Abstract
The role of the DNA damage response protein kinase ataxia telangiectasia-mutated (ATM)- and Rad-3-related (ATR) in the cellular response to DNA damage during the replicative phase of the cell cycle has been extensively studied. However, little is known about ATR kinase function in cells that are not actively replicating DNA and that constitute most cells in the human body. Using small-molecule inhibitors of ATR kinase and overexpression of a kinase-inactive form of the enzyme, I show here that ATR promotes cell death in non-replicating/non-cycling cultured human cells exposed to N-acetoxy-2-acetylaminofluorene (NA-AAF), which generates bulky DNA adducts that block RNA polymerase movement. Immunoblot analyses of soluble protein extracts revealed that ATR and other cellular proteins containing SQ motifs become rapidly and robustly phosphorylated in non-cycling cells exposed to NA-AAF in a manner largely dependent on ATR kinase activity but independent of the essential nucleotide excision repair factor XPA. Although the topoisomerase I inhibitor camptothecin also activated ATR in non-cycling cells, other transcription inhibitors that do not directly damage DNA failed to do so. Interestingly, genetic and pharmacological inhibition of the XPB subunit of transcription factor IIH prevented the accumulation of the single-stranded DNA binding protein replication protein A (RPA) on damaged chromatin and severely abrogated ATR signaling in response to NA-AAF and camptothecin. Together, these results reveal a previously unknown role for transcription factor IIH in ATR kinase activation in non-replicating, non-cycling cells.
Keywords: apoptosis, cell cycle, cell signaling, DNA damage, DNA damage response, DNA repair, genomic instability, RNA polymerase, transcription, transcription factor
Introduction
As one of the major DNA damage response signaling kinases in mammalian cells, the ATM- and Rad3-related (ATR) 2 kinase is primarily thought to respond to DNA polymerase stalling and uncoupling from DNA helicase activity as a result of template lesions or dNTP shortage (1–3). These replicative stress events are characterized by regions of single-stranded DNA (ssDNA) and a junction of ssDNA and dsDNA (5′-primer-template junction), which together serve to recruit ATR and accessory proteins to ultimately activate ATR kinase signaling (2, 4). The functional outcomes of ATR activation in response to replication stress generally involve processes that ultimately promote cell survival, such as replication fork stabilization, cell cycle delay, inhibition of replication origin firing, DNA repair, and homologous recombination (2, 5, 6).
These pro-survival functions of ATR in cells containing replication stress likely limit the therapeutic efficacy of anticancer drugs that damage DNA, and thus small molecule inhibitors of the ATR kinase are being developed as adjuvants in chemotherapy regimens (7–10). Preliminary studies using mouse models of tumor progression have indeed suggested that ATR kinase inhibition can exacerbate the antiproliferative effects of radiation and cisplatin to more effectively slow tumor growth and shrink tumor volume (11, 12).
However, the majority of cells in the body that are exposed to DNA-damaging agents through environmental, dietary, or therapeutic means are in a non-replicating and/or differentiated state. Thus, it is important to determine whether the ATR kinase has any function in the DNA damage response in non-replicating cells, and, if so, how ATR becomes activated to carry out these activities. Using purified proteins and DNA substrates in vitro or growth-arrested, confluent populations of cultured cells, several reports have indicated that ATR may become activated by the direct recognition of bulky DNA adducts by ATR or its interacting proteins (13–18), via ssDNA gaps generated by excision repair (19–23), or by transcription stress caused by RNA polymerase stalling (24–26). Unfortunately, these studies have often restricted their analyses to phosphorylation of substrate proteins, such as H2AX and p53, which are not unique to ATR (27, 28). Moreover, many of these proteins exert complex or undefined roles in cell fate following DNA damage (29–32). Thus, the actual functions of ATR in non-cycling cells have remained largely unexplored.
Nevertheless, a recent report using small molecule inhibitors of ATR kinase activity revealed a pro-apoptotic function for ATR in non-cycling cells exposed to UV light, UV mimetics, and the topoisomerase I poison camptothecin (28). Here I have further extended this finding through the use of a genetic approach in which a kinase-inactive form of ATR is overexpressed in non-cycling cells. Moreover, using the autophosphorylation of ATR and the phosphorylation of SQ motif-containing proteins as biochemical markers of ATR kinase activation, I show that ATR is indeed robustly activated in non-cycling cells exposed to DNA-damaging agents, even at levels of DNA damage that do not yield appreciable cell death. Interestingly, this mode of ATR kinase signaling appears to require overt DNA damage because general inhibitors of RNA polymerase function during transcription failed to induce a significant response. Characterization of the activation mechanism of ATR in non-cycling cells unexpectedly revealed a major role for the XPB DNA translocase subunit of transcription factor IIH (TFIIH) in ATR signaling. This phenotype was correlated with failure to properly load the single-stranded DNA-binding protein RPA on damaged chromatin. Because the DNA unwinding activity of TFIIH is important for transcription and RNA polymerase function, these results implicate a novel function for TFIIH and, specifically, its XPB subunit in ATR activation. Given that the majority of cells in the body are in a quiescent or non-replicating state, these findings have important implications for understanding the physiology of ATR-dependent DNA damage signaling responses in vivo.
Results
Pharmacological and genetic inhibition of the ATR kinase in non-cycling cells demonstrates a pro–cell death function for ATR in response to DNA damage
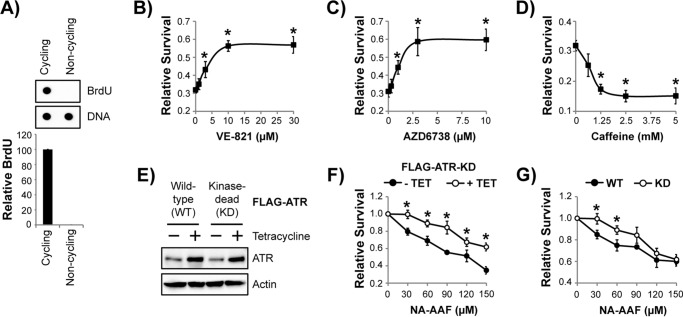
To better characterize the functions of the ATR kinase in non-replicating cells, the human keratinocyte-derived cell line HaCaT was grown to confluence and maintained in a low concentration of serum before treatment with several commonly used small-molecule ATR inhibitors and exposure to a DNA-damaging agent. Immunodot blotting of genomic DNA from cells pulsed with BrdU verified that that the cells were not actively replicating DNA (Fig. 1A) and, hence, can be referred to as being in a non-cycling state. The non-cycling cells were then treated with a small-molecule ATR inhibitor for 30 min prior to treatment with the carcinogenic UV mimetic NA-AAF, which was employed as a model DNA-damaging agent here because it generates bulky adducts on the C8 position of guanines that block RNA polymerase movement when not removed by the nucleotide excision repair system (33–35). Cell survival was then measured 1 day after treatment by crystal violet staining of the remaining adherent cells. As shown in Fig. 1, B–D, in the absence of an ATR inhibitor, nearly 70% of the cells were killed by NA-AAF. In contrast, the highly selective ATR inhibitors VE-821 and AZD6738 provided protection to NA-AAF–treated cells and limited the extent of cell death in a dose-dependent manner, with an EC50 of 3.6 μm and 1.1 μm, respectively (Fig. 1, B and C).
Figure 1.
Pharmacological and genetic inhibition of ATR kinase protects non-replicating cells from the lethal effects of the UV mimetic NA-AAF. A, cycling and non-cycling HaCaT cells were pulsed with 10 μg/ml BrdU for 15 min. Genomic DNA was then purified and analyzed by immunodot blotting with the indicated antibodies. The graph shows the relative level of BrdU incorporation into genomic DNA (normalized to cycling cells) from three independent experiments. B, non-cycling HaCaT cells were treated with the indicated concentration of the ATR inhibitor VE-821 for 30 min prior to treatment with 15 μm NA-AAF. Cells were stained with crystal violet 24 h later to determine relative survival. C, cells were treated with the indicated concentration of AZD6738 and analyzed as described in B. D, cells were treated with caffeine and analyzed as described in B. E, non-cycling U2OS cells containing either a WT or KD FLAG-tagged ATR transgene under the control of a tetracycline-inducible promoter were left untreated or treated with 1 μg/ml tetracycline for 48 h before analysis by immunoblotting. F, non-cycling U2OS cells containing the FLAG-ATR-KD transgene were left untreated (− TET, no tetracycline) or treated with tetracycline (+ TET) for 48 h before exposure to the indicated concentration of NA-AAF. After an additional 48 h, cells were stained with crystal violet to determine relative survival. G, non-cycling U2OS cells induced to express the indicated form of ATR were treated with NA-AAF as in E to determine relative cell survival. *, p < 0.05; indicating a significant difference in survival between the two treatments or cell lines.
