Abstract
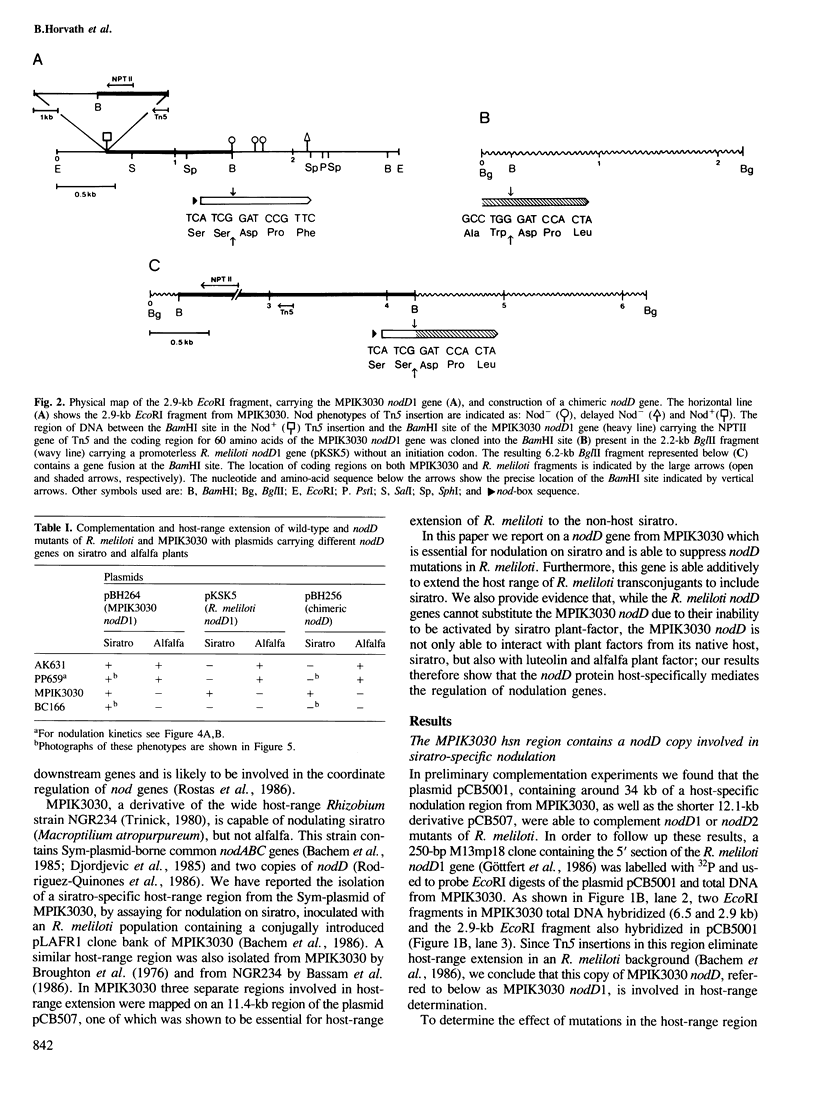
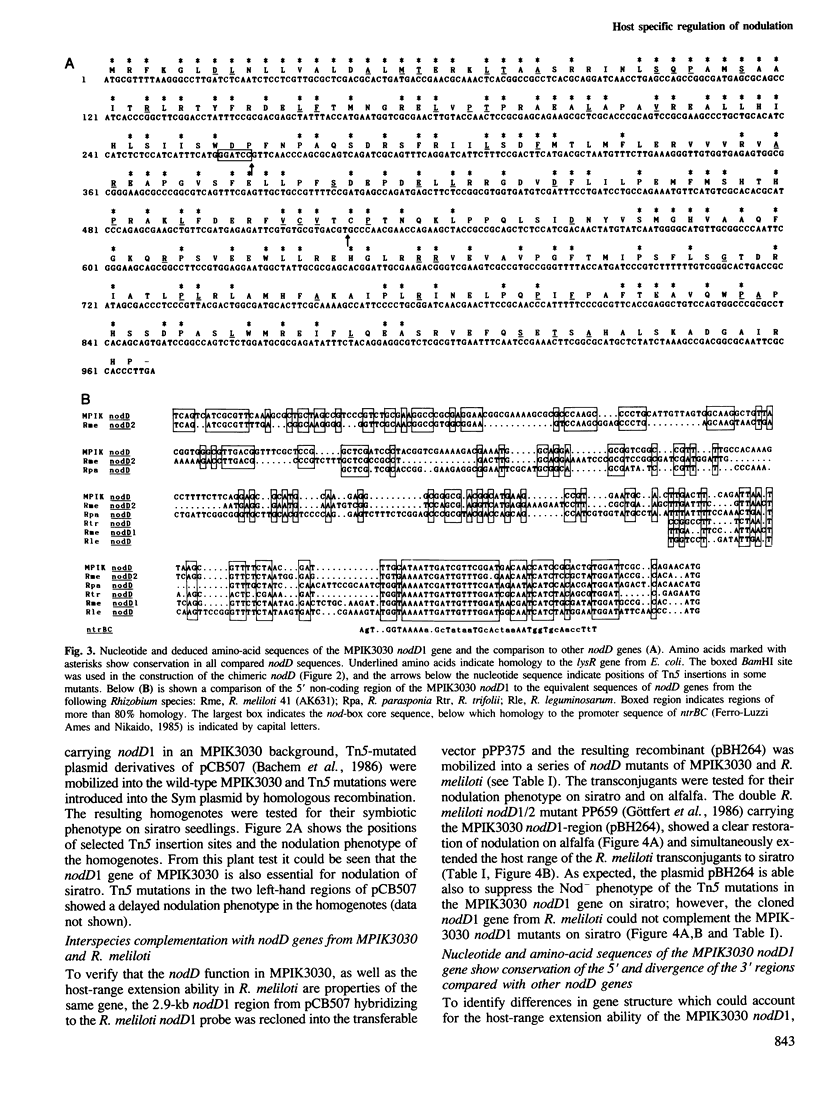
We have identified a nodD gene from the wide host-range Rhizobium strain MPIK3030 (termed nodD1) which is essential for nodulation on Macroptilium atropurpureum (siratro). Experiments with nodA–lacZ gene fusions demonstrate that the MPIK3030 nodD1 regulates expression of the nodABC genes. Additionally, we used nodC–lacZ fusions of Rhizobium meliloti to show that the MPIK3030 nodD1 gene induces expression of these fusions by interacting with plant factors from siratro and from the non-host Medicago sativa (alfalfa). The R. meliloti nodD genes, however, only interact with alfalfa exudate. In line with these results, no complementation of MPIK3030 nodD1 mutants could be obtained on siratro with the R. meliloti nodD genes, while the MPIK3030 nodD1 can complement nodD mutants of R. meliloti on alfalfa. Furthermore, R. meliloti transconjugants harbouring the MPIK3030 nodD1 efficiently nodulate the illegitimate host siratro. When compared with other nodD sequences, the amino acid sequence of the MPIK3030 nodD1 shows a conserved aminoterminus, whereas the carboxy-terminus of the putative gene product diverges considerably. Studies on a chimeric MPIK3030/R. meliloti nodD gene indicates that the carboxy-terminal region is responsible for the interaction with plant factor(s) and may have evolved in different rhizobia specifically to interact with plant–host factors.
