Abstract
Keratinocyte-derived cutaneous squamous cell carcinoma (cSCC) is the most common metastatic skin cancer. Although some of the early events involved in this pathology have been identified, the subsequent steps leading to tumor development are poorly defined. We demonstrate here that the development of mouse tumors induced by the concomitant application of a carcinogen and a tumor promoter (7,12-dimethylbenz[a]anthracene (DMBA) and 12-O-tetradecanoylphorbol-13-acetate (TPA), respectively) is associated with the up-regulation of a previously uncharacterized long noncoding RNA (lncRNA), termed AK144841. We found that AK144841 expression was absent from normal skin and was specifically stimulated in tumors and highly tumorigenic cells. We also found that AK144841 exists in two variants, one consisting of a large 2-kb transcript composed of four exons and one consisting of a 1.8-kb transcript lacking the second exon. Gain- and loss-of-function studies indicated that AK144841 mainly inhibited gene expression, specifically down-regulating the expression of genes of the late cornified envelope-1 (Lce1) family involved in epidermal terminal differentiation and of anticancer genes such as Cgref1, Brsk1, Basp1, Dusp5, Btg2, Anpep, Dhrs9, Stfa2, Tpm1, SerpinB2, Cpa4, Crct1, Cryab, Il24, Csf2, and Rgs16. Interestingly, the lack of the second exon significantly decreased AK144841's inhibitory effect on gene expression. We also noted that high AK144841 expression correlated with a low expression of the aforementioned genes and with the tumorigenic potential of cell lines. These findings suggest that AK144841 could contribute to the dedifferentiation program of tumor-forming keratinocytes and to molecular cascades leading to tumor development.
Keywords: cancer biology; long noncoding RNA (long ncRNA, lncRNA); microarray; RNA; skin
Introduction
Cutaneous squamous cell carcinomas (cSCCs) 3 rank among the most common human malignancies with an annual incidence rate of 150 in 100,000 in the Caucasian population (1). cSCC is a malignant neoplasm of epidermal keratinocytes arising from pretumoral lesions due to sunlight or chemical agent exposure, burns, papillomavirus infection, trauma, or chronic wounds. Most cSCCs are excised without complications. Some, however, present an aggressive profile with a regional spread that is especially prevalent in immunosuppressed patients or in those with repair-deficient genetic conditions (leg ulcers or chronic blistering genodermatoses such as recessive dystrophic epidermolysis bullosa). Some other features of “high-risk” cSCCs include tumor size, thickness or depth, growth rate, and perineural invasion. Taken together, these features make cSCCs responsible for up to 25% of deaths related to skin cancer. cSCCs are highly mutated cancers and display a complex genetic background (2).
At the molecular level, UV-induced mutations in the p53 gene as well as chemical carcinogen-induced mutations in Ha-Ras, associated or not with dysregulations of EGF receptor, CDKN2A, Fyn, STAT3, or Notch-1, have been identified at early stages of cSCC development (3, 4). However, the links between these initial alterations and the development of tumors remain unclear. Additionally, the development and progression of cSCC could also be influenced by alterations in specific noncoding transcripts such as microRNAs and long noncoding RNAs (lncRNAs). In line with this hypothesis, altered expression of miR-21-5p, miR-203, and miR-199a has been reported in cSCCs, suggesting their possible involvement in SCC development (5–7). In a recent study, we reported altered expression for 112 distinct microRNAs in papillomas and/or tumors in a murine model of skin carcinogenesis. We also demonstrated that the miR-193b/miR-365a cluster exerts a tumor-suppressive function through repression of the proliferative, clonogenic, and migratory properties of mouse and human SCC cell lines (8).
The discovery of lncRNA has created a new paradigm in gene expression regulation that has opened up new fields of investigation in embryogenesis, development, and tumorigenesis. Misregulation or mutation affecting several lncRNAs was reported in a variety of malignancies such as prostate (ANRIL and MALAT1), colon (MALAT1 and H19), and breast and liver cancers (HOTAIR, MALAT1, H19, and GAS5) (9, 10). They are either oncogenic (ANRIL, HOTAIR, and MALAT1) or tumor suppressors (GAS5, MEG3, and PR-lncRNA-1) (11). lncRNA expression had previously been profiled in the context of skin differentiation (12, 13) and in pathological situations such as psoriasis (14), keloids (15), systemic sclerosis (16), and basal carcinoma (17). lncRNAs were also suspected to influence the predisposition to skin cancer, for example following vitamin D receptor deletion (18). Finally, a recent study has demonstrated that PICSAR (LINC00162) was strongly overexpressed in cSCC cell lines, suggesting a possible role in the development of human skin carcinoma (19). Surprisingly, so far no studies have characterized the global panorama of lncRNA expression during the development of epidermal squamous cell carcinoma.
The present study was aimed to address this issue. To that end, we have performed a comprehensive gene expression analysis of biological samples collected at different stages of development of cSCC after their chemical induction by 7,12-dimethylbenz[a]anthracene (DMBA) and 12-O-tetradecanoylphorbol-13-acetate (TPA) (20). We identified AK144841, a yet uncharacterized lncRNA, that was strongly up-regulated in cSCC, and we investigated its functional properties. We provide evidence showing that AK144841 inhibits the expression of antitumoral genes in agreement with a possible role in the development of these carcinomas.
Results
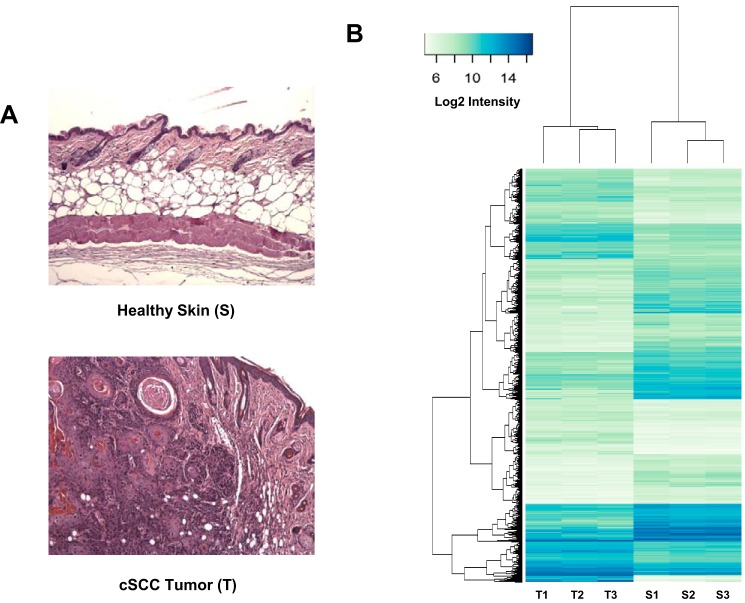
Mouse skin carcinogenesis was initiated by successive topical applications of the chemical mutagen DMBA and the tumor promoter PMA on FVB/N mice. This procedure leads to the development of pretumoral lesions called papillomas, some of which are converted to cSCC tumors (20). Agilent SurePrint microarrays enriched for lncRNA probes were hybridized with RNAs extracted from three tumors and three healthy skin samples (Fig. 1A).
Figure 1.
Gene expression analysis of biopsies of healthy skin and cSCC tumor samples. A, example of tissue sections of healthy mouse skin (S) and DMBA/TPA-induced cSCC tumors (T) used for the transcriptomic analysis. Pictures correspond to hematoxylin/eosin-counterstained formalin-fixed paraffin-embedded sections. B, RNAs extracted from three individual samples of healthy skin treated with acetone only (S1–S3) and three DMBA/TPA-generated cSCC tumors (T1–T3) (see A) were labeled with Cy3 and hybridized with mouse gene expression 8 × 60,000 v1 microarrays from Agilent as described under “Experimental procedures.” The graph shows the heat map comparing the normalized log2 of gene intensity signal in the different conditions. The following threshold values were used to define the set of up- and down-regulated genes: average expression >7.0, absolute log FC >1.0, adjusted p value <0.05.