Although relatively non-selective, caffeine has also been widely used to study ATR signaling, which is based in part on its ability to inhibit the activity of the purified enzyme (36, 37) and abrogate cell cycle checkpoints (38). However, other studies have questioned its utility for studying ATR kinase signaling in cells with DNA damage (39). When caffeine-treated, non-cycling cells were exposed to NA-AAF, I observed that, unlike the specific ATR inhibitors VE-821 and AZD6738, caffeine instead sensitized the cells to the DNA-damaging agent (Fig. 1D).
Because the pharmacological ATR inhibitors may target other kinases besides ATR, I next took a genetic approach to mimic ATR kinase inactivation in non-replicating cells. I therefore took advantage of two U2OS cell lines that can be induced to express either a WT or kinase-inactive (kinase-dead, KD) form of ATR in a tetracycline-inducible manner (40–42). As shown in Fig. 1E, 48-h induction with tetracycline led to a modest increase in total ATR protein levels in the two cell lines.
Two experimental approaches were then used to determine whether the expression of the kinase-inactive form of ATR protects non-cycling cells from NA-AAF in a manner similar to that of the two highly specific ATR kinase inhibitors. In the first method, U2OS cells with the FLAG-ATR KD transgene were grown to confluence and serum-starved prior to induction with tetracycline for 2 days. Non-induced and induced cells were then exposed to increasing concentrations of NA-AAF, and cell survival was measured 2 days later. Although U2OS cells are more resistant to NA-AAF than HaCaT cells (28), increasing concentrations of the drug nonetheless yielded a decreasing fraction of surviving cells (Fig. 1F). Furthermore, cells induced to overexpress ATR-KD with tetracycline were more resistant to NA-AAF than the non-induced cells, which indicates that NA-AAF–induced cell death is dependent in part on ATR kinase activity.
In a second, related approach, I induced ATR-WT and ATR-KD expression with tetracycline in the appropriate cell lines and then monitored cell survival after NA-AAF treatment. The ATR-KD cells were found to be less susceptible to cell death than ATR-WT cells (Fig. 1G). These findings are consistent with the effects of the small-molecule inhibitors of the ATR kinase in non-cycling cells and are strikingly different from the effect of ATR kinase inhibition in asynchronous populations of cells, in which sensitization to DNA-damaging agents has been routinely observed (28, 40, 42).
Together, the pharmacological and genetic approaches for inhibiting ATR kinase function in non-cycling cells complement one another and demonstrate that one function of ATR in non-cycling cells is to promote cell death following NA-AAF treatment. Recent work showed that this pro-death function of ATR occurs in part through stimulation of apoptotic signaling (28). This function of ATR in non-cycling cells is therefore distinct from the pro-survival functions of ATR in replicating, cycling cells.
ATR autophosphorylation on Thr-1989 in non-cycling cells
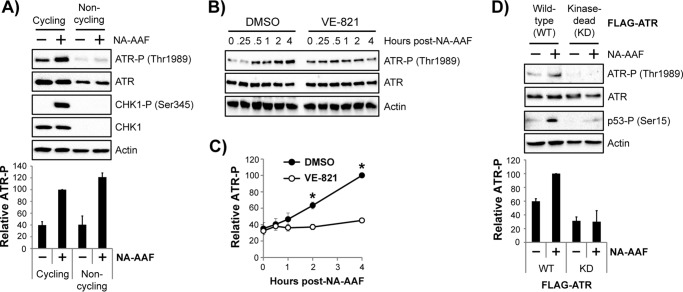
Although the cell survival assays presented in Fig. 1 suggest that ATR is activated in non-cycling cells containing DNA damage, there are currently no established biochemical readouts for ATR activation in non-cycling cells. The checkpoint kinase CHK1 is a canonical substrate for ATR in replicating cells exposed to DNA-damaging agents. However, in non-cycling cells, CHK1 protein is not present (19, 27, 28), and, therefore, no CHK1 phosphorylation is observed following exposure to NA-AAF (Fig. 2A).
Figure 2.
ATR autophosphorylation on Thr-1989 is a marker of ATR activation in non-cycling cells exposed to the UV mimetic NA-AAF. A, cycling and non-cycling HaCaT cells were treated with 20 μm NA-AAF for 1 h. Cell lysates were analyzed by immunoblotting with antibodies targeting the indicated proteins and phosphorylated residues (P). Quantitation of ATR autophosphorylation (average and standard error) from three independent experiments is provided below a representative immunoblot. The phospho-ATR signal was normalized to the total ATR signal for each sample, which was then compared with the NA-AAF–treated cycling cell sample in each experiment (set to an arbitrary value of 100). B, non-cycling HaCaT cells were pretreated with DMSO or 10 μm VE-821 (an ATR inhibitor) for 30 min prior to treatment with 10 μm NA-AAF. Cells were harvested at the indicated time point and analyzed by immunoblotting. C, quantitation of results from at least two independent experiments performed as in B. D, U2OS cells induced with tetracycline for 2 days to express either wild-type or kinase-dead forms of ATR were left untreated or treated with 60 μm NA-AAF for 4 h, and then cell lysates were analyzed by immunoblotting. Quantitation of three independent experiments is provided below the representative immunoblot data. The phospho-ATR signal was normalized to total ATR, and this ratio was set to an arbitrary value of 100 for NA-AAF–treated cells expressing WT ATR. All other samples were compared with this value. *, p < 0.05; indicating a significant difference in NA-AAF–induced ATR phosphorylation in WT and KD cells.
ATR has been shown to phosphorylate itself on Thr-1989 in asynchronous populations of cells exposed to inducers of replication stress (43, 44). To determine whether this residue becomes phosphorylated in non-replicating cells, I exposed both cycling and non-cycling cells to NA-AAF and then monitored Thr-1989 phosphorylation by immunoblotting. As shown in Fig. 2A, a DNA damage-dependent increase in ATR phosphorylation was observed in both the cycling and non-cycling cells. Although the levels of both phosphorylated ATR and total ATR were reduced in non-cycling cells in the absence and presence of NA-AAF, quantitative analyses from several independent experiments comparing the induction of ATR phosphorylation (normalized to total ATR) by NA-AAF showed a similar 2.5-fold increase in ATR phosphorylation in both cycling and non-cycling cells (Fig. 2A, bottom panel).
I next examined the kinetics of ATR phosphorylation and its dependence on ATR kinase activity. As shown in Fig. 2B, ATR became phosphorylated on Thr-1989 in a time-dependent manner following NA-AAF treatment. Importantly, this DNA damage-dependent response was prevented by treatment with the ATR kinase inhibitor VE-821 (Fig. 2, B and C). To further validate the effects of the pharmacological inhibitor on ATR phosphorylation on Thr-1989, the inducible U2OS cell lines expressing either the WT or KD forms of ATR were exposed to NA-AAF. Although an increase in ATR phosphorylation was observed in cells expressing ATR-WT, both background and NA-AAF–dependent Thr-1989 phosphorylation were significantly reduced in cells expressing ATR-KD (Fig. 2D). Moreover, expression of ATR-KD also largely abrogated NA-AAF–induced phosphorylation of the ATR/ATM substrate p53. These results indicate that the phosphorylation of ATR on Thr-1989 can be used as a biomarker for ATR kinase activation in non-cycling cells exposed to DNA-damaging agents and further argue that ATR activation can occur in the absence of canonical replication stress.