Keywords: lacZ-fusions, gene regulation, host specificity, nodulation, Rhizobium
Full text
PDF
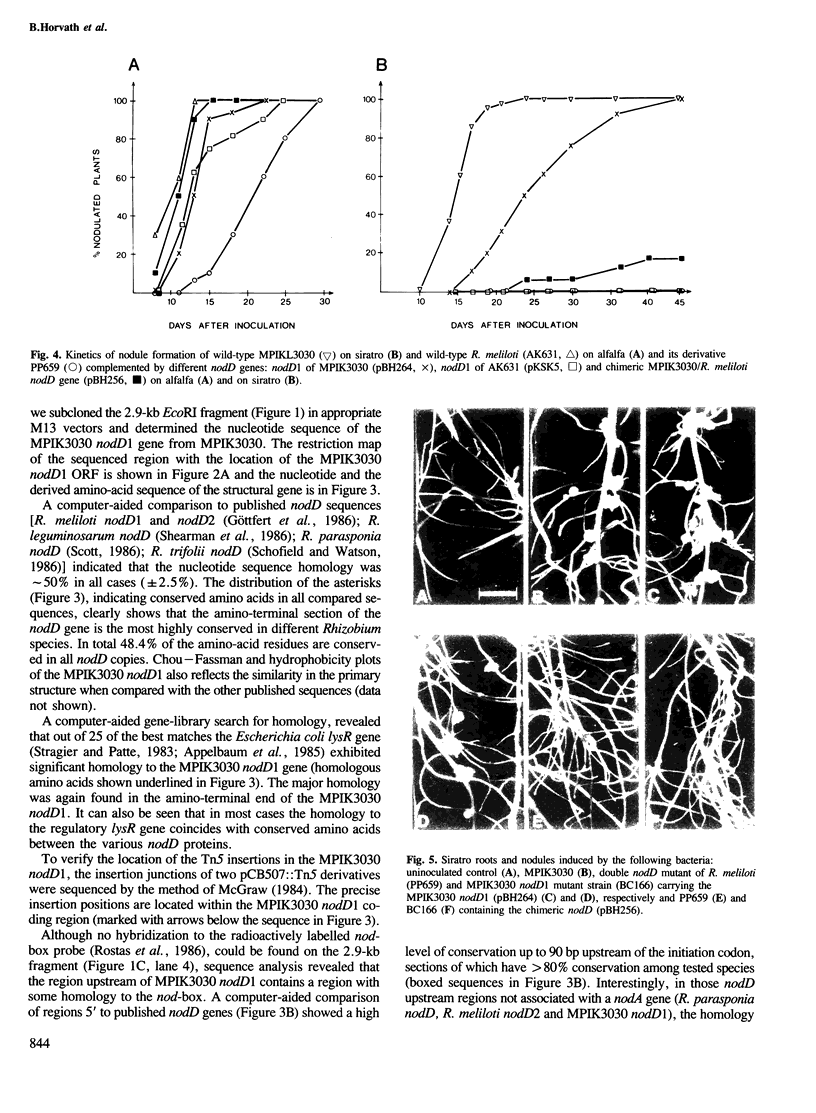




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beringer J. E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974 Sep;84(1):188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Broughton W. J., Wong C. H., Lewin A., Samrey U., Myint H., Meyer H., Dowling D. N., Simon R. Identification of Rhizobium plasmid sequences involved in recognition of Psophocarpus, Vigna, and other legumes. J Cell Biol. 1986 Apr;102(4):1173–1182. doi: 10.1083/jcb.102.4.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bánfalvi Z., Sakanyan V., Koncz C., Kiss A., Dusha I., Kondorosi A. Location of nodulation and nitrogen fixation genes on a high molecular weight plasmid of R. meliloti. Mol Gen Genet. 1981;184(2):318–325. doi: 10.1007/BF00272925. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J. Fusion of the Escherichia coli lac genes to the ara promoter: a general technique using bacteriophage Mu-1 insertions. Proc Natl Acad Sci U S A. 1975 Mar;72(3):809–813. doi: 10.1073/pnas.72.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Castilho B. A., Olfson P., Casadaban M. J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984 May;158(2):488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhoff T. T., Fisher R. F., Jacobs T. W., Mulligan J. T., Long S. R. Nucleotide sequence of Rhizobium meliloti 1021 nodulation genes: nodD is read divergently from nodABC. DNA. 1985 Jun;4(3):241–248. doi: 10.1089/dna.1985.4.241. [DOI] [PubMed] [Google Scholar]
- Ferro-Luzzi Ames G., Nikaido K. Nitrogen regulation in Salmonella typhimurium. Identification of an ntrC protein-binding site and definition of a consensus binding sequence. EMBO J. 1985 Feb;4(2):539–547. doi: 10.1002/j.1460-2075.1985.tb03662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttfert M., Horvath B., Kondorosi E., Putnoky P., Rodriguez-Quiñones F., Kondorosi A. At least two nodD genes are necessary for efficient nodulation of alfalfa by Rhizobium meliloti. J Mol Biol. 1986 Oct 5;191(3):411–420. doi: 10.1016/0022-2836(86)90136-1. [DOI] [PubMed] [Google Scholar]
- Hanau R., Koduri R. K., Ho N., Brenchley J. E. Nucleotide sequence of the control regions for the glnA and glnL genes of Salmonella typhimurium. J Bacteriol. 1983 Jul;155(1):82–89. doi: 10.1128/jb.155.1.82-89.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath B., Kondorosi E., John M., Schmidt J., Török I., Györgypal Z., Barabas I., Wieneke U., Schell J., Kondorosi A. Organization, structure and symbiotic function of Rhizobium meliloti nodulation genes determining host specificity for alfalfa. Cell. 1986 Aug 1;46(3):335–343. doi: 10.1016/0092-8674(86)90654-9. [DOI] [PubMed] [Google Scholar]
- Jacobs T. W., Egelhoff T. T., Long S. R. Physical and genetic map of a Rhizobium meliloti nodulation gene region and nucleotide sequence of nodC. J Bacteriol. 1985 May;162(2):469–476. doi: 10.1128/jb.162.2.469-476.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mulligan J. T., Long S. R. Induction of Rhizobium meliloti nodC expression by plant exudate requires nodD. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6609–6613. doi: 10.1073/pnas.82.19.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Orosz L., Sváb Z., Kondorosi A., Sik T. Genetic studies on rhizobiophage 16-3. I. Genes and functions on the chromosome. Mol Gen Genet. 1973 Sep 27;125(4):341–350. [PubMed] [Google Scholar]