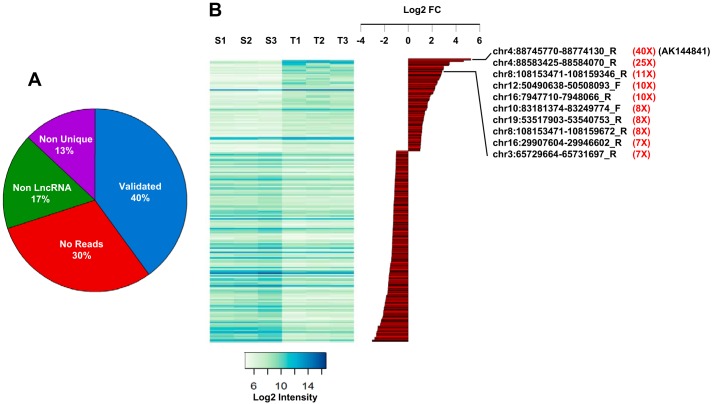
Using thresholds of 0.05 for the adjusted p value and 2-fold for the gene expression ratio between epidermis tumors and the healthy skin controls, we found that 2688 transcripts were up-regulated and 2984 were repressed (Fig. 1B). Among these 5489 selected genes, 507 corresponded to potential lncRNAs based on Agilent probe annotations. To independently validate these results, we performed high-throughput RNA sequencing analyses on biopsies from healthy skin samples, papillomas, and tumors. We quantified RNAseq reads that mapped in the 60-bp regions corresponding to the different Agilent probes. We validated probes when they overlapped with at least four aligned RNAseq reads in one sample (skin, papilloma, or tumor). This comparison between RNAseq and microarray data validated ∼40% (202 of 507) of Agilent lncRNA probes (Fig. 2A). We therefore focused our attention on these validated lncRNA probes. The largest differential expression that was observed for lncRNA probes between normal skin and tumors corresponded to an ncRNA sequence isolated from the RIKEN full-length enriched library named AK144841 (Fig. 2B). According to the microarray data, expression of AK144841 was 40-fold higher in cSCC than in healthy skin. AK144841 encodes a 1502-bp transcript composed of four exons (supplemental Fig. S1). It is located in the locus of the gene ENSMUSG00000085569 (Gm12602) on the reverse strand of chromosome 4 (4qC/88745771–88774130/genome NCBI37/mm9) about 150 kb downstream of the Cdkn2a gene. The Ensembl database revealed that Gm12602 encodes two transcripts, one with four exons (707 bp), ENSMUST00000154352, that is shorter than AK144841 but 100% identical and a second one with two exons (430 bp), ENSMUST00000138655, presenting no identity with AK144841.
Figure 2.
lncRNA expression in healthy skin (S1–S3) and in DMBA/TPA-generated cSCC tumors (T1–T3). A, pie chart summarizing the validation analysis of the lncRNA probes spotted on the Agilent SurePrint microarray (see “Results”). Among the 507 probes up- and down-regulated between healthy skin and tumors, only 202 (40%) have been validated. When considering the 305 transcripts that were excluded, 30% (211) were indeed not detected by RNAseq at a cutoff above four reads, 17.4% (53) showed complementarity with multiple chromosomal regions, and 13.4% (41) were located in exons of protein-coding genes. B, heat map (left) representing the expression intensity of the 202 validated probes in the three samples of normal skin (S1–S3) and tumors (T1–T3). Histograms represent the log2(T/S ratio) (log2 FC) of the validated probes, highlighting the top 10 lncRNAs significantly up-regulated in tumors and the amplitude of their overexpression in tumors (in red). Note that AK144841 is the most up-regulated gene.
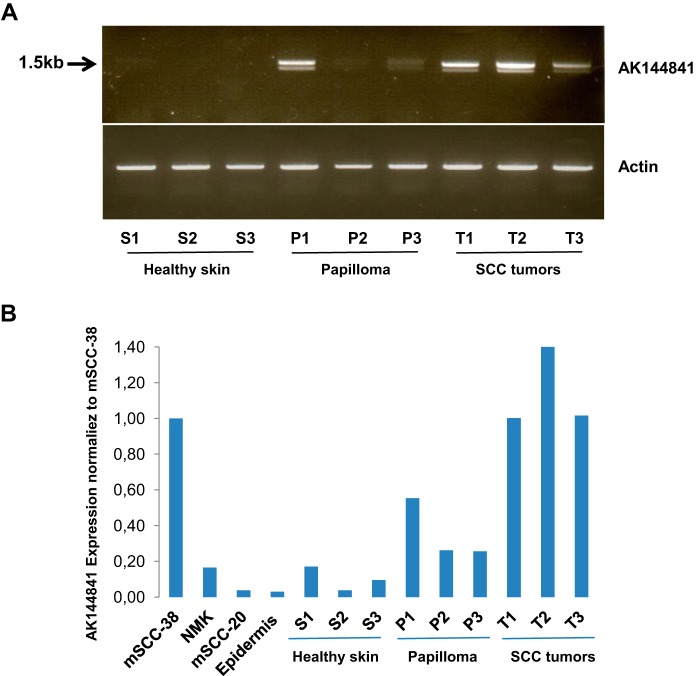
We checked that the transcript corresponding to AK144841 was effectively up-regulated in DMBA/TPA cSCC tumors using primers located in exons 1 and 4 of AK144841. Standard RT-PCR amplifications were done on three independent healthy skin samples (S1–S3), three papillomas (P1–P3), and three tumors (T1–T3). An amplicon corresponding to the expected 1.5-kb size was amplified, validating the presence of AK144841 in the different samples. Supporting the DNA arrays results, AK144841 was strongly up-regulated in the three tumor samples relative to healthy skin (T1–T3 versus S1–S3) (Fig. 3A). In contrast, it was only up-regulated in one of the three analyzed papillomas (P1), suggesting a more heterogeneous expression within pretumoral lesions (Fig. 3, A and B). It is worth noting that PCR was performed on cDNA prepared using oligo(dT), implying that AK144841 is a polyadenylated transcript.
Figure 3.
PCR identification of AK144841 in biological samples. A, RNA was extracted from three individual samples of normal skin (S1–S3) and tumors (T1–T3) and reverse transcribed, and AK144841 was then amplified by classical PCR using specific primer sets located in exons 1 and 4 (upper panel) designed according to the sequence of AK144841 reported in public data. The lower panel represents the amplification of actin used as a control. B, quantification of AK144841 by qPCR in cultured cells (mSCC-38 and mSCC-20), in NMKs, and in samples of normal skin (epidermis; S1–S3), papillomas (P1–P3), and SCC tumors (T1–T3). The histograms represent the expression of AK144841 in the different samples normalized according to its level in mSCC-38. The represented experiment is a typical one chosen among three.
A more precise assessment of AK144841 expression was then performed in tumors, papillomas, and healthy skin samples using quantitative RT-PCR. We also analyzed AK144841 levels in two DMBA/TPA tumor-derived cSCC cell lines, the highly oncogenic mSCC-38 and the non-tumorigenic mSCC-20 (8), as well as in mouse epidermis and normal mouse keratinocytes (NMKs) (Fig. 3B). We observed that AK144841 expression was at least 10-fold higher in tumors and mSCC-38 cells than in epidermis, NMKs, and mSCC-20 cells (Fig. 3B). This analysis also confirms that the expression of AK144841 was more heterogeneous in papilloma.
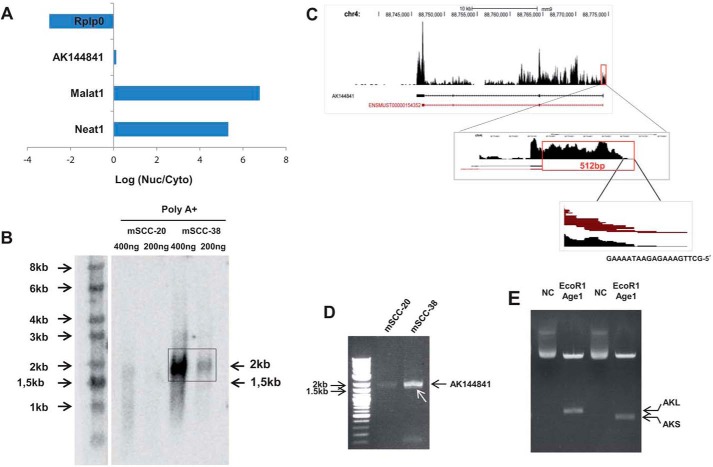
Subcellular localization of AK144841 was characterized after subcellular fractionation of mSCC-38 cells, and expression of AK144841 was assessed by qPCR in cytosolic and nuclear fractions. The expression profile was compared with two nuclear lncRNAs, Malat1 and Neat1, and one cytosolic transcript, Rplp0, which codes for a ribosomal protein. Although Neat1 and Malat1 were exclusively located in the nuclear fraction and Rplp0 was located in the cytosol, AK144841 was equivalently distributed between the nucleus and cytoplasm (log2(cytosol/nucleus) ratio close to zero) (Fig. 4A).
Figure 4.
