed. Brasília, Editora do Ministério da Saúde. [Google Scholar]
- Mulvey M, Bandoni SM. (1994) Genetic variability in the M-Line stock of Biomphalaria glabrata (Mollusca:Planorbidae). Journal of the Helminthological Society of Washington 61(1): 103–108. [Google Scholar]
- Nei M. (1987) Molecular evolutionary genetics. Columbia University Press, 512 pp. [Google Scholar]
- Nylander JAA. (2004) . MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- Ohlweiler FP, Takahashi FY, Guimarães MCA, Gomes SR, Kawano T. (2010) Manual de Gastrópodes límnicos e terrestres do Estado de São Paulo associados às helmintoses. Porto Alegre: Redes Editora 38-59: 141–147. [Google Scholar]
- PAHO [Pan American Health Organization] (1968) A guide for the identificacation of the snail intermediate hosts of schistosomiasis in the Americas. Pan American Sanitary Bureau, Regional Office of the World Health Organization (WHO) Scientific Publication, Washington, DC: 168: 122pp. [Google Scholar]
- Paraense WL, Deslandes N. (1959) The renal ridge as a reliable character for separating Taphius glabratus from Taphius tenagophilus. American Journal of Tropical Medicine and Hygiene 8(4): 456–472. [DOI] [PubMed] [Google Scholar]
- Paraense WL. (1961) Shell versus anatomy in planorbis systematic. I: Australorbis glabratus. Revista Brasileira de Biologia 21(2): 163–170. [PubMed] [Google Scholar]
- Paraense WL. (1966) Biomphalaria amazonica and B. cousini, two new species of neotropical planorbid molluscs. Revista Brasileira de Biologia 26(2): 115–126. [PubMed] [Google Scholar]
- Paraense WL. (1972) Fauna Planorbídica do Brasil. In: Lacaz CS, Baruzzi RG, Siqueira W Jr. (Org.). Introdução à Geografia Médica do Brasil. Edgard Blucher, 568 pp. [Google Scholar]
- Paraense WL. (1974) Biomphalaria oligoza N. N. for Tropicorbis philippianus (Dunker) Sensu Lucena. Revista Brasileira de Biologia 34: 379–86. [Google Scholar]
- Paraense WL. (1975) Estado atual da sistemática dos planorbídeos brasileiros. Arquivos Museu Nacional 55: 105–128. [Google Scholar]
- Paraense WL. (1981) Biomphalaria occidentalis sp. n. from South America (Mollusca Basommatophora Pulmonata). Memórias do Instituto Oswaldo Cruz 76(2): 199–211. http://doi.org/10.1590/S0074-02761981000200011 [DOI] [PubMed] [Google Scholar]
- Paraense WL. (1984) Biomphalaria tenagophila guaibensis ssp.n. from Brazil and Uruguay (Pulmonata: Planorbidae). I. Morphology. Memórias do Instituto Oswaldo Cruz 79(4): 465–469. http://doi.org/10.1590/S0074-02761984000400012 [Google Scholar]
- Paraense WL. (1988) Biomphalaria kuhniana (Clessin, 1883), Planorbid Mollusc from South America. Memória do Instituto Oswaldo Cruz 83(1): 1–12. http://doi.org/10.1590/S0074-02761988000100001 [Google Scholar]
- Paraense WL. (2001) The schistosome vectors in the Americas. Memória Instituto Oswaldo Cruz 96: 7–16. http://doi.org/10.1590/S0074-02762001000900002 [DOI] [PubMed] [Google Scholar]
- Puillandre N, Lambert A, Brouillet S, Achaz G. (2012) ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology 21(8): 1864–1877. http://doi.org/10.1111/j.1365-294X.2011.05239.x [DOI] [PubMed] [Google Scholar]
- Ratnasingham S, Hebert PDN. (2007) Bold: The Barcode of Life Data System (http://www.barcodinglife.org). Molecular Ecology Notes 7 (3): 355–364. http://doi.org/10.1111/j.1471-8286.2007.01678.x [DOI] [PMC free article] [PubMed]
- Ratnasingham S, Hebert PD. (2013) A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PloS one 8(7): e66213. http://doi.org/10.1371/journal.pone.0066213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12): 1572–1574. https://doi.org/10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Scholte RG, Carvalho OS, Malone JB, Utzinger J, Vounatsou P. (2012) Spatial distribution of Biomphalaria spp., the intermediate host snails of Schistosoma mansoni, in Brazil. Geospatial Health 6(3): 95–101. http://doi.org/10.4081/gh.2012.127 [DOI] [PubMed] [Google Scholar]
- Spatz L, Vidigal THDA, Caldeira RL, Neto ED, Cappa SMG, Carvalho OS. (1999) Study of Biomphalaria tenagophila tenagophila, Biomphalaria tenagophila guaibensis and Biomphalaria occidentalis by polymerase chain reaction amplification and restriction enzyme digestion of the ribosomal RNA intergenic spacer regions. Journal of Molluscan Studies 65(2): 143–149. https://doi.org/10.1093/mollus/65.2.143 [Google Scholar]