ATR plays a major role in DNA damage-induced protein phosphorylation events in non-cycling cells
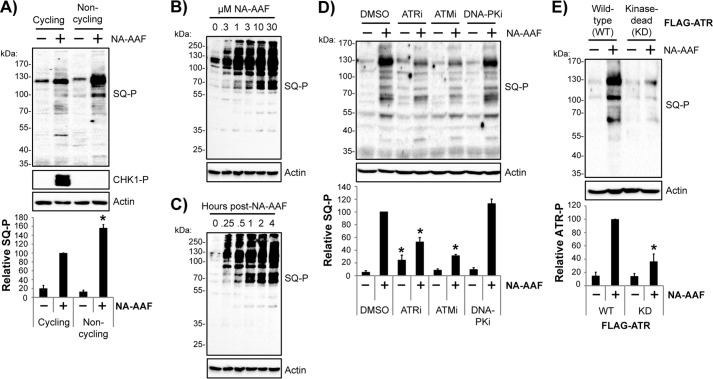
Mass spectrometric studies have demonstrated that hundreds of proteins become phosphorylated by ATM and/or ATR in response to DNA damage in proliferating cells (45, 46). To determine whether ATR makes a significant contribution to protein phosphorylation events in non-cycling cells, I made use of a mixture of monoclonal antibodies targeting phosphorylated SQ motifs common to ATR and ATM substrate proteins (47) in immunoblotting experiments of lysates from NA-AAF–treated cells. As shown in Fig. 3A, NA-AAF induced the phosphorylation of many SQ motif-containing proteins in both cycling and non-cycling cells. Consistent with earlier results, CHK1 phosphorylation was only observed in cycling cells. Quantification of the total SQ motif phosphorylation from several independent experiments demonstrated a 5- to 10-fold increase in protein phosphorylation and a slightly stronger total response in non-cycling cells than in cycling cells (Fig. 3A, bottom panel).
Figure 3.
Analysis of DNA damage–induced protein phosphorylation events in non-cycling cells reveals a major role for the ATR kinase. A, cycling and non-cycling HaCaT cells left untreated or treated for 1 h with 20 μm NA-AAF before harvesting and analysis by immunoblotting with the indicated antibodies, which included an antibody mixture that recognizes phosphorylated SQ motifs (SQ-P) common to ATR and ATM kinase substrates. The graph shows the quantitation of three independent experiments. The total phosphoprotein signals on the blots were normalized first to the Ponceau S stain and then to the NA-AAF–treated cycling cell sample, which was set to an arbitrary value of 100. *, p < 0.05; indicating a significant difference in protein phosphorylation between cycling and non-cycling cells. B, non-cycling HaCaT cells were treated with the indicated concentration of NA-AAF. Cells were harvested 1 h later and analyzed by immunoblotting. C, non-cycling cells were treated with 10 μm NA-AAF, and cells were harvested at the indicated time points and analyzed by immunoblotting. D, cells were treated with the indicated DNA damage response kinase inhibitor for 30 min prior to addition of 20 μm NA-AAF. Cells were harvested 2 h later and analyzed by immunoblotting. The graph shows the relative SQ motif phosphorylation (normalized to the DMSO control) from three independent experiments. *, p < 0.05; indicating a significant difference in SQ motif phosphorylation between DMSO- and ATR inhibitor/ATM inhibitor–treated cells. E, U2OS cells expressing either wild-type or kinase-dead ATR were left untreated or treated with 20 μm NA-AAF for 2 h, and then cell lysates were analyzed by immunoblotting. The graph quantifies the SQ motif phosphorylation from three independent experiments, which was significantly reduced (*, p < 0.05) in NA-AAF-treated cells expressing the kinase-dead form of ATR.
Additional analyses demonstrate that the degree of SQ motif phosphorylation in non-cycling cells was dependent on NA-AAF concentration and occurred at low doses of NA-AAF that do not lead to detectable cell death (28, 48, 49) (Fig. 3B). Similarly, analysis of SQ motif phosphorylation kinetics revealed robust signaling as early as 15 min after drug administration and continued phosphorylation over the course of at least 4 h (Fig. 3C).
To clarify the dependence of SQ motif phosphorylation on ATR kinase activity, cells were pretreated with specific inhibitors of ATR and the related DNA damage response kinases ATM and DNA-PK. ATR inhibition reduced the level of SQ motif phosphorylation by ∼50% (Fig. 3D). However, the ATR inhibitor alone also induced modest protein phosphorylation, which indicates that ATR inhibition may induce genomic stress that activates other DNA damage response kinases in non-replicating cells. Nevertheless, and consistent with previous evidence that ATM is activated in non-replicating cells exposed to UV mimetics (23, 27, 28, 50), ATM inhibition also partially reduced the extent of SQ motif phosphorylation. In contrast, the DNA-PK inhibitor failed to significantly affect protein phosphorylation. These results demonstrate that both ATR and ATM contribute to SQ motif phosphorylation in non-replicating cells exposed to NA-AAF.
USOS cells expressing the WT and KD forms of ATR were next employed to validate that ATR kinase activity contributes to DNA damage-dependent SQ motif phosphorylation in non-cycling cells. As shown in Fig. 3E, NA-AAF–induced protein phosphorylation was significantly attenuated in ATR-KD cells in comparison with the cells expressing ATR-WT.
In summary, these results demonstrate that SQ motif phosphorylation occurs rapidly in non-cycling cells containing bulky, transcription-stalling DNA adducts at non-lethal concentrations of NA-AAF and is mediated in large part via the ATR kinase. Thus, in addition to ATR autophosphorylation on Thr-1989, SQ motif phosphorylation can also serve as a convenient marker for DNA damage-dependent ATR signaling in non-cycling cells.
Reduced expression of the essential nucleotide excision repair factor XPA does not significantly affect NA-AAF–induced ATR kinase signaling in non-replicating cells
The nucleotide excision repair system can excise NA-AAF-induced DNA lesions from the genome (51), and experiments with cultured cells and defined in vitro assays with purified proteins have indicated that excision gaps enlarged by the endonucleolytic action of ExoI are stimuli for ATR kinase activation (19, 20, 52, 53). However, these analyses of ATR activation have utilized a rather limited number of protein substrates, such as p53 and RPA, which are not necessarily specific to ATR. Indeed, I recently showed that the simultaneous inhibition of both the ATR and ATM kinases was necessary to eliminate p53, H2AX, and KAP-1 phosphorylation in non-cycling human cells exposed to either UV light or the UV mimetic NA-AAF (28). Thus, the extent to which excision gaps versus other stimuli activate ATR in non-replicating cells is not known.
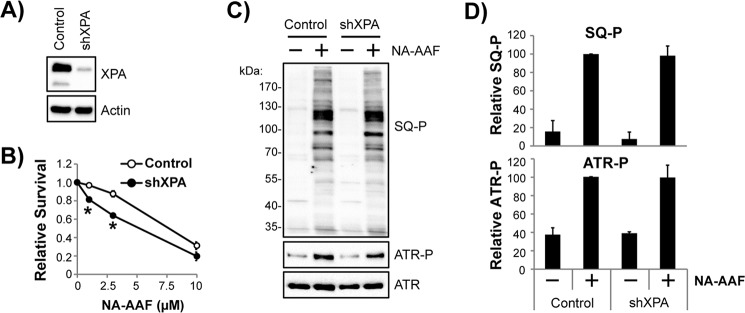
To determine whether ATR kinase signaling in non-cycling cells is dependent on nucleotide excision repair, expression of the core excision repair factor XPA was reduced by RNA interference. As shown in Fig. 4A, the use of a lentivirus shRNA targeting XPA mRNA efficiently lowered XPA protein levels by ∼95%. Importantly, this degree of knockdown was sufficient to modestly sensitize non-cycling shXPA-expressing HaCaT cells to the short-term, toxic effects of NA-AAF (Fig. 4B).
Figure 4.
Depletion of the nucleotide excision repair factor XPA does not prevent ATR kinase signaling in non-cycling cells exposed to NA-AAF. A, HaCaT cells transduced with either a control or XPA shRNA–expressing vector were analyzed by Western blotting to verify knockdown of the XPA protein. B, control or XPA shRNA-expressing HaCaT cells maintained in a non-cycling state were exposed to the indicated concentration of NA-AAF and then stained with crystal violet to validate that the loss of XPA increases the sensitivity of the cells to NA-AAF. *, p < 0.01; indicating a significant difference in survival between the two cell lines at the indicated doses of NA-AAF. C, control and XPA shRNA–expressing HaCaT cells were left untreated or treated with 20 μm NA-AAF and harvested for immunoblot analysis 2 h later. SQ-P, phosphorylated SQ motif. D, quantitation of three independent experiments performed as in C. Phosphoprotein signals were normalized to the NA-AAF–treated control cell sample, which was set to an arbitrary value of 100.