Left, cytolocalization of AK144841 and Northern blot analysis. A, cellular localization of AK144841. The cytoplasmic (Cyto) and nuclear (Nuc) fractions of mSCC-38 cells were separated using PARISTM. RNA was extracted, and the expression of the indicated genes was measured by qPCR. The figure represents the ratio of the expression of each gene between the cytosol and the nucleus. Neat1 and Malat1 are specific markers of the nuclear fraction, whereas Rplp0 is exclusively cytoplasmic. B, AK144841 expression in mSCC-38 and mSCC-20 cells determined by Northern blotting. Northern blotting was performed using 400 or 200 ng of poly(A)+ RNAs purified using oligo(dT)-coupled magnetic beads and hybridized with a radiolabeled probe specific for AK144841. The arrows indicate the size of AK144841 (2 kb) detected in mSCC-38 cells and the position of the expected AK144841 according to the sequence available in public data set (1.5 kb). Right, structural characterization of AK144841. C, characterization of the 5′-extremity of AK144841 by RNAseq. Upper panel, RNAseq read coverage of the AK144841 region on chromosome 4 (chr4). Middle panel, coverage graph of the 5′-end of AK144841showing a magnification of the new extremity of the first exon. The red box is drawn around the 512-bp region that maps beyond the initially reported 5′-extremity of AK144841. The bottom panel shows the 20-bp sequence corresponding to the 5′-extremity of AK144841 determined from the analysis of our RNAseq data set. The mouse genome reference was NCBI37/mm9. D, PCR amplification of AK144841 from cDNA of mSCC-38 and mSCC-20 using primer sets corresponding to the 5′- and 3′-ends determined by RNAseq as illustrated in A. Note the presence of the two adjacent fragments with the more intense band corresponding to AKL (∼2 kb), whereas the weaker band corresponds to AKS (white arrow). Note the weak expression of AK144841 in mSCC-20. E, cloning of AKL and AKS. PCR-amplified AKS + AKL were subcloned into pLJM1 as described under “Experimental procedures,” and then each isoform was identified after cleavage of the purified plasmids with both AgeI and EcoRI. Lane NC represents the migration of the non-cleaved plasmid.
To assess the precise structure of AK144841, we performed a Northern blot experiment using poly(A)+ RNAs from mSCC-20 and mSCC-38 cells. RNA was separated on an agarose gel, transferred onto nitrocellulose, and hybridized with a 32P-radiolabeled PCR-amplified DNA probe corresponding to exon 4 of AK144841. The Northern blot revealed the presence of a unique transcript at 2 kb in mSCC-38 cells that was not detected in mSCC-20 cells (Fig. 4B).
The discrepancy with the size predicted by the RIKEN database (1.5 kb; as indicated by the arrow in Fig. 4B) was further clarified by the RNAseq analysis of tumors and mSCC-38 cells. Reads aligned to the genomic region corresponding to AK144841 (4qC/88745771–88774130) revealed a perfect match between the 3′-end of the transcript detected in mSCC-38 cells and tumors and the full-length sequence first identified in the RIKEN library. In contrast, a quite different situation was observed for the 5′-end, which started 512 bp upstream of the reported sequence of AK144841 (Fig. 4C). We indeed confirmed by RT-PCR that mSCC-38 and to a lesser extent mSCC-20 cells do express a 2017-bp variant of AK144841 characterized by a longer exon 1 (Fig. 4D). We also noted the presence of a smaller transcript at a size of about 1807 bp (1.8 kb) (Fig. 4D, white arrow). Molecular cloning and sequencing of the long and short forms (Fig. 4E) revealed that both transcripts fully matched with the RIKEN AK144841 except that exon 2 (210 bp) was missing in the short fragment (supplemental Fig. S1). The two isoforms were thereafter named AKL and AKS, for the 2- and 1.8-kb forms, respectively.
To elucidate the functional role of AK144841, we developed gain- and loss-of-function experiments. Two siRNAs directed against distinct regions of AK144841 decreased its expression by more than 80% after 48 h in mSCC-38 cells (supplemental Fig. S2). The impact of AK144841 knockdown on the transcriptome of mSCC-38 cells was then assessed using whole genome microarray analysis (Fig. 5). Genes whose expression was affected by both siRNAs were selected based on the following thresholds: log2(average expression) >7, absolute log2(-fold change) >0.6, and adjusted p value <0.05. AK144841 knockdown significantly altered the expression of 129 annotated genes (112 up- and 17 down-regulated).
Figure 5.
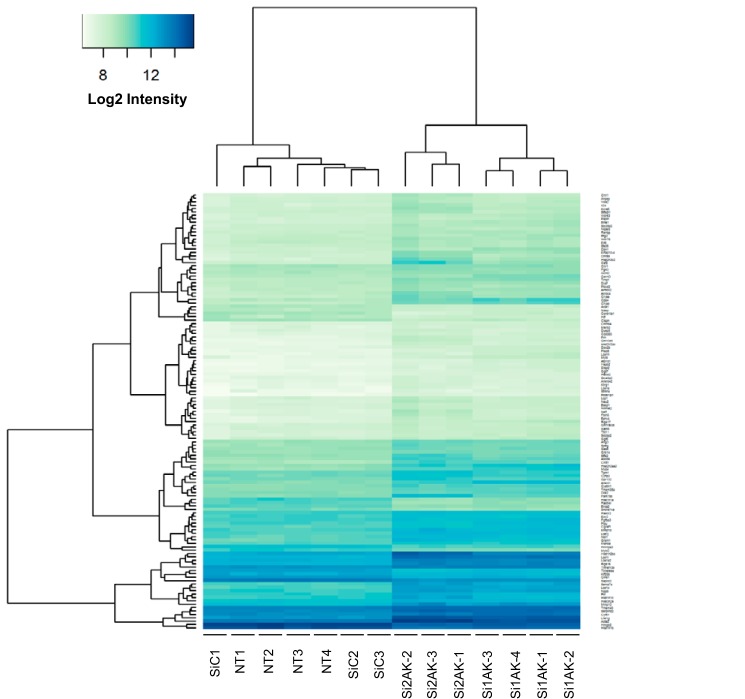
Effects of AK144841 silencing on the mSCC-38 cell transcriptome. mSCC-38 cells were transfected with two distinct siRNAs directed against AK144841 (Si1AK and Si2AK) and a control siRNA (SiC). The gene expression profiles were determined at 48 h post-transfection using a pan genomic microarrays (gene expression 8 × 60,000 microarrays from Agilent). The graph shows the heat map representing the normalized log2 intensity of the 129 genes whose expression is statistically modulated by AK144841 siRNA in the different experimental conditions. Threshold values used to select the 129 up- and down-regulated genes represented on the heat map are average expression >7.0, absolute log FC >0.6, and adjusted p value <0.05. NT1–4, four samples of non-transfected mSCC-38 cells. SiC1–3, three samples of mSCC-38 cells transfected with a non-relevant siRNA (SiC). Si1AK1–4, four samples of mSCC-38 cells transfected with Si1AK, the first AK144841-specific siRNA. Si2AK1–3, three samples of mSCC-38 cells transfected with Si2AK, the second AK144841 siRNA.
Interestingly, about 35% of the genes that were significantly up-regulated by both siRNAs were also less expressed in mSCC-38 cells compared with mSCC-20 cells, consistent with a lower level of expression of AK144841 in these latter cells (not shown). A functional enrichment analysis of this gene list using Ingenuity Pathway Analysis (IPA) software (QIAGEN Bioinformatics) indicated that the genes modulated by AK144841 knockdown were associated with “cancer” and “cytoskeleton and microtubule organization,” consistent with the increase of AK14484 in tumor cells (supplemental Table S1). We also observed that AK144841 silencing strongly increased the expression of some genes involved in the keratinocyte differentiation program and including several members of the late cornified envelope-1 (Lce1) family.
