Abstract
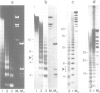
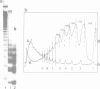
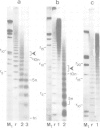
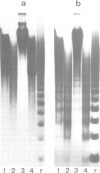
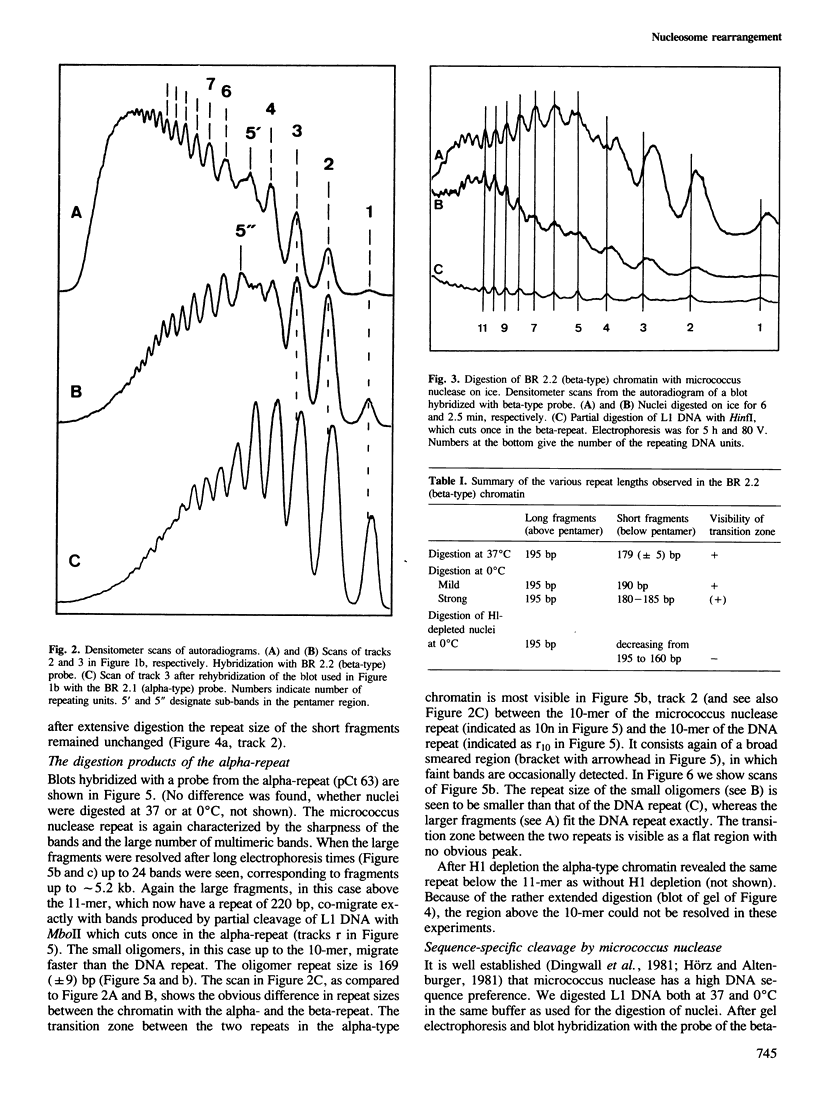
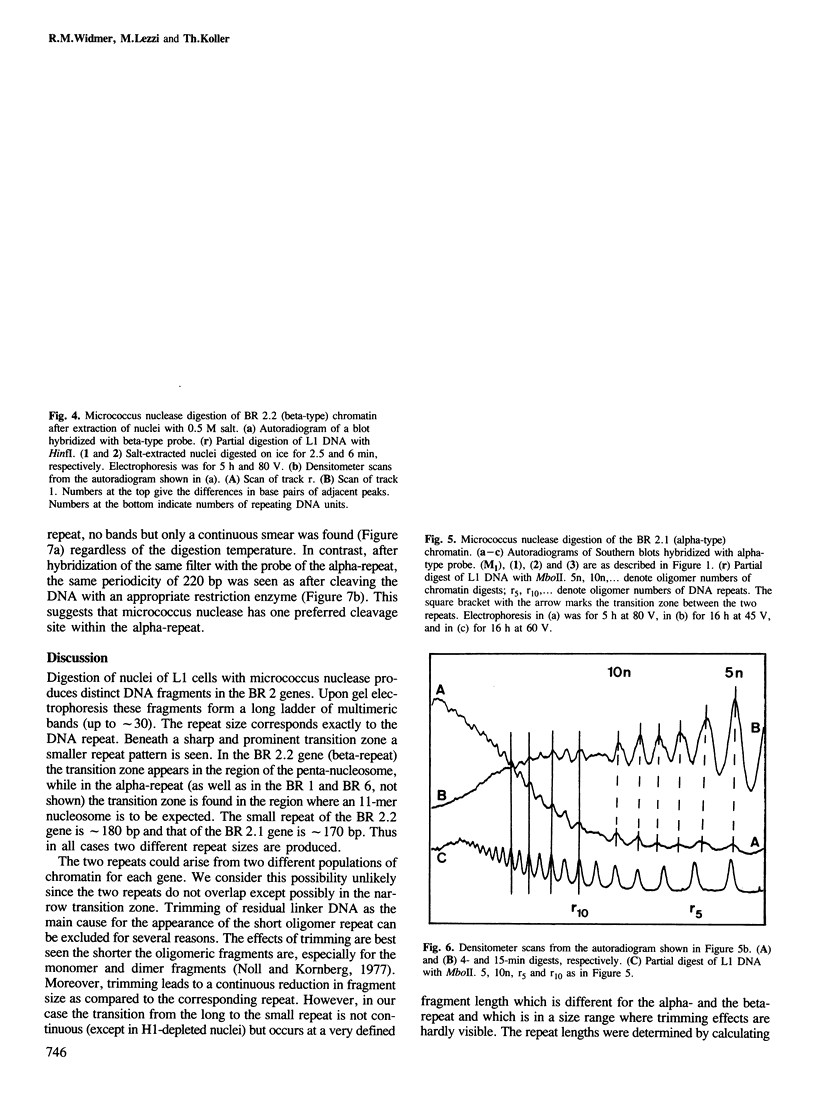
We have analysed by micrococcus nuclease digestion the chromatin structure of genes in the Balbiani ring (BR) regions of a Chironomus cell line. Gel electrophoresis of the DNA fragments reveals a repeating structure which consists of two repeat sizes, a long repeat seen in the large fragments and a small repeat seen in the small fragments. The two repeats hardly overlap, except in a narrow transition zone which is at a different fragment size in the BR 2.2 and the BR 2.1 gene. The sizes of the large repeats fit the repeat of the underlying DNA sequence. The short repeats are between 170 and 180 bp, and after H1 depletion the short repeat in the BR 2.2 gene is 160 bp. Our most favoured interpretation of these data is that in intact chromatin the nucleosomes in the BR genes are phased with respect to the repeating DNA sequence, whereas micrococcus nuclease digestion leads to loss of a nucleosome-positioning constraint and hence to rearrangement of the nucleosomes. Our results imply a possible artefact of nuclease digestion of chromatin, which has to be taken into account in mapping nucleosome positions.