I next examined the induction of ATR kinase signaling in NA-AAF-treated cells by monitoring the phosphorylation status of ATR and SQ motif-containing proteins in the cell lysates. As shown in Fig. 4C, strong phosphorylation was observed for both readouts of ATR activation in both control and shXPA-expressing cells. Quantitation of four independent experiments failed to detect a significant difference in protein phosphorylation between the two cell lines (Fig. 4D). These results suggest that the bulk of ATR kinase signaling in NA-AAF-treated non-replicating cells may not be the result of gaps generated by nucleotide excision repair. Consistent with this interpretation, ATR kinase inhibition was shown previously to protect cells depleted of XPA from the lethal effects of NA-AAF (28). Thus, some other stimulus is likely responsible for the majority of ATR kinase signaling in non-replicating cells exposed to NA-AAF.
Direct DNA damage, but not general transcription stress, leads to robust activation of ATR kinase signaling in non-cycling cells
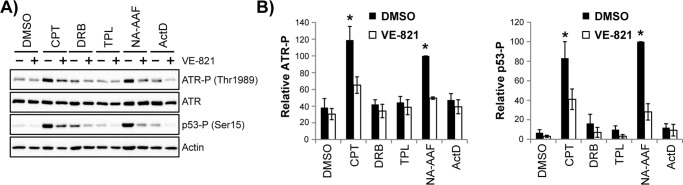
To further examine the mechanism of ATR kinase activation in non-replicating cells, non-cycling HaCaT cells were treated with various compounds that interfere with transcription. Camptothecin (CPT) causes direct DNA damage through the stabilization of transient topoisomerase I–DNA cleavage complexes that normally help to resolve superhelical tension that is generated in DNA during gene transcription. As shown in Fig. 5A, camptothecin induced a nearly 3-fold increase in ATR phosphorylation on Thr-1989 in a manner similar to NA-AAF. Treatment with the ATR inhibitor VE-821 largely blocked this response. In contrast, additional transcription inhibitors that ultimately cause cell death (28) but do not directly cause DNA damage failed to stimulate ATR autophosphorylation. These transcription inhibitors included triptolide (TPL), 5,6-dichloro-1-β-d-ribofuranosyl-1H-benzimidazole (DRB), and actinomycin D (ActD). Importantly, TPL, DRB, and ActD act via different mechanisms and at different stages of transcription (54). As a DNA intercalator, ActD directly inhibits the movement of RNA polymerases (54). In contrast, DRB specifically inhibits the CDK9 kinase activity of the positive transcription elongation factor P-TEFb, which normally phosphorylates RNA polymerase II and facilitates the transition of the polymerase from its initiated to its elongating state. Lastly, TPL forms a covalent complex with the XPB subunit of TFIIH, which inactivates the ATPase activity of the enzyme and prevents the initiation of transcription (55).
Figure 5.
Direct DNA damage, but not general transcription stress, leads to robust activation of ATR kinase signaling in non-cycling cells. A, non-cycling HaCaT cells were treated with DMSO or the ATR inhibitor VE-821 before exposure to 2.5 μm CPT, 100 μm DRB, 300 nm TPL, 20 μm NA-AAF, or 30 ng/ml ActD. Cells were harvested 4 h later and analyzed by immunoblotting. B, quantitation of ATR and p53 phosphorylation from four independent experiments performed as in A. Signals were compared with the NA-AAF–treated samples, which were set to an arbitrary value of 100. *, p < 0.05; indicating a significant difference in protein phosphorylation between drug-treated and DMSO-treated cells.
Because some of these agents have been shown to lead to either p53 phosphorylation and/or stabilization in fibroblasts (24, 56–59), I also monitored the phosphorylation of p53 on Ser-15 (an ATM/ATR target site) in non-cycling HaCaT cells. Strong ATR-dependent p53 phosphorylation was observed in cells treated with CPT and NA-AAF (Fig. 5A). However, the extent of p53 phosphorylation induced by DRB, TPL, and ActD was relatively weak in comparison with that induced by CPT and NA-AAF. These data, which are quantified in Fig. 5B, suggest that direct damage to the DNA template is necessary for robust ATR activation in non-cycling cells and that general inhibition of transcription is insufficient to elicit a significant response.
Inhibition of the XPB subunit of TFIIH abrogates ATR kinase activation and prevents RPA loading onto chromatin
Although the general transcription inhibitors failed to induce significant ATR kinase signaling in non-cycling cells, the results allowed me to examine whether the collision of RNA polymerases with a DNA lesion may be required to induce ATR activation (24–26, 60). Thus, according to this hypothesis, the stalling of RNA polymerase movement prior to exposure to NA-AAF or CPT should prevent the activation of ATR.
Therefore, HaCaT cells were treated with different transcription inhibitors before exposure to NA-AAF. As shown in Fig. 6A, NA-AAF treatment resulted in a clear increase in ATR, p53, and SQ motif protein phosphorylation in cells treated with DMSO, ActD, or DRB. Furthermore, quantitation of several independent experiments revealed that ActD and DRB actually caused modest potentiation of NA-AAF–induced ATR signaling (Fig. 6B). These results indicate that interfering with transcription elongation prior to DNA damage formation does not negatively impact the subsequent activation of the ATR kinase.
Figure 6.
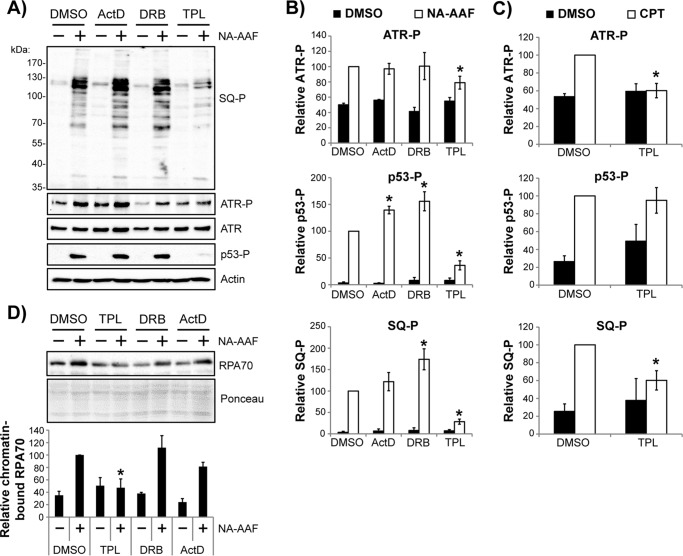
Inhibition TFIIH with triptolide abrogates the bulk of ATR kinase signaling in response to DNA damage in non-cycling cells. A, non-cycling HaCaT cells were pretreated with DMSO or the indicated transcription inhibitor for 30 min prior to exposure to 20 μm NA-AAF. Cells were harvested 2 h later and analyzed by immunoblotting. B, quantitation of ATR, p53, and ATM/ATR substrate (SQ-P) phosphorylation from four independent experiments performed as in A. The phosphoprotein samples from cells treated with DMSO + NA-AAF were set to an arbitrary value of 100, and all other samples were compared with this value. C, non-cycling HaCaT cells were treated and analyzed as in A and B, except that cells were treated with camptothecin instead of NA-AAF. *, p < 0.05l; indicating a significant difference in protein phosphorylation between drug-treated and DMSO-treated cells. D, cells were treated as in A, except that cells were harvested 1 h after NA-AAF administration and then fractionated to isolate chromatin-associated proteins. The graph shows the relative level of chromatin-associated RPA70 (normalized to Ponceau staining) from three independent experiments. *, p < 0.05; indicating a significant difference in RPA chromatin level between TPL- and DMSO-treated cells.
In striking contract, TPL treatment instead caused strong inhibition of ATR, p53, and SQ motif phosphorylation following exposure to NA-AAF (Fig. 6, A and B), which indicates that the effect of TPL on DNA damage processing and ATR activation occurs through a distinctly different mechanism than that of the transcription inhibitors DRB and ActD. Nevertheless, to further confirm this finding, I next examined the effect of TPL on CPT-induced ATR kinase activation. Quantitation of these results is provided in Fig. 6C and shows that the activation of ATR signaling in non-cycling cells treated with CPT is largely attenuated by prior treatment with the TFIIH inhibitor TPL.