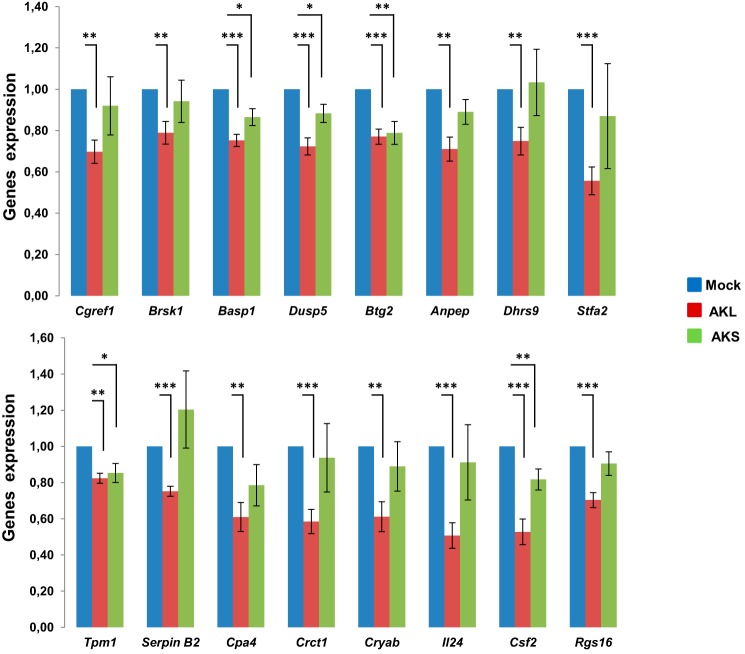
We then analyzed the effect of AK144841 gain of function on the expression of a specific subset of 19 candidate genes selected from the list of 112 genes up-regulated after AK144841 silencing and mostly based on their potential association with the tumoral process. mSCC-38 cells were transfected with the pLJM1 plasmids containing either the AKL or AKS isoform, and expression of Ankrd1, Cgref1, Brsk1, Basp1, Cnn1, Dusp5, Btg2, Anpep, Nefl, Dhrs9, Stfa2, Tpm1, SerpinB2, Cpa4, Crct1, Cryab, Il24, Csf2, and Rgs16 mRNA was assessed by qPCR 72 h after transfection. We observed that the overexpression of AKL consistently inhibited the expression of 16 of the 19 analyzed transcripts (84%) (Fig. 6) with Ankrd1, Cnn1, and Nefl mRNA being unaffected following AK144841 overexpression. This alteration was low (∼30%) but statistically significant. In contrast, overexpression of AKS has only a limited effect on the above mentioned genes because only Basp1, Dusp5, Btg2, Tpm1, and Csf2 mRNA was down-regulated upon AKS overexpression. Moreover, although statistically relevant, the inhibitions of these genes were weaker than those observed with AKL, suggesting a strong contribution of exon 2 of AK144841 in its biological effect. The modest level of inhibition of the different genes was likely linked to the efficiency of mSCC-38 transfection, estimated at ∼30%, as assessed in a parallel assay using a pLJM1-EGFP construct that allowed visualization and quantification of the cells that have been efficiently transfected using our protocol. Thus, a 30% inhibition implies an almost total repression of AK14841 expression in the fraction of transfected cells.
Figure 6.
Effect of AK144841 overexpression on genes whose expression was up-regulated upon AK144841 silencing. mSCC-38 cells were transfected with plasmids pLJM1-AKL and -AKS or an empty pLJM1 using Lipofectamine 3000 and harvested after 72 h. Total RNA was extracted, and the expression of Cgref1, Brsk1, Basp1, Dusp5, Btg2, Anpep, Dhrs9, Stfa2, Tpm1, SerpinB2, Cpa4, Crct1, Cryab, Il24, Csf2, and Rgs16 mRNA was measured by qPCR using specific primers. The histograms represent the expression of each gene normalized to their expression in cells transfected with the empty pLJM1 (Mock). Error bars represent the S.E. of five individual experiments, and asterisks represent the Student's t test p value (*, p < 0.05; **, p < 0.01; ***, p < 0.001). The transfection efficiency estimated using a pLJM1-GFP vector was ∼30%. The primer sequences can be obtained by request.
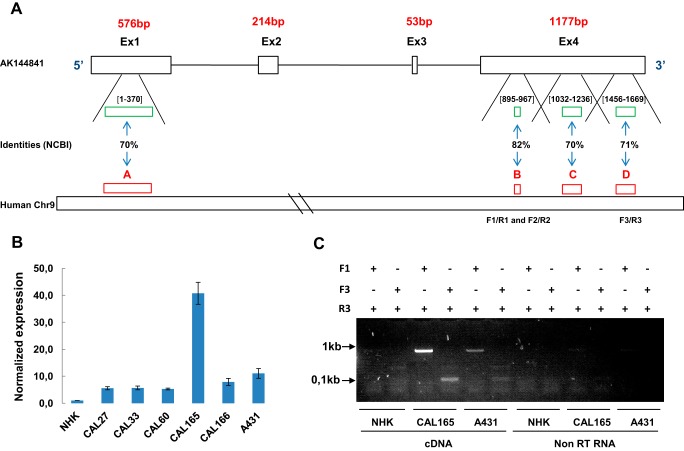
lncRNA sequences are not as highly conserved among species as protein-coding genes (21). However, we searched for a potential human ortholog by aligning the AK144841 sequence with the human genome to identify a potential human ortholog. The AK144841 locus was indeed conserved in humans with identities above 70% within a region located on human chromosome 9 downstream of CDKN2A and between the MTAP and IFNE genes. Interestingly, strong identities, ranging from 70 to 82%, were found in four domains contained in the first and fourth exons of AK144841 (Fig. 7A). We then designed primers in the human locus and performed qPCR on cDNAs from normal human keratinocytes (NHKs) as well as human cSCC (A431) and different head and neck SCC cell lines. Amplification of a specific PCR product using three different primer sets supports the existence of a human transcript that was not expressed in NHKs. In contrast, it was detected in six tumor cell lines with a maximal expression in CAL165 (Fig. 7B). The amplification by standard PCR of a unique product of about 800 bp in CAL165 and A431 cells but not in NHKs (Fig. 7C) supports the existence of a human ortholog of AK144841 (hAK144841) associated with the tumoral phenotype. Amplification of this fragment was not due to genomic DNA contamination (Fig. 7C).
Figure 7.
Identification of hAK144841. A, representation of the four regions on human chromosome 9 (A, B, C, and D) showing identities >70% with mouse AK144841. A, Chr9:21788348–21788715; B, Chr9:21754756–21754828; C, Chr9:21754518–21754740; D, Chr9:21753686–21753902. The green boxes and the numbers between brackets schematize the position of these identities on Exon 1 and Exon 4 of AK144841. B, research and quantification of a putative hAK144841 transcript in different SCC cell lines by qPCR. Experiments were performed using oligo(dT)-synthesized cDNA of the indicated cells lines and the three different primer sets (F1/R1, F2/R2, and F3/R3) designed in the regions presenting homologies with AK144841 as indicated on the scheme in A. The expression of AK144841 was normalized to its level in NHKs. The graph represents the mean of two different experiments. Error bars represent S.E. of three experiments. C, classical PCR experiments performed using cDNA and non-reverse-transcribed RNAs of CAL165 and A431 cells. Two primers sets, F1/R3 and F3/R3, were used, generating amplicons of about 800 and 100 bp, respectively. Note the absence of amplification when the PCRs were carried out with non-reverse-transcribed (non-RT) RNAs.
Discussion
The present study focuses on the functional characterization of a transcript previously annotated AK144841 and originating from the cloning and sequencing of a cDNA RIKEN fragment from a highly metastatic and drug-resistant mouse tumor (RCB-0558: http://www.ncbi.nlm.nih.gov/nucleotide/AK144841) (22, 23). We demonstrated here that the sequence of this initially reported AK144841 was incomplete. Using RNAseq experiments and PCR approaches, we identified a transcript of 2 kb that is 100% identical to the previously reported AK144841 but 512 bp longer in its 5′-extremity. Interestingly we show that two splicing variants of this new AK144841 exist, one of 2 kb with four exons and one 1.8-kb fragment lacking exon 2. In line with this observation, a recent automatic annotation pipeline of NCBI based on RNAseq from mouse liver and bladder tissues confirmed our findings in terms of AK144841 transcript exon structures and lengths (NCBI Reference Sequence XR_390705.1).
AK144841 is classified as an lncRNA (Ensembl). It is absent in normal skin cells and strongly up-regulated in all tumors generated by the DMBA/TPA treatment, and it is expressed in only some papillomas. There is a good correlation between expression levels of AK144841 and the capacity of cells to generate tumors when injected in mice. Indeed, AK144841 is highly expressed in cells with high tumorigenic capacity such as mSCC-38 but absent in the non-tumorigenic mSCC-20 cells. Interestingly, we found that AK144841 is expressed in another model of skin tumorigenesis resulting from the constitutive expression of oncogenic Hras1G12V (24) (data accessible in the NCBI Gene Expression Omnibus (GEO) under accession number GSE64867). Conversely, it was not detected in tumors of a UV-induced Trp53-dependent mouse model of cutaneous SCC, 4 suggesting that AK144841 deregulation could result from the oncogenic mutation of the Hras1 gene.
Gain- and loss-of-function experiments clearly show that alterations of the endogenous expression of AK144841 affects the mSCC-38 transcriptome and demonstrate that it is likely endowed with gene silencer activity. Qualitative analysis of the genes whose expression is altered following AK144841 silencing or overexpression suggests that AK144841 could effectively play a functional role in tumor development. Indeed, 14 of the 112 genes targeted by AK144841 have indeed been identified as potential tumor suppressors (Rgs16, cgref1, brsk1, Cryab, IL24, Basp1, Dusp5, and Csf2) and/or prognosis markers down-regulated in tumors of different origins (Btg2, Anpep, Dhrs9, Stfa2, Tpm1, SerpinB2, and Dusp5) (supplemental Table S2). Some characteristics of these genes are summarized hereafter.
