Plants have been estimated to collectively synthesize more than 30,000 different terpenoids, of which many have useful applications in the manufacture of foods, industrial compounds, and pharmaceuticals. Terpenoids are synthesized from the condensation, in a head to tail fashion, of 5-carbon isoprene (or hemiterpene) units. Major terpenoid classes include mono-, sesqui-, and diterpenes, which are mostly secondary metabolites, as well as tri- and tetraterpenes, which are generally primary metabolites. This large family of compounds includes essential molecules such as carotenoids, gibberellins, abscissic acid and brassinosteroids, sterols, and the phytol chains of chlorophylls, tocopherols, and quinones. However, the vast majority are secondary metabolites, such as the volatile constituents of essential oils, and complex molecules like the anticancer drug paclitaxel that are thought to act as defensive agents (1).
The large number of useful terpenoids offers many potentially attractive targets for genetic engineering. In one of the first successful genetic modifications of a plant terpenoid pathway, Mahmoud and Croteau (2) report increasing flux through the monoterpene pathway in mint plants, resulting in an increased essential oil yield. They also improved the quality of the oil by expressing an antisense derivative of the menthofuran synthase gene to down-regulate synthesis of the undesirable constituent menthofuran. Their work builds on a recent major revision in understanding plant terpenoid metabolism and represents a useful example of the state of the art and future directions in metabolic engineering.
Until recently, it was thought that the synthesis of terpenoids in higher plants was by a cytosolic route that is derived from mevalonate. However, during the past few years it has become clear that plants also use a parallel plastid pathway that converts pyruvate and glyceraldehyde3- phosphate to 1-deoxyxyulose5-phosphate (DXP), which is metabolized in a series of steps to isopentenyl diphosphate and dimethylallyl diphosphate—the common precursors of all terpenoids (3). This latter pathway, termed the DXPS pathway, is prevalent in bacteria but has not been found in fungi or most animals. Plants use the mevalonate-dependent pathway to synthesize sesquiterpenes and triterpenes, whereas other major terpenoids derive from the DXPS pathway (3). Because discovery of the plastidial route in plants is relatively recent, little is known of the mechanisms that limit flux through the DXPS pathway. The gene encoding the first step enzyme 1-deoxy-d-xylulose-5-phosphate synthase (DXPS), has been constitutively overexpressed in bacteria and Arabidopsis (4–6). In both cases, increased enzyme activity caused an increase in accumulation of downstream terpenoids, indicating that DXPS is rate-limiting. In Arabidopsis plants, abscisic acid and α-tocopherol were most affected, increasing 4- and 2-fold, respectively (6).
In mint, it is thought that essential oil monoterpenes, which accumulate in glandular trichomes (Fig. 1), derive from the DXPS pathway (7). Because it is generally undesirable to alter the balance of monoterpenes in the oil, manipulations designed to increase the amount of oil are limited to enzymes downstream of DXPS, but upstream of geranyl diphosphate synthase, the committed step in monoterpene biosynthesis. Mahmoud and Croteau (2) exploited a gene that they had previously isolated, encoding deoxyxylulose phosphate reductoisomerase (DXR), which converts DXP to 2-C-methylerythritol4-phosphate, and constitutes the first committed step in the DXPS pathway of terpenoid biosynthesis (8). They substituted a strong constitutive promoter for the DXR promoter, and introduced the modified DXR gene into peppermint plants. The result was striking: Most transgenic plants accumulated more oil than control plants, with increases of up to 50%. Oil increases were also proportional to DXR activity in the plants and with few exceptions, the chemical composition of the oil was indistinguishable from that of control plants. As a by-product of the experiment, Mahmoud and Croteau also obtained plants where DXR expression was undetectable, presumably because of cosuppression. These cosuppressed plants had low chlorophyll and decreased oil terpene accumulation, a phenotype similar to that of Arabidopsis cla1 mutants, which are defective in DXPS activity (9). The work of Mahmoud and Croteau is the first success, to our knowledge, at increasing yield in an essential oil crop, and the first demonstration, to our knowledge, in plants that DRX is a rate-limiting enzyme of the DXPS pathway. This observation has implications for increasing the accumulation of useful terpenoids in other species.
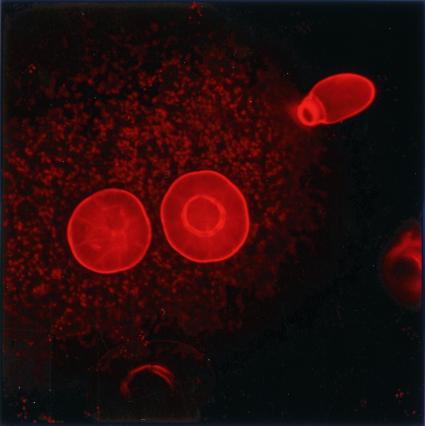
Figure 1.
Confocal image of some glandular trichomes on the surface of mint leaves (looking top down). The leaf was treated with Nile red, which fluoresces red when it is in a lipophilic environment such as the oil in the trichome. The faint red background is from chlorophyll fluorescence. Image coutesy of Gert-jan de Boer.