Keywords: chromatin structure, micrococcus nuclease digestion, nucleosome phasing, nucleosome rearrangement
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bloom K. S., Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982 Jun;29(2):305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Bonven B. J., Gocke E., Westergaard O. A high affinity topoisomerase I binding sequence is clustered at DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell. 1985 Jun;41(2):541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- Böck H., Abler S., Zhang X. Y., Fritton H., Igo-Kemenes T. Positioning of nucleosomes in satellite I-containing chromatin of rat liver. J Mol Biol. 1984 Jun 15;176(1):131–154. doi: 10.1016/0022-2836(84)90385-1. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Abmayr S. M., Fleischmann G., Lowenhaupt K., Elgin S. C., Keene M. A., Howard G. C. Chromatin structure and gene activity: the role of nonhistone chromosomal proteins. CRC Crit Rev Biochem. 1982;13(1):1–86. doi: 10.3109/10409238209108709. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Lomonossoff G. P., Laskey R. A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., Cartwright I. L., Thomas G. H., Elgin S. C. Selected topics in chromatin structure. Annu Rev Genet. 1985;19:485–536. doi: 10.1146/annurev.ge.19.120185.002413. [DOI] [PubMed] [Google Scholar]
- Fleischmann G., Pflugfelder G., Steiner E. K., Javaherian K., Howard G. C., Wang J. C., Elgin S. C. Drosophila DNA topoisomerase I is associated with transcriptionally active regions of the genome. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6958–6962. doi: 10.1073/pnas.81.22.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick J. T., Eissenberg J. C., Elgin S. C. Micrococcal nuclease as a DNA structural probe: its recognition sequences, their genomic distribution and correlation with DNA structure determinants. J Mol Biol. 1986 Aug 20;190(4):619–633. doi: 10.1016/0022-2836(86)90247-0. [DOI] [PubMed] [Google Scholar]
- Galler R., Rydlander L., Riedel N., Kluding H., Edström J. E. Balbiani ring induction in phosphate metabolism. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1448–1452. doi: 10.1073/pnas.81.5.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galler R., Saiga H., Widmer R. M., Lezzi M., Edström J. E. Two genes in Balbiani ring 2 with metabolically different 75S transcripts. EMBO J. 1985 Nov;4(11):2977–2982. doi: 10.1002/j.1460-2075.1985.tb04032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D. S., Pflugfelder G., Wang J. C., Lis J. T. Topoisomerase I interacts with transcribed regions in Drosophila cells. Cell. 1986 Feb 14;44(3):401–407. doi: 10.1016/0092-8674(86)90461-7. [DOI] [PubMed] [Google Scholar]
- Hozier J., Renz M., Nehls P. The chromosome fiber: evidence for an ordered superstructure of nucleosomes. Chromosoma. 1977 Jul 18;62(4):301–317. doi: 10.1007/BF00327030. [DOI] [PubMed] [Google Scholar]
- Hörz W., Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörz W., Fittler F., Zachau H. G. Sequence specific cleavage of African green monkey alpha-satellite DNA by micrococcal nuclease. Nucleic Acids Res. 1983 Jul 11;11(13):4275–4285. doi: 10.1093/nar/11.13.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo-Kemenes T., Hörz W., Zachau H. G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- Jäckle H., Almeida J. C., Galler R., Kluding H., Lehrach H., Edström J. E. Constant and variable parts in the Balbiani ring 2 repeat unit and the translation termination region. EMBO J. 1982;1(7):883–888. doi: 10.1002/j.1460-2075.1982.tb01264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec E. B., Worcel A. The positive transcription factor of the 5S RNA gene induces a 5S DNA-specific gyration in Xenopus oocyte extracts. Cell. 1985 Jul;41(3):945–953. doi: 10.1016/s0092-8674(85)80075-1. [DOI] [PubMed] [Google Scholar]
- Linxweller W., Hörz W. Reconstitution experiments show that sequence-specific histone-DNA interactions are the basis for nucleosome phasing on mouse satellite DNA. Cell. 1985 Aug;42(1):281–290. doi: 10.1016/s0092-8674(85)80123-9. [DOI] [PubMed] [Google Scholar]
- Muller M. T., Pfund W. P., Mehta V. B., Trask D. K. Eukaryotic type I topoisomerase is enriched in the nucleolus and catalytically active on ribosomal DNA. EMBO J. 1985 May;4(5):1237–1243. doi: 10.1002/j.1460-2075.1985.tb03766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness P. J., Labhart P., Banz E., Koller T., Parish R. W. Chromatin structure along the ribosomal DNA of Dictyostelium. Regional differences and changes accompanying cell differentiation. J Mol Biol. 1983 May 25;166(3):361–381. doi: 10.1016/s0022-2836(83)80090-4. [DOI] [PubMed] [Google Scholar]