CRYAB is down-regulated in nasopharyngeal carcinoma (NPC) (25) and associated with its progression (26). Further studies were done in testicular (27) and breast (28) cancers. Re-expression of CRYAB in NPC cell lines reduced their tumorigenicity in vivo. NPC progression-associated phenotypes as well as epithelial-mesenchymal transition-associated markers were suppressed upon CRYAB re-expression, strongly suggesting that CRYAB is a tumor suppressor gene in this type of cancer (25).
BRSK1 is a cell cycle inhibitor, inducing a G2/M arrest through the inhibition of Cdk1 (29). BRSK1 has also been recently identified as a tumor suppressor in breast cancer where it regulates the cell cycle via p27kip and the PI3K/Akt pathway (30).
CGREF1 (CGR11) also clearly exhibits growth-suppressive properties. It is regulated by p53 and is able to inhibit the growth of many cell lines (31). Its overexpression significantly inhibits the phosphorylation of ERK and p38 MAPK and suppresses the proliferation of HEK293T and HCT116 cells (32).
RGS16 is a regulator of the G protein signaling family. Its expression is reduced in some breast tumors as well as in pancreatic cancer. The loss of RGS16 in breast tumors promotes PI3K signaling induced by growth factors and thereby induces cell proliferation and resistance to targeted chemotherapies (33). RGS16 has also been shown to inhibit migration and invasion of pancreatic cancer cells (34).
IL24 is a well characterized multifunctional tumor suppressor gene triggering apoptosis and inhibiting angiogenesis and cellular invasion (35, 36). Similar to RGS16 and BRSK1, IL24 has some antitumoral properties through inhibition of the PI3K pathway in lung and breast cancer cells.
CSF2 (GM-CSF) is an antitumor adjuvant that enhances immune responses mainly through the activation of dendritic cells and has been used in the treatment of many cancers including melanoma (37–39). In mouse, it has been shown that lowering Csf2 concentrations in tumor-bearing lungs increases cell invasion and lung colonization (40).
ANPEP (CD13) mRNA expression is down-regulated in invasive tumors such as prostate and colorectal cancers (41, 42).
Down-regulation of BASP1 is a necessary event for Myc-induced oncogenesis (43).
BTG2 is a p53-dependent inhibitor of the proliferation and invasion of gastric cancer cells. It also has a negative effect on cancer cell metastasis by inhibiting Src–focal adhesion kinase signaling (44–46).
DUSP5 functions as a tumor suppressor gene by causing proliferation arrest in the G1/S phase of the cell cycle by dephosphorylating phospho-ERK1/2. It inhibits the progression of prostate carcinoma, and its down-regulation may accurately predict poor prognosis in prostate carcinoma patients. DUSP5 is also down-regulated in gastric cancer following methylation of its promoter (47, 48).
STFA2 (Stefin A) was described to be a promising candidate marker for prognosis in patients with carcinoma of the head and neck where down-regulation of STFA2 expression was noticed compared with the nontumoral mucosa (49).
DHRS9 down-regulation strongly correlates with tumor progression and may serve as a potential prognostic biomarker in colorectal cancer (50).
TPM1 mRNA was consistently lost in breast cancer cells and in squamous cell carcinoma, indicating that this decrease could be a common biochemical event in carcinogenesis (51, 52).
Finally, clinical results reveal that a lower level of SERPINB2 expression is associated with poor prognosis and outcome in head and neck, gastric, breast, and lung cancers (53–55). In contrast, overexpression of SERPINB2 was found to result in suppression of invasion of cancer cells (56, 57).
Currently, we do not know whether or not these above mentioned genes are direct targets of AK144841, but interestingly we observed that the mRNA expressions of 12 of them (75%) (Cryab, Cgref1, Rgs16, Il24, Csf2, Anpep, Tpm1, Basp1, Btg2, SerpinB2, Stfa2, and Dusp5) were lower in mSCC-38 cells that express a high level of AK144841 than in mSCC-20 cells lacking AK144841 (supplemental Fig. S3), supporting the model of an anticorrelation existing between expression of AK144841 and that of the above mentioned genes. The fact that expression of these tumor inhibitors was lower in mSCC-38 cells than in mSCC-20 cells could also explain the higher tumorigenicity of mSCC-38 compared with mSCC-20 and highlight the impact of AK144841 on the tumoral phenotype in these cells. It is worth noting that this effect does not likely result from an alteration of the survival and proliferative properties of the SCC cells because in vitro overexpression or silencing of AK144841 has no obvious effect on these parameters. Moreover, siRNA-mediated AK144841 silencing also did not impair tumor engraftment (not shown).
Besides, AK144841 silencing also stimulated the expression of genes involved in the terminal differentiation of keratinocytes, among which are members of the late cornified envelop group 1 cluster (Lce1-i/g/c/a2/j/e) and Crct1. This suggests that the up-regulation AK144841 that we observed in tumors can prevent the differentiation program. In mice, the Lce1 cluster comprises 14 genes (Lce1a–Lce1m) located on chromosome 3. This region, called the epidermal differentiation complex, also includes genes encoding loricrin (Lor), involucrin (Ivl), filagrin (Flg), and the small proline-rich protein family members (Sprrs). Lce1s are expressed in the later stages of epithelial differentiation and are incorporated into the cornified envelope through cross-linking by transglutaminases (58). In association with other differentiation genes such as Lor, Ivl, Flg, and Sprrs, Lce1 family members contribute to the barrier function of the skin (59). The alteration of AK144841 expression clearly impacts the level of seven of the 14 Lce1 family members (Lce1a2, -c, -e, -g, -h, -i, and -j) but has no effect on Lor, Ivl, Flg, or Elovl3 expression, thus implying that AK144841 only controls a part of the differentiation program.
The differentiation status of tumors often influences their aggressiveness. Immature tumors are generally more aggressive than differentiated tumors (60). In line with this concept, we observed that the highly tumorigenic mSCC-38 cell line expressed lower levels of differentiation markers such as Lor, Ivl, Flg, Abca12, Elovl3, and Sprr than the non-tumorigenic mSCC-20 cells (supplemental Fig. S3). By targeting the Lce1 family genes, AK144841 could thus partake in the dedifferentiation program and cSCC tumor growth.
It is also worth noting that AK144841 was up-regulated in all DMBA/TPA-induced tumors, whereas it was only up-regulated in some papillomas. Because about 10% of the DMBA/TPA-induced papillomas progress into tumors, it would be particularly interesting to understand the exact contribution of AK144841 to such tumoral transformation.
The regulation of skin differentiation by ncRNA is a recent concept. Some microRNAs have been reported to play a role in epidermal differentiation (61, 62), and it has been recently demonstrated that skin differentiation could also be regulated by two lncRNAs. The first one, TINCR, is required for high messenger RNA abundance of key differentiation genes such as FLG, LOR, KRT1 ALOXE3, ALOX12B, CASP14, and ELOVL3 (12) but was not reported to control Lce1 family gene expression. The second one, DANCR (anti-differentiation ncRNA), suppresses differentiation by down-regulating FLG, LOR, KRT1, KRT10, and LCE1s (13).
We examined the expression of Tincr and Dancr in our murine tumorigenesis model (RNAseq data) and observed that, although easily detectable, Dancr does not significantly vary between normal and tumor tissues. In contrast and as reported in human SCC (12), Tincr was at least 5-fold lower in cSCC tumors and in mSCC-38 cells than in normal skin and in mSCC-20 cells, respectively. The mirror expression of Tincr and AK144841 could therefore contribute to the loss of differentiation in tumor and in mSCC-38 through their respective effects on Lor, Ivl, Flg, Abca12, and Elovl3 on one hand and on Lce1s and Crct1 on the other hand (supplemental Fig. S3).
Interestingly, along with their involvement in keratinocyte differentiation, LCE1 members also exert a tumor suppressor action by regulating the activity of the arginine methylase PRMT5 (63). By targeting Lce1s, AK144841 could thus influence both the differentiation status of tumors and their growth.
To date, the mechanism of action of AK144841 still remains to be identified. However, we can already hypothesize that its four exons are critical for mediating its biological effect. Indeed, the deletion of the second exon (observed in AKS overexpression experiments) does impair the biological function of AK144841 even if it does not fully inhibit it. These aspects are beyond the scope of the present study and will be the matter of future investigations.
We provide experimental data supporting the existence of a transcript potentially encoding hAK144841 in SCC of different origins, but a further structural characterization is also necessary. hAK144841 is absent from normal keratinocytes, suggesting a possible role in tumoral progression. To date, one human lncRNA (PICSAR) has been suspected to be involved the development of sSCC (19), but our RNAseq data experiments did not reveal any PICSAR expression in CAL165, i.e. the cell type expressing the highest level of hAK144841. This supports the idea of additional lncRNAs contributing to this pathology. In this context, hAK144841 could represent an interesting candidate.