Based on the success of this experiment, the question arises as to what extent such an approach can be extrapolated to the engineering of other aspects of metabolism in plants. Are single-enzyme manipulations generally useful for increasing or decreasing accumulation of selected metabolites? Generally, decreasing the accumulation of a compound has been easier to achieve. One such experiment was aimed at increasing the oleic acid content of seed storage lipids to obtain improved oil quality. In this experiment, Hitz et al. (10) used cosuppression to reduce the activity of oleate desaturase in soybean seeds. As a result, oleic acid accumulated to almost 90% of the fatty acid seed content, with a concomitant decrease in linoleic acid content. In a complementary example, Shintani and DellaPenna (11) increased expression of a gene encoding γ-tocopherol methyltransferase, the enzyme that converts γ-tocopherol (low vitamin E activity) to α-tocopherol (high vitamin E activity) in Arabidopsis. Seeds of the transgenic plants inverted the ratio of the α to the γ form, thereby increasing 10-fold the seed vitamin E activity (11).
Attempts at increasing flux by manipulating the activity of single enzymes have met with mixed success. Most efforts aimed at increasing flux through biochemical pathways have targeted slow steps, where enzyme concentration is theoretically limiting, or regulatory enzymes that catalyze irreversible reactions and are regulated by specific effector molecules. Such enzymes often catalyze pathway branchpoints. A good example of such efforts is the manipulation of phytoene synthase activity in plants. Overexpression of the gene encoding this enzyme, which catalyzes the first committed step in carotenoid biosynthesis, has had very different biochemical outcomes depending on where and when the gene was expressed. Constitutive expression of a tomato cDNA in tomato resulted in dwarfism and lower lycopene levels in the fruit (12). These deleterious effects were apparently caused by a reduction in flux through the competing gibberellin pathway, which had negative effects on the hormonal balance of transgenic plants. By contrast, significant increases in fruit carotenoids were obtained when the bacterial phytoene synthase gene crtB was expressed in a fruit-specific manner (13). Perhaps the most spectacular success was obtained when the same crtB gene was expressed in canola seeds, resulting in a 50-fold increase of α- and β-carotene (14). However, in rice, only phytoene increased when the daffodil phytoene synthase gene was expressed in seeds (15). These results highlight the importance of understanding the adjoining aspects of metabolism when manipulating a metabolic pathway.
One of the reasons why overexpression of rate-limiting enzymes may not result in enhanced flux through the pathway is that catalysis may be down-regulated by feedback inhibition. In some cases, it has been possible to circumvent this level of control. In one notable experiment, an effector-insensitive bacterial homolog of a key enzyme of starch biosynthesis, ADP-glucose pyrophosphorylase, was introduced into potato plants. The transgenic potatoes accumulated up to 60% more starch than untransformed potatoes, or tubers transformed with an effector-sensitive version of the enzyme (16). Other attempts were not as successful. For instance, a feedback-insensitive regulatory enzyme of the lysine biosynthetic pathway, dihydrodipicolinate synthase, was overexpressed in transgenic plants. Flux through the pathway increased, but increases in lysine accumulation were limited by enhanced breakdown (17). In another case, a feedback-insensitive cytosolic form of acetyl-CoA carboxylase, a regulatory enzyme of the fatty acid biosynthesis pathway, was overexpressed in seeds of transgenic canola plants. A relatively slight increase in seed oil was obtained (about 5%), which suggests the existence of additional mechanisms controlling product accumulation (18).
Clearly, although single enzymes can enhance flux, control of multiple steps may be necessary to achieve net gains in product accumulation. The principle of multigenic control of flux is illustrated by the work of Ye et al. (19) on increasing carotenoid accumulation in rice. Because overexpression in rice seeds of the daffodil phytoene synthase gene PSY resulted only in phytoene accumulation, the authors produced transgenic plants overexpressing PSY, as well as two additional genes, the daffodil lycopene β-cyclase gene, LCY-B, and the Erwinia phytoene desaturase gene, crtI. The resulting rice seeds accumulated high levels of β-carotene (19). Production of the nutritionally enhanced rice, or “golden rice,” has been hailed as a significant advance in the world fight against vitamin A deficiency.
Because the activity of multiple enzymes may need to be increased, to increase flux through biochemical pathways, there is interest in manipulating regulatory genes such as kinases or transcription factors to up-regulate entire pathways. The unresolved issue, in this respect, concerns the degree to which all genes in biosynthetic pathways are coordinately controlled. Several examples suggest that at least some pathways are under coordinate control. The accumulation of anthocyanins in maize pericarp requires the activity of two genes, R and C1, which encode myc- and myb-type transcription factors, respectively. Simultaneous expression of R and C1 in Arabidopsis and tobacco caused a dramatic increase in anthocyanin accumulation, a process that requires the expression of many genes (20, 21). More recently, a jasmonate-regulated transcription factor gene was isolated from the rosy periwinkle (Catharantus roseus), a species that produces the anticancer terpenoid indole alkaloids vincristine and vinblastine. High-level expression of this gene in C. roseus suspension cells increased the expression of multiple genes for alkaloid biosynthesis, and media supplementation with a precursor of the monoterpenoid moiety secologanin resulted in a significant increase in accumulation of indole alkaloids (22). Of relevance to the work of Mahmoud and Croteau (2), a recent study suggests that induction of monoterpene biosynthesis genes is coordinated in developing glandular trichomes of mint, and that oil accumulation is largely controlled at the level of transcription (23). Thus, it seems likely that metabolic engineering of plants is poised to enter a new phase that will draw on many previously separate lines of enquiry.