Triptolide forms a covalent complex with the XPB subunit of TFIIH (55), which inhibits the ATPase activity of XPB that is required for TFIIH to unwind DNA during the initiation of transcription so that previously loaded RNA polymerase II can synthesize mRNA. Thus, TPL acts in a different manner than ActD or DRB, which instead inhibit transcriptional elongation via intercalation into DNA and by preventing RNA polymerase II phosphorylation (54), respectively. Given that ssDNA coated with RPA is generally considered to be a prerequisite for ATR kinase recruitment and activation in response to replication stress and other genotoxic stimuli (2, 3, 61, 62), I next examined whether TPL affected the accumulation of RPA on the chromatin fraction of cells following generation of DNA damage. Although NA-AAF treatment led to a 2.5- to 3-fold increase in RPA protein levels on chromatin in cells treated with DMSO, DRB, or ActD (Fig. 6D), TPL instead completely blocked the DNA damage–dependent enrichment of RPA on chromatin. These findings therefore complement the ATR kinase signaling defects induced by TPL and indicate an important role for TFIIH in generating a most widely recognized signal for ATR kinase activation in non-replicating cells.
Although the use of TPL specifically implicates a role for the XPB subunit of TFIIH in ATR activation, TFIIH is a multisubunit enzyme with several distinct biochemical activities that are potentially relevant to transcription and associated genotoxic stress responses (63). Thus, to further determine whether the XPB subunit of TFIIH is specifically required for ATR kinase activation in response to DNA damage in non-replicating cells, I next examined how two additional small-molecule inhibitors of TFIIH affected DNA damage-induced ATR activation in non-replicating cells. These compounds included spironolactone and THZ1. Spironolactone (SP) induces rapid and specific proteolytic degradation of the XPB subunit of TFIIH while leaving the remaining subunits of TFIIH largely intact (64). THZ1 is a specific inhibitor of the CDK7 kinase component of TFIIH (65) that phosphorylates RNA polymerase II to promote transcription elongation. As shown in Fig. 7A, TPL and SP abrogated SQ motif phosphorylation by 55–70% in non-replicating cells treated with NA-AAF. In contrast, THZ1 modestly stimulated SQ motif phosphorylation following NA-AAF exposure. This potentiation is analogous to the effect of DRB (Fig. 6B), which similarly inhibits RNA polymerase II phosphorylation (54). Importantly, similar results were obtained when cells were treated with the DNA-damaging agent CPT (data not shown).
Figure 7.
The XPB subunit of TFIIH is required for ATR kinase activation in response to DNA damage in non-cycling cells. A, non-cycling HaCaT cells were pretreated with DMSO, 1 μm TPL, 10 μm SP, or 1 μm THZ1 for 30 min prior to exposure to 15 μm NA-AAF. Cells were harvested 2 h later and analyzed by immunoblotting. The graph shows the quantitation of SQ motif protein phosphorylation (SQ-P) from at least three independent experiments. *, p < 0.05; indicating a significant difference in protein phosphorylation between drug-treated and DMSO-treated cells. B, HaCaT cells were transfected with control or XPB siRNAs prior to treatment with NA-AAF as in A. Cell lysates were analyzed by immunoblotting to verify XPB knockdown and SQ motif phosphorylation. The graph shows the relative level of SQ motif–containing protein phosphorylation from three independent experiments. *, p < 0.05; indicating a significant difference between control and XPB siRNA–transfected cells.
To further validate that XPB is important for ATR activation in response to DNA damage in non-replicating cells, RNA interference was used to reduce XPB protein levels prior to exposure of cells to NA-AAF. Similar to the effects of TPL and SP, the XPB siRNAs partially blocked NA-AAF–induced SQ motif phosphorylation in non-replicating cells (Fig. 7B). Together, the genetic and pharmacological approaches reveal an unanticipated role for the XPB DNA translocase component of TFIIH in the activation of ATR in non-replicating cells exposed to DNA-damaging agents.
Discussion
The functions of the ATR kinase in promoting cell survival in response to replication stress are well documented (2, 3, 5, 6). In contrast, little is known regarding the role of ATR in response to DNA damage in cells that are not actively replicating DNA. Here I have extended the previous finding that ATR can promote an apoptotic form of cell death in response to UV light, UV mimetics, and other transcriptional stressors (28) by providing complementary pharmacological and genetic data showing that inhibition of ATR kinase activity protects non-cycling cells from DNA damage–induced lethality (Fig. 1). Thus, ATR kinase inhibition can have completely opposite functional effects on cell survival that depend not on the DNA-damaging agent but, instead, on the proliferation status of the cell and the related genomic stress that is encountered. Because the overwhelming majority of cells in the human body are in a differentiated, quiescent, or slowly cycling state, this phenomenon has important implications regarding our understanding of physiological responses to DNA damage, including in epithelial cells, which are at greatest risk of exposure to dietary, occupational, and environmental carcinogens.
These findings are also relevant to the use of ATR kinase inhibitors in cancer chemotherapy regimens (7, 8, 10). In addition to facilitating cell death of rapidly proliferating cancer cells, the results suggest that ATR inhibitors may provide protection to other cell types that are not actively undergoing DNA synthesis. Consistent with this hypothesis, a recent study indicated that the ATR inhibitor AZD6738 may be radioprotective in certain contexts within intestinal crypt cells in mice exposed to total-body ionizing irradiation (66).
Determining the mechanism of ATR activation in non-cycling cells and its downstream functional targets are therefore important issues for improving cancer chemotherapy protocols and for understanding how DNA damage promotes mutagenesis and carcinogenesis. The use of ATR autophosphorylation and SQ motif phosphorylation shown here (Figs. 2 and 3) should facilitate such analyses of ATR signaling by providing readily employable biochemical readouts for ATR activation in cells that are not actively replicating DNA.
The predominant model for ATR kinase activation involves its recruitment to ssDNA coated by RPA (62), which, during the replicative phase of the cell cycle, is thought to occur when DNA damage or a lack of dNTP precursors causes DNA helicase and DNA polymerase activities to become uncoupled (1). Whether a similar scenario takes place in non-replicating cells in response to RNA polymerase stalling is not clear, and it was therefore somewhat surprising that the inhibition of transcriptional elongation with DRB and ActD did not lead to robust ATR activation (Fig. 6). This finding indicates that the structural requirements for ATR recruitment and activation (4) are not satisfied in non-replicating cells in the absence of overt DNA damage caused by compounds such as NA-AAF and CPT.
Interestingly, the dramatic abrogation of DNA damage-dependent ATR signaling by the TFIIH/XPB inhibitors triptolide and spironolactone (Fig. 6 and 7) was also surprising. However, given the role of TFIIH, and specifically its XPB subunit, in unwinding DNA during transcription initiation (63, 67), its apparent function in promoting ATR kinase activation may therefore be analogous to that of the minichromosome maintenance helicase when DNA damage is encountered during DNA synthesis (1). TPL is known to form a covalent complex with Cys-342 of XPB and inhibits its ATPase activity (55, 68), which is required for DNA translocation. Thus, the demonstration that TPL interferes with RPA accumulation on damaged chromatin (Fig. 6D) is consistent with such a role for XPB/TFIIH in generating ssDNA necessary for ATR recruitment and/or activation. However, further studies are needed to test this hypothesis. Although XPB ATPase activity is also necessary for nucleotide excision repair (67), the demonstration that reduced expression of the essential excision repair factor XPA does not significantly impact ATR kinase signaling in non-cycling cells (Fig. 4) and the fact that topoisomerase I inhibition also activates ATR (Fig. 5 and 6) suggest that a transcription-associated function of TFIIH is more relevant to ATR signaling here. Whether this is strictly a transcription initiation event or is instead associated with TFIIH acting at RNA polymerases stalled at DNA lesions to generate ssDNA in other contexts will need to be more clearly resolved. Detailed biochemical studies with purified protein components and defined DNA substrates will therefore be critical to characterizing this new mode of ATR kinase activation. Finally, interfering with XPB expression and function does not completely eliminate DNA damage–dependent ATR signaling, and thus there are likely other modes of ATR activation in non-cycling cells.
Furthermore, although the use of non-cycling cells in this report was borne out of a desire to uncover replication- and cell cycle–independent functions of the ATR kinase, this TFIIH-dependent mode of ATR activation probably also occurs in cycling and replicating cells to some extent. Consistent with this hypothesis, TFIIH subunits were found by mass spectrometry to accumulate on nascent DNA at stalled replication forks (69). Preliminary studies using a sensitive assay for detecting the small excised oligonucleotide products of nucleotide excision repair (70–73) have so far indicated that TFIIH function in this context is likely independent of nucleotide excision repair. 3 Thus, I speculate that TFIIH may help to resolve transcription problems or replication–transcription collisions in replicating cells in part through activation of a specific ATR signaling cascade.