The study of the biological structure and function of hAK144841 is still underway. However, our functional findings relative to its mouse homolog and particularly its effect on antitumoral genes suggest that this novel actor could possibly contribute to cSCC in humans as well, opening the way for new diagnostic and/or therapeutic solutions. Early identification of hAK144841 in pretumoral lesions such as actinic keratosis could help preempt tumor development and improve response to treatment.
Experimental procedures
DMBA/TPA treatment
The tumor induction procedure was carried out as described previously (8) in accordance with the Declaration of Helsinki and was approved by the local ethics committee. Six-week-old female FVB/N mice (n = 45) were subjected to the DMBA two-hit multistage skin carcinogenesis protocol as described previously (20). Mice were topically treated with 200 nmol of DMBA in 0.2 ml of acetone and then twice weekly for 6 weeks with 5 nmol of PMA. A second dose of DMBA was administered at the 8th week followed by the resumption of PMA treatment for 14 more weeks. Control mice (n = 20) were only topically treated with 0.2 ml of acetone. Mice were monitored weekly for papillomas and tumors. Healthy skin samples (acetone-treated), papillomas, and tumors were harvested throughout the protocol, and biopsies were frozen for RNA extraction.
Cell cultures
Murine cSCC cell lines mSCC-20 and mSCC-38
mSCC-20 and mSCC-38 cell lines were established from DMBA/PMA-induced cSCCs. Tumors were dissected, cut into 1–2-mm3 pieces, and incubated with collagenase A (1.5 mg/ml) and Dispase II (5 mg/ml) for 1 h at 37 °C under orbital shaking. After filtration on a 70-μm mesh and centrifugation at 1800 rpm for 5 min, cells were plated on culture flasks coated with type I collagen in the presence of lethally irradiated feeder cells in William's medium E without calcium but containing antibiotics (100 IU/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml Fungizone). Cells were progressively cultured under classical conditions (in Dulbecco's modified Eagle's medium containing 10% FCS without antibiotics, collagen coating, and feeder cells) and used between passages 30 and 50 in our experiments. As reported previously (8), mSCC-38 generated tumors when injected subcutaneously in nude mice, whereas mSCC-20 did not.
Human keratinocytes
Human keratinocytes isolated from healthy neonatal foreskin as described (64) were cultured on a feeder of lethally irradiated 3T3 fibroblasts.
Human oral squamous cell carcinoma cell lines
CAL27, CAL33, CAL60, CAL165, and CAL166 (65) cell lines were kindly provided by Dr. J. L. Fischel (Centre Antoine Lacassagne, Nice, France), and the A431 cell line was obtained from ATCC. All these cell lines were grown in DMEM containing 10% fetal calf serum (Hyclone).
Cloning procedures
Because of their close proximity, the short (AKS) and long (AKL) isoforms of AK144841 could not be individually isolated after PCR amplification. To circumvent this hurdle, AKS and AKL were amplified together by standard PCR from poly(A)+ cDNA of mSCC-38 cells and ligated into pLJM1 vector between AgeI (5′) and EcoRI (3′) sites. STBL2 bacteria were transformed with pLJM1-AKL+AKS, and colonies were screened by PCR for the presence of either AKL or AKS after cleavage with AgeI and EcoRI. The clones containing AKS or AKL were expanded, and the plasmids were purified using the NucleoBond Xtra Maxi kit (Macherey Nagel).
Northern blot analysis
Northern blotting was carried out using 400 and 200 ng of mSCC-20 and mSCC-38 poly(A)+ purified RNA following the procedure described previously (66). Membranes were hybridized with a 32P-radiolabeled AK144841-specific probe (879 bp) amplified by polymerase chain reaction from cDNA of mSCC-38 using the following primer set: forward, 5′GGTGAGACACAAATGGACCTGC-3′; reverse, 5′-TGGGAACTCGGGTTTCACTCC-3′.
Gene expression analyses by PCR
Total RNA was isolated with Qiazol reagent (Qiagen) according to the manufacturer's instructions. For gene expression, RNAs were reverse transcribed using the SuperScript II reverse transcriptase kit (Life Technologies) in the presence of oligo(dT). mRNA levels of candidate genes were quantified by one-step real-time qPCR performed with 10 ng of cDNA and using the SYBR Green I Master Mix (Roche Applied Science). Gene expression levels were normalized against Rplp0. All reactions were done in triplicate using the Light Cycler 480 Sequence detection system (Roche Applied Science) and expression levels were calculated using the comparative threshold cycle (CT) method (2−ΔΔCT).
Gene expression was also measured by standard PCR. In this condition, genes were amplified by GoTaq DNA polymerase using 100 ng of cDNA. The full-length AK144841 (AKL and AKS) cloned in pLJM1 was obtained by standard PCR amplification using the high fidelity PrimeSTAR DNA polymerase (Takara). All primer sequences corresponding to the different genes analyzed in this study are available upon request.
Transfection experiments
mSCC-38 cells were transiently transfected with pLJM1 (Addgene) containing either the AKL or AKS isoform of AK144841 using Lipofectamine 3000 (Gibco) according to the manufacturer's protocol. Cells were harvested after 48 or 72 h and directly lysed in Qiazol reagent to extract RNA. A condition where cells were transfected with empty PLJM1 (mock) was also included in the experiments.
The siRNA transfection experiments were carried out in mSCC-38 cells using the RNAiMAX Lipofectamine reagent as devised by the manufacturer (Invitrogen). Two different siRNAs raised against AK144841 (Si1AK and Si2AK) and a control (SiC) were used at 20 nm each. Cells were directly lysed in Qiazol 48 or 72 h after transfection. The different siRNAs were designed and synthesized by Dharmacon: Si1AK, CCAAGAGAATAGAGATTAT; Si2AK, GCTGGGAGAAGAAGAGATT.
Subcellular fractionation
The nucleus and cytoplasm of mSCC-38 were separated using the Ambion Protein and RNA Isolation System, PARISTM. RNA was purified from each cellular compartment using Trizol LS solution (Ambion) according to the manufacturer's procedure and reverse transcribed using SuperScript II reverse transcriptase. The expression of AK144841, Neat1, and Malat1 (markers of the nuclear compartment) and Rplp0 (cytoplasm reporter) was measured by qPCR as described above.
Gene expression analysis by microarrays
For gene expression arrays, RNA samples (RNA integrity number >9) were labeled with Cy3 dye using the Low Input Quick Amp kit (Agilent) as recommended by the supplier. 600 ng of Cy3-labeled cRNA probes was hybridized on 8 × 60,000 high density Agilent SurePrint G3 gene expression mouse microarrays.
Gene expression profiles in healthy skin and DMBA-TPA tumors
Three biological replicates (in vivo derived healthy and tumor skin samples) were analyzed for each condition.
Effect of siRNA on the transcriptome of mSCC-38 cells
Fourteen samples from siRNA-transfected mSCC-38 cells (three control siRNAs (SiC), four Si1AK, three Si2AK, and four nontransfected cell samples) were hybridized on the Agilent microarrays.
Microarray data analyses were performed using R (The R Project for Statistical Computing). Quality control of expression arrays was performed using the Bioconductor package arrayQualityMetrics and custom R scripts. Additional analyses of expression arrays were performed using the Bioconductor package limma. Briefly, data were normalized using the quantile method. No background subtraction was performed. Replicated probes were averaged after normalization and control probes were removed. Statistical significance was assessed using the limma moderated t statistic. p values were adjusted for multiple testing using the Benjamini-Hochberg procedure, which controls the false discovery rate.
RNA sequencing
2 μg of total RNAs was extracted from mouse sSCC tumors, normal skin samples, and papillomas as well as cultured mSCC-38 and mSCC-20 cells and human head and neck carcinoma cells (CAL165). Poly(A) RNAs were purified using a Dynabeads mRNA purification kit (Invitrogen) and tagmented for 9 min at 95 °C. RNA libraries were then generated with the NEBNext Small Library Prep Set for SOLiD (New England Biolabs) and sequenced on the Applied Biosystems SOLiD 5500 Wildfire system following the manufacturer's instructions. All data generated from RNA sequencing were stored on the microarray information system Mediante (67).
Reads from the SOLiD system were aligned in color space to mouse genome release mm9 for mouse samples and human release hg19 for CAL165 with LifeScope software (Life Technologies) using default parameters for RNA sequencing. For a given gene, the mapped sequence reads were counted against the current gene annotation general transfer format files for mouse and human (mm9.gtf and hg19.gtf). For each gene, the raw read data were normalized and expressed as reads per kilobase per million reads.
Author contributions
G. P. carried out most of the experiments. R. R. developed the biological model and isolated the biopsies and the mSCC38 and mSCC20 cell lines. I. B. performed the PCR experiments to validate the existence of AK114841 and to measure its expression in different biological samples. R. A. carried out all plasmid constructions used in the study. V. M. and G. R. performed the high-throughput sequencing and the microarrays experiments, respectively. N. N. and A. P. performed all bioinformatics analyses. B. M. and P. B. read the manuscript and contributed to design of the experiments and discussion of the results.