- Standley CJ, Pointier JP, Issia L, Wisnivesky-Colli C, Stothard JR. (2011) Identification and characterization of Biomphalaria peregrina (Orbignyi, 1835) from Agua Escondida in northern Patagonia, Argentina. Journal of Natural History 45(5-6): 347–356. http://doi.org/10.1080/00222933.2010.531153 [Google Scholar]
- Standley CJ, Goodacre SL, Wade CM, Stothard JR. (2014) The population genetic structure of Biomphalaria choanomphala in Lake Victoria, East Africa: implications for schistosomiasis transmission. Parasites & Vectors 7(1): 524 http://doi.org/10.1186/s13071-014-0524-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard JR, Sousa-Figueiredo JC, Betson M, Bustinduy A, Reinhard-Rupp J. (2013) Schistosomiasis in African infants and preschool children: let them now be treated! Trends in Parasitology 29(4): 197–205. http://doi.org/10.1016/j.pt.2013.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30(12): 2725–2729. https://doi.org/10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics, 105(2): 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles HMS. (2005) Distribuição geográfica das espécies dos caramujos transmissores de Schistosoma mansoni no Estado de São Paulo. Revista da Sociedade Brasileira de Medicina Tropical 38(5): 426–32. https://doi.org/10.1590/S0037-86822005000500013 [DOI] [PubMed] [Google Scholar]
- Teodoro TM, Janotti-Passos LK, Carvalho OS, Caldeira RL. (2010) Occurrence of Biomphalaria cousini (Mollusca: Gastropoda) in Brazil and its susceptibility to Schistosoma mansoni (Platyhelminths: Trematoda). Molecular Phylogenetics and Evolution 57(1): 144–151. http://dx.doi.org/10.1016/j.ympev.2010.05.019 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25(24): 4876–4882. https://doi.org/10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan R, Santos P. (2007) ITS2 variability of Biomphalaria (Mollusca, Planorbidae) species from the Paranapanema Valley (São Paulo State, Brazil): diversity patterns, population structure, and phylogenetic relationships. Genetics and Molecular Biology 30(1): 139–144. http://doi.org/10.1590/S1415-47572007000100024 [Google Scholar]
- Tuan R, Ohlweiler FP, Palasio RGS, Zanna RD, Guimarães MCA. (2012) Pattern of genetic divergence of mitochondrial DNA sequences in Biomphalaria tenagophila complex species based on barcode and morphological analysis. InTech Open Access Publisher; (eds) Rijeka Croatia, 293–310. https://doi.org/10.5772/25550 [Google Scholar]
- Vaz JF. (1989) Distribuição e dispersão de Biomphalaria tenagophila (d’Orbigny, 1835) (Gastropoda-Pulmonata). Ciência e Cultura (São Paulo) 41(1): 14–27. [Google Scholar]
- Vidigal THDA, Dias Neto E, Spatz L, Nunes ND, Pires RE, Simpson AJG, Carvalho OS. (1998) Genetic variability and identification of the intermediate snail hosts of Schistosoma mansoni. Memórias do Instituto Oswaldo Cruz 93: 103–110. http://doi.org/10.1590/S0074-02761998000700014 [DOI] [PubMed] [Google Scholar]
- Vidigal THDA, Kissinger JC, Caldeira RL, Pires ECR, Monteiro E, Simpson AJG, Carvalho OS. (2000) Phylogenetic relationships among Brazilian Biomphalaria species (Mollusca: Planorbidae) based upon analysis of ribosomal ITS2 sequences. Parasitology 121(6): 611–620. https://doi.org/10.1017/S0031182000006831 [DOI] [PubMed] [Google Scholar]
- Wethington AR, Zavodna M, Smith MK, Oliveira G, Lewis F, Minchella DJ. (2007) Population genetic structure of Biomphalaria glabrata in a schistosomiasis-endemic region in Brazil. Journal of Molluscan Studies 73(1): 45–52. https://doi.org/10.1093/mollus/eyl028 [Google Scholar]
- WHO [World Health Organization] (2013) Schistosomiasis: progress report 2001-2011, strategic plan 2012-2020. Department of Control of Neglected Tropical Diseases (NTD), Geneva, 74 pp http://www.who.int/iris/handle/10665/78074 [Google Scholar]
- Woolhouse MEJ, Chandiwana SK. (1989) Spatial and temporal heterogeneity in the population dynamics of Bulinus globosus and Biomphalaria pfeifferi and in the epidemiology of their infection with schistosomes. Parasitology 98(01): 21–34. https://doi.org/10.1017/S0031182000059655 [DOI] [PubMed] [Google Scholar]
- Zhang J, Kapli P, Pavlidis P, Stamatakis A. (2013) A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29(22): 2869–2876. https://doi.org/10.1093/bioinformatics/btt49 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Author: Roseli Tuan
Data type: molecular data
Explanation note: ML and NJ trees generated with MEGA 6 software from the alignment of the 104 Biomphalaria COI sequences.