In summary, the results presented here indicate that ATR activation occurs in non-cycling cells through a mechanism that may be analogous to the one that takes place in response to replication stress, in which ssDNA generated by DNA unwinding and coated by RPA plays a crucial role in recruiting and activating ATR (1, 2, 4, 61). So far, the only known function for ATR in non-replicating cells is promotion of an apoptotic form of cell death. However, it is also possible that the reduced apoptosis and cell death that is observed in cells exposed to ATR inhibitors is an indirect consequence of an abnormal DNA damage response that causes alterations to global gene expression. Along these lines, a recent report demonstrated that ATR regulates alternative splicing in UV-irradiated cells and that more than 80 genes involved in apoptosis undergo significant alternative splicing in response to UV–induced cyclobutane pyrimidine dimers (74). Nevertheless, given the broad diversity of ATR and ATM substrate proteins that have been identified by phosphoproteomics (45, 46), other functions for ATR in non-cycling cells are expected. Additional studies will therefore be necessary to fully define this new mechanism of ATR activation and its functions in non-replicating cells.
Experimental procedures
Cell lines
U2OS cell lines expressing wild-type and kinase-dead forms of FLAG-ATR (GW33 and GK41) in a tetracycline-inducible manner were obtained from Paul Nghiem (40–42). The U2OS cell lines and HaCaT keratinocytes were maintained in DMEM supplemented with 10% FBS, 6 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in a 5% CO2 humidified incubator. Subconfluent cells grown under these conditions are referred to as cycling cells throughout this manuscript. Cells were brought to a non-cycling state following plating at 40–60% confluence, growth for 2 days in normal medium until the cells reached confluence, and then a medium change to DMEM containing 0.5% FBS for 2–3 days prior to experimentation. Measurement of BrdU incorporation into the genomic DNA of the cells grown under these two conditions was performed as described previously (28) and was used to validate that the cells were either cycling (replicating DNA) or non-cycling (not replicating DNA). FLAG-ATR expression was induced in GW33 and GK41 U2OS cell lines by addition of 1 μg/ml of tetracycline to the culture medium for 48 h.
Chemicals and reagents
NA-AAF was purchased from the MRIGlobal Chemical Carcinogen Repository and resuspended in 95% ethanol. BrdU, TPL, DRB, CPT, ActD, caffeine, and tetracycline were obtained from Sigma. Inhibitors of the DNA damage response kinases ATR (VE-821 and AZD6738), ATM (KU55933), and DNA-PK (NU7441) were purchased from Selleckchem. SP and THZ1 were obtained from APExBio.
RNA interference
The plasmid pLKO.1 and an XPA shRNA-containing derivative were from the Open Biosystems TRC1 shRNA library (75). HEK293T cells were used to generate lentiviral DNA particles by co-transfection of the packaging plasmid psPAX2 and the envelope plasmid pMD2.G with the appropriate pLKO.1 vector and Lipofectamine 2000. Control siRNA-A and TFIIH p89 (XPB) siRNA were purchased from Santa Cruz Biotechnology and diluted in Opti-MEM for transfection with Lipofectamine RNAiMAX (Invitrogen) at a final concentration of 25 nm. Cells were transfected once when the cells were ∼60% confluent and then again 24 h later. Culture medium was replaced with low serum–containing medium 6 h after the second transfection, and experiments were performed 36 h later.
Immunoblotting
Cells were washed with cold PBS, scraped from the plate, and pelleted by gentle centrifugation. Cells were then lysed for 20 min on ice in 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, and 1% Triton X-100. Following centrifugation in a microcentrifuge for 10–15 min at maximum speed, the soluble cell lysates were transferred to new tubes. Chromatin-associated proteins were enriched from cells following two extractions with a modified cytoskeletal buffer (10 mm Tris-HCl (pH 7.4), 100 mm NaCl, 3 mm MgCl2, 1 mm EDTA, 1 mm Na3VO4, 10 mm NaF, and 0.1% Triton X-100). Equal amounts of cell lysates were separated by SDS-PAGE, transferred to nitrocellulose, and then probed by immunoblotting using standard procedures. All blots were stained with Ponceau S, and images were obtained for later quantitative purposes. Primary antibodies included antibodies against ATR (sc-1887), CHK1 (sc-8408), actin (I-19), and XPA (sc-853) from Santa Cruz Biotechnology and phospho-CHK1 (Ser-345, 2348), phospho-p53 (Ser-15, 9284), and phospho-ATM/ATR substrate (SQ, 9607). The phospho-ATR (Thr-1989, GTX128145) antibody was from GeneTex, and the RPA70 antibody was from Bethyl Laboratories (A300-421A). All primary antibodies were used at 1:1000 or 1:2000 dilution in 1× TBST (50 mm Tris-HCl (pH 7.4), 135 mm NaCl, and 0.1% Tween 20). Secondary antibodies included horseradish peroxidase-linked anti-rabbit IgG, anti-mouse IgG, and anti-goat IgG. Chemiluminescence was visualized with Clarity Western ECL substrate (Bio-Rad), West Femto substrate (Thermo Scientific), or ECL Prime Western blotting detection reagent (GE Healthcare/Amersham Biosciences) using the Molecular Imager Chemi-Doc XRS+ or MP imaging systems (Bio-Rad). Ponceau-stained membranes and chemiluminescent signals within the linear range of detection were quantified using Image Lab (Bio-Rad) or ImageQuant software (GE Healthcare). For each immunoblot, the phosphoprotein signal was quantified and normalized to ATR or the total Ponceau S stain. The maximum signal for each blot was set to an arbitrary value of 100, and all other phosphoprotein/total protein ratios were then normalized to this value for each immunoblot. All experiments analyzing DNA damage response signaling were repeated two to four times, as indicated, and the average (and standard error) of the phosphoprotein/total protein ratios were determined and plotted. t tests were used to determine statistically significant differences between treatment groups.
Cell survival assays
Cell survival assays were performed as described previously using crystal violet staining and quantitation of the solubilized dye with a spectrophotometer (28). The absorbance values of the untreated samples were set to an arbitrary value of 1 for each experiment, and all treatment samples were normalized to this value. All cell survival experiments were performed at least three times.
Author contributions
M. G. K. conceived the idea for this project, carried out the experiments, and wrote the paper.
Acknowledgments
I thank the WSU Proteome Analysis Laboratory for use of the Bio-Rad ChemiDoc MP imaging system, Paul Nghiem for providing the GW33 and GK41 cell lines, Yanzhe Gao for technical help, and Aziz Sancar, Jeffrey B. Travers, and Chris M. Rapp for support while carrying out this work. I also thank Özdemirhan Serçin, Eric Romer, Yong-jie Xu, Michael Leffak, and Jeffrey B. Travers for comments on this manuscript.
This work was supported in part by a pilot project award (to M. G. K.) from the University of North Carolina Center for Environmental Health and Susceptibility (National Institutes of Health Grant P30ES010126). The author declares that he has no conflicts of interest with the contents of this article. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
M. G. Kemp, unpublished data.
- ATR
- ATM- and Rad3-related
- ATM
- ataxia telangiectasia-mutated
- ssDNA
- single-stranded DNA
- TFIIH
- transcription factor IIH
- RPA
- replication protein A
- NA-AAF
- N-acetyoxy-2-acetylaminofluorene
- KD
- kinase-dead
- CPT
- camptothecin
- TPL
- triptolide
- DRB
- 5,6-dichloro-1-β-d-ribofuranosyl-1H-benzimidazole
- ActD
- actinomycin D
- SP
- spironolactone
- DNA-PK
- DNA-dependent protein kinase.