Supplementary Material
Acknowledgments
Sequencing was performed by “UCA GenomiX” Functional Genomics Platform of the University of Nice Sophia Antipolis. We also thank Liliane Michalik (Center for Integrative Genomic, University of Lausanne, Switzerland) and Annabelle Mouchotte for skillful discussions and technical help.
This work was supported by the CNRS, INSERM, the French Government (Agence Nationale de la Recherche (ANR)) through the “Investments for the Future” LABEX SIGNALIFE (Grant ANR-11-LABX-0028-01) and FRANCE GENOMIQUE (Grants ANR-10-INBS-09-03 and ANR-10-INBS-09-02), Canceropôle Provence Alpes Côte d'Azur (PACA), and la Ligue contre le cancer. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S3 and Tables S1 and S2.
The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO SuperSeries accession number GSE89516 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE89516).
L. Michalik, personal communication.
- cSCC
- cutaneous squamous cell carcinoma
- SCC
- squamous cell carcinoma
- DMBA
- 7,12-dimethylbenz[a]anthracene
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- lncRNA
- long noncoding RNA
- Lce1
- late cornified envelope-1
- miR
- microRNA
- ANRIL
- antisense noncoding RNA in the INK4 locus
- PICSAR
- p38-inhibited cutaneous squamous cell carcinoma-associated long intervening noncoding RNA
- RNAseq
- RNA sequencing
- ncRNA
- noncoding RNA
- NMK
- normal mouse keratinocyte
- qPCR
- quantitative PCR
- AKS
- AK144841 short
- AKL
- AK144841 long
- NHK
- normal human keratinocyte
- hAK144841
- human ortholog of AK144841
- NPC
- nasopharyngeal carcinoma
- Lor
- loricrin
- Ivl
- involucrin
- Flg
- filagrin
- Sprr
- small proline-rich protein family member
- TINCR
- tissue differentiation-inducing non-protein-coding RNA
- DANCR
- differentiation-antagonizing non-protein-coding RNA
- FC
- -fold change
- Chr
- chromosome.
References
- 1. Madan V., Lear J. T., and Szeimies R. (2010) Non-melanoma skin cancer. Lancet 375, 673–685 [DOI] [PubMed] [Google Scholar]
- 2. Durinck S., Ho C., Wang N. J., Liao W., Jakkula L. R., Collisson E. A., Pons J., Chan S.-W., Lam E. T., Chu C., Park K., Hong S. W., Hur J. S., Huh N., Neuhaus I. M., et al. (2011) Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 1, 137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Emmert S., Schön M. P., and Haenssle H. A. (2014) Molecular biology of basal and squamous cell carcinomas. Adv. Exp. Med. Biol. 810, 234–252 [DOI] [PubMed] [Google Scholar]
- 4. Ratushny V., Gober M. D., Hick R., Ridky T. W., and Seykora J. T. (2012) From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J. Clin. Investig. 122, 464–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rentoft M., Fahlén J., Coates P. J., Laurell G., Sjöström B., Rydén P., and Nylander K. (2011) miRNA analysis of formalin-fixed squamous cell carcinomas of the tongue is affected by age of the samples. Int. J. Oncol. 38, 61–69 [PubMed] [Google Scholar]
- 6. Dziunycz P., Iotzova-Weiss G., Eloranta J. J., Läuchli S., Hafner J., French L. E., and Hofbauer G. F. (2010) Squamous cell carcinoma of the skin shows a distinct microRNA profile modulated by UV radiation. J. Invest. Dermatol. 130, 2686–2689 [DOI] [PubMed] [Google Scholar]
- 7. Wang S. H., Zhou J. D., He Q. Y., Yin Z. Q., Cao K., and Luo C. Q. (2014) miR-199a inhibits the ability of proliferation and migration by regulating CD44-Ezrin signaling in cutaneous squamous cell carcinoma cells. Int. J. Clin. Exp. Pathol. 7, 7131–7141 [PMC free article] [PubMed] [Google Scholar]
- 8. Gastaldi C., Bertero T., Xu N., Bourget-Ponzio I., Lebrigand K., Fourre S., Popa A., Cardot-Leccia N., Meneguzzi G., Sonkoly E., Pivarcsi A., Mari B., Barbry P., Ponzio G., and Rezzonico R. (2014) miR-193b/365a cluster controls progression of epidermal squamous cell carcinoma. Carcinogenesis 35, 1110–1120 [DOI] [PubMed] [Google Scholar]
- 9. Qi P., and Du X. (2013) The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod. Pathol. 26, 155–165 [DOI] [PubMed] [Google Scholar]
- 10. Fatima R., Akhade V. S., Pal D., and Rao S. M. (2015) Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol. Cell. Ther. 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huarte M. (2015) The emerging role of lncRNAs in cancer. Nat. Med. 21, 1253–1261 [DOI] [PubMed] [Google Scholar]
- 12. Kretz M., Siprashvili Z., Chu C., Webster D. E., Zehnder A., Qu K., Lee C. S., Flockhart R. J., Groff A. F., Chow J., Johnston D., Kim G. E., Spitale R. C., Flynn R. A., Zheng G. X., et al. (2013) Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493, 231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kretz M., Webster D. E., Flockhart R. J., Lee C. S., Zehnder A., Lopez-Pajares V., Qu K., Zheng G. X., Chow J., Kim G. E., Rinn J. L., Chang H. Y., Siprashvili Z., and Khavari P. A. (2012) Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 26, 338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gupta R., Ahn R., Lai K., Mullins E., Debbaneh M., Dimon M., Arron S., and Liao W. (2016) Landscape of long noncoding RNAs in psoriatic and healthy skin. J. Invest. Dermatol. 136, 603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liang X., Ma L., Long X., and Wang X. (2015) LncRNA expression profiles and validation in keloid and normal skin tissue. Int. J. Oncol. 47, 1829–1838 [DOI] [PubMed] [Google Scholar]
- 16. Wang Z., Jinnin M., Nakamura K., Harada M., Kudo H., Nakayama W., Inoue K., Nakashima T., Honda N., Fukushima S., and Ihn H. (2016) Long non-coding RNA TSIX is upregulated in scleroderma dermal fibroblasts and controls collagen mRNA stabilization. Exp. Dermatol. 25, 131–136 [DOI] [PubMed] [Google Scholar]
- 17. Sand M., Bechara F. G., Sand D., Gambichler T., Hahn S. A., Bromba M., Stockfleth E., and Hessam S. (2016) Long-noncoding RNAs in basal cell carcinoma. Tumour Biol. 37, 10595–10608 [DOI] [PubMed] [Google Scholar]
- 18. Jiang Y. J., and Bikle D. D. (2014) LncRNA profiling reveals new mechanism for VDR protection against skin cancer formation. J. Steroid Biochem. Mol. Biol. 144, 87–90 [DOI] [PubMed] [Google Scholar]
- 19. Piipponen M., Nissinen L., Farshchian M., Riihilä P., Kivisaari A., Kallajoki M., Peltonen J., Peltonen S., and Kähäri V. M. (2016) Long noncoding RNA PICSAR promotes growth of cutaneous squamous cell carcinoma by regulating ERK1/2 activity. J. Invest. Dermatol. 136, 1701–1710 [DOI] [PubMed] [Google Scholar]
- 20. Owens D. M., Wei S., and Smart R. C. (1999) A multihit, multistage model of chemical carcinogenesis. Carcinogenesis 20, 1837–1844 [DOI] [PubMed] [Google Scholar]
- 21. Johnsson P., Lipovich L., Grandér D., and Morris K. V. (2014) Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 1840, 1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carninci P., and Hayashizaki Y. (1999) High-efficiency full-length cDNA cloning. Methods Enzymol. 303, 19–44 [DOI] [PubMed] [Google Scholar]
- 23. Carninci P., Shibata Y., Hayatsu N., Sugahara Y., Shibata K., Itoh M., Konno H., Okazaki Y., Muramatsu M., and Hayashizaki Y. (2000) Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discover of new genes. Genome Res. 10, 1617–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oshimori N., Oristian D., and Fuchs E. (2015) TGF-β promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 160, 963–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang Z., Cheng Y., Chiu P. M., Cheung F. M., Nicholls J. M., Kwong D. L., Lee A. W., Zabarovsky E. R., Stanbridge E. J., Lung H. L., and Lung M. L. (2012) Tumor suppressor αB-crystallin (CRYAB) associates with the cadherin/catenin adherens junction and impairs NPC progression-associated properties. Oncogene 31, 3709–3720 [DOI] [PubMed] [Google Scholar]