Footnotes
See companion article on page 8915 in issue 15 of volume 98.
References
- 1.Harborne J B. In: Ecological Chemistry and Biochemistry of Plant Terpenoids. Harborne J B, Tomas-Barberan R A, editors. Oxford: Clarendon; 1991. pp. 399–426. [Google Scholar]
- 2.Mahmoud S S, Croteau R B. Proc Natl Acad Sci USA. 2001;98:8915–8920. doi: 10.1073/pnas.141237298. .(First Published June 26, 2001; 10.1073/pnas.141237298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lichtenthaler H K. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:47–66. doi: 10.1146/annurev.arplant.50.1.47. [DOI] [PubMed] [Google Scholar]
- 4.Harker M, Bramley P M. FEBS Lett. 1999;448:115–119. doi: 10.1016/s0014-5793(99)00360-9. [DOI] [PubMed] [Google Scholar]
- 5.Matthews P D, Wurtzel E T. Appl Microbiol Biotechnol. 2000;53:396–400. doi: 10.1007/s002530051632. [DOI] [PubMed] [Google Scholar]
- 6.Estevez J M, Cantero A, Reindl A, Reichler S, Leon P. J Biol Chem. 2001;276:22901–22909. doi: 10.1074/jbc.M100854200. [DOI] [PubMed] [Google Scholar]
- 7.McCaskill D, Croteau R. Planta. 1995;197:49–56. [Google Scholar]
- 8.Lange B M, Croteau R. Arch Biochem Biophys. 1999;365:170–174. doi: 10.1006/abbi.1999.1168. [DOI] [PubMed] [Google Scholar]
- 9.Estevez J M, Cantero A, Romero C, Kawaide H, Jimenez L F, Kuzuyama T, Seto H, Kamiya Y, Leon P. Plant Physiol. 2000;124:95–103. doi: 10.1104/pp.124.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hitz W D, Yadav N S, Reiter R S, Mauvais C J, Kinney A J. In: Plant Lipid Metabolism. Kader J C, Mazliak P, editors. London: Kluwer; 1995. pp. 506–508. [Google Scholar]
- 11.Shintani D, DellaPenna D. Science. 1998;282:2098–2100. doi: 10.1126/science.282.5396.2098. [DOI] [PubMed] [Google Scholar]
- 12.Fray R G, Wallace A, Fraser P D, Valero D, Hedden P, Bramley P M, Grierson D. Plant J. 1995;8:693–701. [Google Scholar]
- 13.Hirschberg J. Curr Opin Plant Biol. 2001;4:210–218. doi: 10.1016/s1369-5266(00)00163-1. [DOI] [PubMed] [Google Scholar]
- 14.Shewmaker C K, Sheehy J A, Daley M, Colburn S, Ke D Y. Plant J. 1999;20:401–412. doi: 10.1046/j.1365-313x.1999.00611.x. [DOI] [PubMed] [Google Scholar]
- 15.Burkhardt P K, Beyer P, Wunn J, Kloti A, Armstrong G A, Schledz M, von Lintig J, Potrykus I. Plant J. 1997;11:1071–1078. doi: 10.1046/j.1365-313x.1997.11051071.x. [DOI] [PubMed] [Google Scholar]
- 16.Stark D M, Timmermann K P, Barry G F, Preiss J, Kishore G M. Science. 1992;258:287–292. doi: 10.1126/science.258.5080.287. [DOI] [PubMed] [Google Scholar]
- 17.Falco S C, Guida T, Locke M, Mauvais J, Sanders C, Ward R T, Webber P. Biotechnology. 1995;13:577–582. doi: 10.1038/nbt0695-577. [DOI] [PubMed] [Google Scholar]
- 18.Somerville C R, Browse J B, Jaworski J, Ohlrogge J. In: Biochemistry and Molecular Biology of Plants. Buchanan B, Gruissem W, Jones R, editors. Rockville, MD: Am. Soc. Plant Physiol.; 2000. p. 1367. [Google Scholar]
- 19.Ye X, Al Babili S, Kloti A, Zhang J, Lucca P, Bayer P, Potrykus I. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- 20.Lloyd A M, Walbot V, Davis R W. Science. 1992;258:1773–1775. doi: 10.1126/science.1465611. [DOI] [PubMed] [Google Scholar]
- 21.Bruce W, Folkerts O, Garnaat C, Crasta O, Roth B, Bowen B. Plant Cell. 2000;12:65–80. doi: 10.1105/tpc.12.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van der Fits L, Memelink J. Science. 2000;289:295–297. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- 23.McConkey M E, Gershenzon J, Croteau R. Plant Physiol. 2000;122:215–224. doi: 10.1104/pp.122.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]