References
- 1. Byun T. S., Pacek M., Yee M. C., Walter J. C., and Cimprich K. A. (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19, 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cimprich K. A., and Cortez D. (2008) ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nam E. A., and Cortez D. (2011) ATR signalling: more than meeting at the fork. Biochem. J. 436, 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. MacDougall C. A., Byun T. S., Van C., Yee M. C., and Cimprich K. A. (2007) The structural determinants of checkpoint activation. Genes Dev. 21, 898–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., and Linn S. (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 6. Ciccia A., and Elledge S. J. (2010) The DNA damage response: making it safe to play with knives. Mol. Cell. 40, 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karnitz L. M., and Zou L. (2015) Molecular pathways: targeting ATR in cancer therapy. Clin. Cancer Res. 21, 4780–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Toledo L. I., Murga M., and Fernandez-Capetillo O. (2011) Targeting ATR and Chk1 kinases for cancer treatment: a new model for new (and old) drugs. Mol. Oncol. 5, 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Llona-Minguez S., Höglund A., Jacques S. A., Koolmeister T., and Helleday T. (2014) Chemical strategies for development of ATR inhibitors. Expert Rev. Mol. Med. 16, e10. [DOI] [PubMed] [Google Scholar]
- 10. Fokas E., Prevo R., Hammond E. M., Brunner T. B., McKenna W. G., and Muschel R. J. (2014) Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treat. Rev. 40, 109–117 [DOI] [PubMed] [Google Scholar]
- 11. Fokas E., Prevo R., Pollard J. R., Reaper P. M., Charlton P. A., Cornelissen B., Vallis K. A., Hammond E. M., Olcina M. M., Gillies McKenna W., Muschel R. J., and Brunner T. B. (2012) Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell. Death Dis. 3, e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vendetti F. P., Lau A., Schamus S., Conrads T. P., O'Connor M. J., and Bakkenist C. J. (2015) The orally active and bioavailable ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of cisplatin to resolve ATM-deficient non-small cell lung cancer in vivo. Oncotarget 6, 44289–44305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Unsal-Kaçmaz K., Makhov A. M., Griffith J. D., and Sancar A. (2002) Preferential binding of ATR protein to UV-damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 99, 6673–6678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang G., and Sancar A. (2006) Recruitment of DNA damage checkpoint proteins to damage in transcribed and nontranscribed sequences. Mol. Cell. Biol. 26, 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi J. H., Lindsey-Boltz L. A., and Sancar A. (2007) Reconstitution of a human ATR-mediated checkpoint response to damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 104, 13301–13306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choi J. H., Lindsey-Boltz L. A., and Sancar A. (2009) Cooperative activation of the ATR checkpoint kinase by TopBP1 and damaged DNA. Nucleic Acids Res. 37, 1501–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu Y., Fang Y., Shao H., Lindsey-Boltz L., Sancar A., and Modrich P. (2010) Interactions of human mismatch repair proteins MutSα and MutLα with proteins of the ATR-Chk1 pathway. J. Biol. Chem. 285, 5974–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yilmaz S., Sancar A., and Kemp M. G. (2011) Multiple ATR-Chk1 pathway proteins preferentially associate with checkpoint-inducing DNA substrates. PLoS ONE 6, e22986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lindsey-Boltz L. A., Kemp M. G., Reardon J. T., DeRocco V., Iyer R. R., Modrich P., and Sancar A. (2014) Coupling of human DNA excision repair and the DNA damage checkpoint in a defined in vitro system. J. Biol. Chem. 289, 5074–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sertic S., Pizzi S., Cloney R., Lehmann A. R., Marini F., Plevani P., and Muzi-Falconi M. (2011) Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proc. Natl. Acad. Sci. U.S.A. 108, 13647–13652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Willis J., Patel Y., Lentz B. L., and Yan S. (2013) APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 110, 10592–10597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vrouwe M. G., Pines A., Overmeer R. M., Hanada K., and Mullenders L. H. (2011) UV-induced photolesions elicit ATR-kinase-dependent signaling in non-cycling cells through nucleotide excision repair-dependent and -independent pathways. J. Cell Sci. 124, 435–446 [DOI] [PubMed] [Google Scholar]
- 23. Ray A., Blevins C., Wani G., and Wani A. A. (2016) ATR- and ATM-mediated DNA damage response is dependent on excision repair assembly during G1 but not in S phase of cell cycle. PLoS ONE 11, e0159344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Derheimer F. A., O'Hagan H. M., Krueger H. M., Hanasoge S., Paulsen M. T., and Ljungman M. (2007) RPA and ATR link transcriptional stress to p53. Proc. Natl. Acad. Sci. U.S.A. 104, 12778–12783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ljungman M. (2007) The transcription stress response. Cell Cycle 6, 2252–2257 [DOI] [PubMed] [Google Scholar]
- 26. Lindsey-Boltz L. A., and Sancar A. (2007) RNA polymerase: the most specific damage recognition protein in cellular responses to DNA damage? Proc. Natl. Acad. Sci. U.S.A. 104, 13213–13214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wakasugi M., Sasaki T., Matsumoto M., Nagaoka M., Inoue K., Inobe M., Horibata K., Tanaka K., and Matsunaga T. (2014) Nucleotide excision repair-dependent DNA double-strand break formation and ATM signaling activation in mammalian quiescent cells. J. Biol. Chem. 289, 28730–28737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kemp M. G., and Sancar A. (2016) ATR Kinase inhibition protects non-cycling cells from the lethal effects of DNA damage and transcription stress. J. Biol. Chem. 291, 9330–9342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Revet I., Feeney L., Bruguera S., Wilson W., Dong T. K., Oh D. H., Dankort D., and Cleaver J. E. (2011) Functional relevance of the histone γH2Ax in the response to DNA damaging agents. Proc. Natl. Acad. Sci. U.S.A. 108, 8663–8667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cleaver J. E. (2011) γH2Ax: biomarker of damage or functional participant in DNA repair “all that glitters is not gold!” Photochem. Photobiol. 87, 1230–1239 [DOI] [PubMed] [Google Scholar]
- 31. Meek D. W. (2015) Regulation of the p53 response and its relationship to cancer. Biochem. J. 469, 325–346 [DOI] [PubMed] [Google Scholar]
- 32. McKay B. C., Chen F., Perumalswami C. R., Zhang F., and Ljungman M. (2000) The tumor suppressor p53 can both stimulate and inhibit ultraviolet light-induced apoptosis. Mol. Biol. Cell 11, 2543–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen Y. H., Matsumoto Y., Shibutani S., and Bogenhagen D. F. (1991) Acetylaminofluorene and aminofluorene adducts inhibit in vitro transcription of a Xenopus 5S RNA gene only when located on the coding strand. Proc. Natl. Acad. Sci. U.S.A. 88, 9583–9587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Y. H., and Bogenhagen D. F. (1993) Effects of DNA lesions on transcription elongation by T7 RNA polymerase. J. Biol. Chem. 268, 5849–5855 [PubMed] [Google Scholar]
- 35. Donahue B. A., Fuchs R. P., Reines D., and Hanawalt P. C. (1996) Effects of aminofluorene and acetylaminofluorene DNA adducts on transcriptional elongation by RNA polymerase II. J. Biol. Chem. 271, 10588–10594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hall-Jackson C. A., Cross D. A., Morrice N., and Smythe C. (1999) ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene 18, 6707–6713 [DOI] [PubMed] [Google Scholar]
- 37. Sarkaria J. N., Busby E. C., Tibbetts R. S., Roos P., Taya Y., Karnitz L. M., and Abraham R. T. (1999) Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59, 4375–4382 [PubMed] [Google Scholar]
- 38. Kaufmann W. K., Heffernan T. P., Beaulieu L. M., Doherty S., Frank A. R., Zhou Y., Bryant M. F., Zhou T., Luche D. D., Nikolaishvili-Feinberg N., Simpson D. A., and Cordeiro-Stone M. (2003) Caffeine and human DNA metabolism: the magic and the mystery. Mutat. Res. 532, 85–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cortez D. (2003) Caffeine inhibits checkpoint responses without inhibiting the ataxia-telangiectasia-mutated (ATM) and ATM- and Rad3-related (ATR) protein kinases. J. Biol. Chem. 278, 37139–37145 [DOI] [PubMed] [Google Scholar]