- Peters N. K., Frost J. W., Long S. R. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science. 1986 Aug 29;233(4767):977–980. doi: 10.1126/science.3738520. [DOI] [PubMed] [Google Scholar]
- Putnoky P., Kondorosi A. Two gene clusters of Rhizobium meliloti code for early essential nodulation functions and a third influences nodulation efficiency. J Bacteriol. 1986 Sep;167(3):881–887. doi: 10.1128/jb.167.3.881-887.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg C., Boistard P., Dénarié J., Casse-Delbart F. Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol Gen Genet. 1981;184(2):326–333. doi: 10.1007/BF00272926. [DOI] [PubMed] [Google Scholar]
- Rossen L., Johnston A. W., Downie J. A. DNA sequence of the Rhizobium leguminosarum nodulation genes nodAB and C required for root hair curling. Nucleic Acids Res. 1984 Dec 21;12(24):9497–9508. doi: 10.1093/nar/12.24.9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossen L., Shearman C. A., Johnston A. W., Downie J. A. The nodD gene of Rhizobium leguminosarum is autoregulatory and in the presence of plant exudate induces the nodA,B,C genes. EMBO J. 1985 Dec 16;4(13A):3369–3373. doi: 10.1002/j.1460-2075.1985.tb04092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostas K., Kondorosi E., Horvath B., Simoncsits A., Kondorosi A. Conservation of extended promoter regions of nodulation genes in Rhizobium. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1757–1761. doi: 10.1073/pnas.83.6.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield P. R., Watson J. M. DNA sequence of Rhizobium trifolii nodulation genes reveals a reiterated and potentially regulatory sequence preceding nodABC and nodFE. Nucleic Acids Res. 1986 Apr 11;14(7):2891–2903. doi: 10.1093/nar/14.7.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. F. Conserved nodulation genes from the non-legume symbiont Bradyrhizobium sp. (Parasponia). Nucleic Acids Res. 1986 Apr 11;14(7):2905–2919. doi: 10.1093/nar/14.7.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman C. A., Rossen L., Johnston A. W., Downie J. A. The Rhizobium leguminosarum nodulation gene nodF encodes a polypeptide similar to acyl-carrier protein and is regulated by nodD plus a factor in pea root exudate. EMBO J. 1986 Apr;5(4):647–652. doi: 10.1002/j.1460-2075.1986.tb04262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stragier P., Patte J. C. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. III. Nucleotide sequence and regulation of the lysR gene. J Mol Biol. 1983 Aug 5;168(2):333–350. doi: 10.1016/s0022-2836(83)80022-9. [DOI] [PubMed] [Google Scholar]
- Török I., Kondorosi E., Stepkowski T., Pósfai J., Kondorosi A. Nucleotide sequence of Rhizobium meliloti nodulation genes. Nucleic Acids Res. 1984 Dec 21;12(24):9509–9524. doi: 10.1093/nar/12.24.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]