- Ness P. J., Parish R. W., Koller T. Mapping of endogenous nuclease-sensitive regions and of putative topoisomerase sites of action along the chromatin of Dictyostelium ribosomal RNA genes. J Mol Biol. 1986 Apr 5;188(3):287–300. doi: 10.1016/0022-2836(86)90155-5. [DOI] [PubMed] [Google Scholar]
- Neubauer B., Linxweiler W., Hörz W. DNA engineering shows that nucleosome phasing on the African green monkey alpha-satellite is the result of multiple additive histone-DNA interactions. J Mol Biol. 1986 Aug 20;190(4):639–645. doi: 10.1016/0022-2836(86)90249-4. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Pauli U. H., Seebeck T., Braun R. Sequence-specific cleavage of chromatin by staphylococcal nuclease can generate an atypical nucleosome pattern. Nucleic Acids Res. 1982 Jul 24;10(14):4121–4133. doi: 10.1093/nar/10.14.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning in vivo and in vitro. Bioessays. 1986 Apr;4(4):172–176. doi: 10.1002/bies.950040408. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spadafora C., Oudet P., Chambon P. Rearrangement of chromatin structure induced by increasing ionic strength and temperature. Eur J Biochem. 1979 Oct;100(1):225–235. doi: 10.1111/j.1432-1033.1979.tb02053.x. [DOI] [PubMed] [Google Scholar]
- Sundin O., Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell. 1981 Sep;25(3):659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- Sümegi J., Wieslander L., Daneholt B. A hierarchic arrangement of the repetitive sequences in the Balbiani ring 2 gene of Chironomus tentans. Cell. 1982 Sep;30(2):579–587. doi: 10.1016/0092-8674(82)90254-9. [DOI] [PubMed] [Google Scholar]
- Thoma F., Bergman L. W., Simpson R. T. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol. 1984 Aug 25;177(4):715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Koller T. Unravelled nucleosomes, nucleosome beads and higher order structures of chromatin: influence of non-histone components and histone H1. J Mol Biol. 1981 Jul 15;149(4):709–733. doi: 10.1016/0022-2836(81)90354-5. [DOI] [PubMed] [Google Scholar]
- Thoma F. Protein-DNA interactions and nuclease-sensitive regions determine nucleosome positions on yeast plasmid chromatin. J Mol Biol. 1986 Jul 20;190(2):177–190. doi: 10.1016/0022-2836(86)90291-3. [DOI] [PubMed] [Google Scholar]
- Thoma F., Simpson R. T. Local protein-DNA interactions may determine nucleosome positions on yeast plasmids. Nature. 1985 May 16;315(6016):250–252. doi: 10.1038/315250a0. [DOI] [PubMed] [Google Scholar]
- Weaver D. T., Fields-Berry S. C., DePamphilis M. L. The termination region for SV40 DNA replication directs the mode of separation for the two sibling molecules. Cell. 1985 Jun;41(2):565–575. doi: 10.1016/s0092-8674(85)80029-5. [DOI] [PubMed] [Google Scholar]
- Widmer R. M., Lucchini R., Lezzi M., Meyer B., Sogo J. M., Edström J. E., Koller T. Chromatin structure of a hyperactive secretory protein gene (in Balbiani ring 2) of Chironomus. EMBO J. 1984 Jul;3(7):1635–1641. doi: 10.1002/j.1460-2075.1984.tb02022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieslander L., Sümegi J., Daneholt B. Evidence for a common ancestor sequence for the Balbiani ring 1 and Balbiani ring 2 genes in Chironomus tentans. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6956–6960. doi: 10.1073/pnas.79.22.6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Gargiulo G., Jessee B., Udvardy A., Louis C., Schedl P. Chromatin fine structure of the histone gene complex of Drosophila melanogaster. Nucleic Acids Res. 1983 Jan 25;11(2):421–439. doi: 10.1093/nar/11.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss C. Chironomus tentans epithelial cell lines sensitive to ecdysteroids, juvenile hormone, insulin and heat shock. Exp Cell Res. 1982 Jun;139(2):309–319. doi: 10.1016/0014-4827(82)90255-5. [DOI] [PubMed] [Google Scholar]
- Zentgraf H., Franke W. W. Differences of supranucleosomal organization in different kinds of chromatin: cell type-specific globular subunits containing different numbers of nucleosomes. J Cell Biol. 1984 Jul;99(1 Pt 1):272–286. doi: 10.1083/jcb.99.1.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y., Fittler F., Hörz W. Eight different highly specific nucleosome phases on alpha-satellite DNA in the African green monkey. Nucleic Acids Res. 1983 Jul 11;11(13):4287–4306. doi: 10.1093/nar/11.13.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y., Hörz W. Nucleosomes are positioned on mouse satellite DNA in multiple highly specific frames that are correlated with a diverged subrepeat of nine base-pairs. J Mol Biol. 1984 Jun 15;176(1):105–129. doi: 10.1016/0022-2836(84)90384-x. [DOI] [PubMed] [Google Scholar]