- 40. Nghiem P., Park P. K., Kim Y., Vaziri C., and Schreiber S. L. (2001) ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc. Natl. Acad. Sci. U.S.A. 98, 9092–9097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Casper A. M., Nghiem P., Arlt M. F., and Glover T. W. (2002) ATR regulates fragile site stability. Cell 111, 779–789 [DOI] [PubMed] [Google Scholar]
- 42. Nghiem P., Park P. K., Kim Ys Y. S., Desai B. N., and Schreiber S. L. (2002) ATR is not required for p53 activation but synergizes with p53 in the replication checkpoint. J. Biol. Chem. 277, 4428–4434 [DOI] [PubMed] [Google Scholar]
- 43. Liu S., Shiotani B., Lahiri M., Maréchal A., Tse A., Leung C. C., Glover J. N., Yang X. H., and Zou L. (2011) ATR autophosphorylation as a molecular switch for checkpoint activation. Mol. Cell. 43, 192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nam E. A., Zhao R., Glick G. G., Bansbach C. E., Friedman D. B., and Cortez D. (2011) Thr-1989 phosphorylation is a marker of active ataxia telangiectasia-mutated and Rad3-related (ATR) kinase. J. Biol. Chem. 286, 28707–28714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R. 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., and Elledge S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 46. Stokes M. P., Rush J., Macneill J., Ren J. M., Sprott K., Nardone J., Yang V., Beausoleil S. A., Gygi S. P., Livingstone M., Zhang H., Polakiewicz R. D., and Comb M. J. (2007) Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 104, 19855–19860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Traven A., and Heierhorst J. (2005) SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. BioEssays 27, 397–407 [DOI] [PubMed] [Google Scholar]
- 48. van Oosterwijk M. F., Filon R., Kalle W. H., Mullenders L. H., and van Zeeland A. A. (1996) The sensitivity of human fibroblasts to N-acetoxy-2-acetylaminofluorene is determined by the extent of transcription-coupled repair, and/or their capability to counteract RNA synthesis inhibition. Nucleic Acids Res. 24, 4653–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van Oosterwijk M. F., Filon R., de Groot A. J., van Zeeland A. A., and Mullenders L. H. (1998) Lack of transcription-coupled repair of acetylaminofluorene DNA adducts in human fibroblasts contrasts their efficient inhibition of transcription. J. Biol. Chem. 273, 13599–13604 [DOI] [PubMed] [Google Scholar]
- 50. Tresini M., Warmerdam D. O., Kolovos P., Snijder L., Vrouwe M. G., Demmers J. A., van IJcken W. F., Grosveld F. G., Medema R. H., Hoeijmakers J. H., Mullenders L. H., Vermeulen W., and Marteijn J. A. (2015) The core spliceosome as target and effector of non-canonical ATM signalling. Nature 523, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hess M. T., Gunz D., and Naegeli H. (1996) A repair competition assay to assess recognition by human nucleotide excision repair. Nucleic Acids Res. 24, 824–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Giannattasio M., Follonier C., Tourrière H., Puddu F., Lazzaro F., Pasero P., Lopes M., Plevani P., and Muzi-Falconi M. (2010) Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol. Cell 40, 50–62 [DOI] [PubMed] [Google Scholar]
- 53. Kemp M. G., and Hu J. (2017) Post excision events in human nucleotide excision repair. Photochem. Photobiol. 93, 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bensaude O. (2011) Inhibiting eukaryotic transcription: which compound to choose? How to evaluate its activity? Transcription 2, 103–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Titov D. V., Gilman B., He Q. L., Bhat S., Low W. K., Dang Y., Smeaton M., Demain A. L., Miller P. S., Kugel J. F., Goodrich J. A., and Liu J. O. (2011) XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat. Chem. Biol. 7, 182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van Gijssel H. E., Mullenders L. H., van Oosterwijk M. F., and Meerman J. H. (2003) Blockage of transcription as a trigger for p53 accumulation by 2-acetylaminofluorene DNA-adducts. Life Sci. 73, 1759–1771 [DOI] [PubMed] [Google Scholar]
- 57. Klibanov S. A., O'Hagan H. M., and Ljungman M. (2001) Accumulation of soluble and nucleolar-associated p53 proteins following cellular stress. J. Cell Sci. 114, 1867–1873 [DOI] [PubMed] [Google Scholar]
- 58. Ljungman M., O'Hagan H. M., and Paulsen M. T. (2001) Induction of ser15 and lys382 modifications of p53 by blockage of transcription elongation. Oncogene 20, 5964–5971 [DOI] [PubMed] [Google Scholar]
- 59. O'Hagan H. M., and Ljungman M. (2004) Nuclear accumulation of p53 following inhibition of transcription is not due to diminished levels of MDM2. Oncogene 23, 5505–5512 [DOI] [PubMed] [Google Scholar]
- 60. Ljungman M., and Lane D. P. (2004) Transcription: guarding the genome by sensing DNA damage. Nat. Rev. Cancer. 4, 727–737 [DOI] [PubMed] [Google Scholar]
- 61. You Z., Kong L., and Newport J. (2002) The role of single-stranded DNA and polymerase α in establishing the ATR, Hus1 DNA replication checkpoint. J. Biol. Chem. 277, 27088–27093 [DOI] [PubMed] [Google Scholar]
- 62. Zou L., and Elledge S. J. (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 63. Egly J. M., and Coin F. (2011) A history of TFIIH: two decades of molecular biology on a pivotal transcription/repair factor. DNA Repair 10, 714–721 [DOI] [PubMed] [Google Scholar]
- 64. Alekseev S., Ayadi M., Brino L., Egly J. M., Larsen A. K., and Coin F. (2014) A small molecule screen identifies an inhibitor of DNA repair inducing the degradation of TFIIH and the chemosensitization of tumor cells to platinum. Chem. Biol. 21, 398–407 [DOI] [PubMed] [Google Scholar]
- 65. Kwiatkowski N., Zhang T., Rahl P. B., Abraham B. J., Reddy J., Ficarro S. B., Dastur A., Amzallag A., Ramaswamy S., Tesar B., Jenkins C. E., Hannett N. M., McMillin D., Sanda T., Sim T., et al. (2014) Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511, 616–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vendetti F. P., Leibowitz B. J., Barnes J., Schamus S., Kiesel B. F., Abberbock S., Conrads T., Clump D. A., Cadogan E., O'Connor M. J., Yu J., Beumer J. H., and Bakkenist C. J. (2017) Pharmacologic ATM but not ATR kinase inhibition abrogates p21-dependent G1 arrest and promotes gastrointestinal syndrome after total body irradiation. Sci. Rep. 7, 41892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Coin F., Oksenych V., and Egly J. M. (2007) Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol. Cell 26, 245–256 [DOI] [PubMed] [Google Scholar]
- 68. He Q. L., Titov D. V., Li J., Tan M., Ye Z., Zhao Y., Romo D., and Liu J. O. (2015) Covalent modification of a cysteine residue in the XPB subunit of the general transcription factor TFIIH through single epoxide cleavage of the transcription inhibitor triptolide. Angew. Chem. Int. Ed. Engl. 54, 1859–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dungrawala H., Rose K. L., Bhat K. P., Mohni K. N., Glick G. G., Couch F. B., and Cortez D. (2015) The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol. Cell 59, 998–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Song J., Kemp M. G., and Choi J. H. (2017) Detection of the excised, damage-containing oligonucleotide products of nucleotide excision repair in human cells. Photochem. Photobiol. 93, 192–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Choi J. H., Kim S. Y., Kim S. K., Kemp M. G., and Sancar A. (2015) An integrated approach for analysis of the DNA damage response in mammalian cells: nucleotide excision repair, DNA damage checkpoint, and apoptosis. J. Biol. Chem. 290, 28812–28821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Choi J. H., Gaddameedhi S., Kim S. Y., Hu J., Kemp M. G., and Sancar A. (2014) Highly specific and sensitive method for measuring nucleotide excision repair kinetics of ultraviolet photoproducts in human cells. Nucleic Acids Res. 42, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hu J., Choi J. H., Gaddameedhi S., Kemp M. G., Reardon J. T., and Sancar A. (2013) Nucleotide excision repair in human cells: fate of the excised oligonucleotide carrying DNA damage in vivo. J. Biol. Chem. 288, 20918–20926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Muñoz M. J., Nieto Moreno N., Giono L. E., Cambindo Botto A. E., Dujardin G., Bastianello G., Lavore S., Torres-Méndez A., Menck C. F., Blencowe B. J., Irimia M., Foiani M., and Kornblihtt A. R. (2017) Major roles for pyrimidine dimers, nucleotide excision repair, and ATR in the alternative splicing response to UV irradiation. Cell. Rep. 18, 2868–2879 [DOI] [PubMed] [Google Scholar]
- 75. Yang X., Boehm J. S., Yang X., Salehi-Ashtiani K., Hao T., Shen Y., Lubonja R., Thomas S. R., Alkan O., Bhimdi T., Green T. M., Johannessen C. M., Silver S. J., Nguyen C., Murray R. R., et al. (2011) A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659–661 [DOI] [PMC free article] [PubMed] [Google Scholar]