- 26. Lung H. L., Lo C. C., Wong C. C., Cheung A. K., Cheong K. F., Wong N., Kwong F. M., Chan K. C., Law E. W., Tsao S. W., Chua D., Sham J. S., Cheng Y., Stanbridge E. J., Robertson G. P., et al. (2008) Identification of tumor suppressive activity by irradiation microcell-mediated chromosome transfer and involvement of αB-crystallin in nasopharyngeal carcinoma. Int. J. Cancer 122, 1288–1296 [DOI] [PubMed] [Google Scholar]
- 27. Takashi M., Katsuno S., Sakata T., Ohshima S., and Kato K. (1998) Different concentrations of two small stress proteins, αB crystallin and HSP27 in human urological tumor tissues. Urol. Res. 26, 395–399 [DOI] [PubMed] [Google Scholar]
- 28. Lin D. I., Barbash O., Kumar K. G., Weber J. D., Harper J. W., Klein-Szanto A. J., Rustgi A., Fuchs S. Y., and Diehl J. A. (2006) Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-αB crystallin) complex. Mol. Cell 24, 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu R., Niida H., and Nakanishi M. (2004) Human SAD1 kinase is involved in UV-induced DNA damage checkpoint function. J. Biol. Chem. 279, 31164–31170 [DOI] [PubMed] [Google Scholar]
- 30. Wang H., Liu X.-B., Chen J.-H., Wang Q.-Q., Chen J.-P., Xu J.-F., Sheng C.-Y., and Ni Q.-C. (2014) Decreased expression and prognostic role of cytoplasmic BRSK1 in human breast carcinoma: correlation with Jab1 stability and PI3K/Akt pathway. Exp. Mol. Pathol. 97, 191–201 [DOI] [PubMed] [Google Scholar]
- 31. Madden S. L., Galella E. A., Riley D., Bertelsen A. H., and Beaudry G. A. (1996) Induction of cell growth regulatory genes by p53. Cancer Res. 56, 5384–5390 [PubMed] [Google Scholar]
- 32. Deng W., Wang L., Xiong Y., Li J., Wang Y., Shi T., and Ma D. (2015) The novel secretory protein CGREF1 inhibits the activation of AP-1 transcriptional activity and cell proliferation. Int. J. Biochem. Cell Biol. 65, 32–39 [DOI] [PubMed] [Google Scholar]
- 33. Liang G., Bansal G., Xie Z., and Druey K. M. (2009) RGS16 inhibits breast cancer cell growth by mitigating phosphatidylinositol 3-kinase signaling. J. Biol. Chem. 284, 21719–21727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carper M. B., Denvir J., Boskovic G., Primerano D. A., and Claudio P. P. (2014) RGS16, a novel p53 and pRb cross-talk candidate inhibits migration and invasion of pancreatic cancer cells. Genes Cancer 5, 420–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Menezes M. E., Bhatia S., Bhoopathi P., Das S. K., Emdad L., Dasgupta S., Dent P., Wang X.-Y., Sarkar D., and Fisher P. B. (2014) MDA-7/IL-24: multifunctional cancer killing cytokine. Adv. Exp. Med. Biol. 818, 127–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Panneerselvam J., Jin J., Shanker M., Lauderdale J., Bates J., Wang Q., Zhao Y. D., Archibald S. J., Hubin T. J., and Ramesh R. (2015) IL-24 inhibits lung cancer cell migration and invasion by disrupting the SDF-1/CXCR4 signaling axis. PLoS One 10, e0122439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dranoff G. (2003) GM-CSF-secreting melanoma vaccines. Oncogene 22, 3188–3192 [DOI] [PubMed] [Google Scholar]
- 38. Urdinguio R. G., Fernandez A. F., Moncada-Pazos A., Huidobro C., Rodriguez R. M., Ferrero C., Martinez-Camblor P., Obaya A. J., Bernal T., Parra-Blanco A., Rodrigo L., Santacana M., Matias-Guiu X., Soldevilla B., Dominguez G., et al. (2013) Immune-dependent and independent antitumor activity of GM-CSF aberrantly expressed by mouse and human colorectal tumors. Cancer Res. 73, 395–405 [DOI] [PubMed] [Google Scholar]
- 39. Golden E. B., Chhabra A., Chachoua A., Adams S., Donach M., Fenton-Kerimian M., Friedman K., Ponzo F., Babb J. S., Goldberg J., Demaria S., and Formenti S. C. (2015) Local radiotherapy and granulocyte-macrophage colony-stimulating factor to generate abscopal responses in patients with metastatic solid tumours: a proof-of-principle trial. Lancet Oncol. 16, 795–803 [DOI] [PubMed] [Google Scholar]
- 40. Cheng H., Wang A., Meng J., Zhang Y., and Zhu D. (2015) Enhanced metastasis in RNF13 knockout mice is mediated by a reduction in GM-CSF levels. Protein Cell 6, 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sørensen K. D., Abildgaard M. O., Haldrup C., Ulhøi B. P., Kristensen H., Strand S., Parker C., Høyer S., Borre M., and Ørntoft T. F. (2013) Prognostic significance of aberrantly silenced ANPEP expression in prostate cancer. Br. J. Cancer 108, 420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wiese A. H., Auer J., Lassmann S., Nährig J., Rosenberg R., Höfler H., Rüger R., and Werner M. (2007) Identification of gene signatures for invasive colorectal tumor cells. Cancer Detect. Prev. 31, 282–295 [DOI] [PubMed] [Google Scholar]
- 43. Hartl M., Nist A., Khan M. I., Valovka T., and Bister K. (2009) Inhibition of Myc-induced cell transformation by brain acid-soluble protein 1 (BASP1). Proc. Natl. Acad. Sci. U.S.A. 106, 5604–5609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mao B., Zhang Z., and Wang G. (2015) BTG2: a rising star of tumor suppressors (review). Int. J. Oncol. 46, 459–464 [DOI] [PubMed] [Google Scholar]
- 45. Lim S. K., Choi Y. W., Lim I. K., and Park T. J. (2012) BTG2 suppresses cancer cell migration through inhibition of Src-FAK signaling by downregulation of reactive oxygen species generation in mitochondria. Clin. Exp. Metastasis. 29, 901–913 [DOI] [PubMed] [Google Scholar]
- 46. Zhang L., Huang H., Wu K., Wang M., and Wu B. (2010) Impact of BTG2 expression on proliferation and invasion of gastric cancer cells in vitro. Mol. Biol. Rep. 37, 2579–2586 [DOI] [PubMed] [Google Scholar]
- 47. Cai C., Chen J.-Y., Han Z. D., He H. C., Chen J.-H., Chen Y. R., Yang S.-B., Wu Y. D., Zeng Y. R., Zou J., Liang Y. X., Dai Q. S., Jiang F. N., and Zhong W.-D. (2015) Down-regulation of dual-specificity phosphatase 5 predicts poor prognosis of patients with prostate cancer. Int. J. Clin. Exp. Med. 8, 4186–4194 [PMC free article] [PubMed] [Google Scholar]
- 48. Shin S. H., Park S. Y., and Kang G. H. (2013) Down-regulation of dual-specificity phosphatase 5 in gastric cancer by promoter CpG island hypermethylation and its potential role in carcinogenesis. Am. J. Pathol. 182, 1275–1285 [DOI] [PubMed] [Google Scholar]
- 49. Strojan P., Anicin A., Svetic B., Pohar M., Smid L., and Kos J. (2007) Stefin A and stefin B: markers for prognosis in operable squamous cell carcinoma of the head and neck. Int. J. Radiat. Oncol. Biol. Phys. 68, 1335–1341 [DOI] [PubMed] [Google Scholar]
- 50. Hu L., Chen H.-Y., Han T., Yang G.-Z., Feng D., Qi C.-Y., Gong H., Zhai Y.-X., Cai Q.-P., and Gao C.-F. (2016) Downregulation of DHRS9 expression in colorectal cancer tissues and its prognostic significance. Tumour Biol. 37, 837–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zare M., Jazii F. R., Soheili Z.-S., and Moghanibashi M.-M. (2012) Downregulation of tropomyosin-1 in squamous cell carcinoma of esophagus, the role of Ras signaling and methylation. Mol. Carcinog. 51, 796–806 [DOI] [PubMed] [Google Scholar]
- 52. Bharadwaj S., and Prasad G. L. (2002) Tropomyosin-1, a novel suppressor of cellular transformation is downregulated by promoter methylation in cancer cells. Cancer Lett. 183, 205–213 [DOI] [PubMed] [Google Scholar]
- 53. Chou R. H., Wen H. C., Liang W. G., Lin S. C., Yuan H. W., Wu C. W., and Chang W. S. (2012) Suppression of the invasion and migration of cancer cells by SERPINB family genes and their derived peptides. Oncol. Rep. 27, 238–245 [DOI] [PubMed] [Google Scholar]
- 54. Huang Z., Li H., Huang Q., Chen D., Han J., Wang L., Pan C., Chen W., House M. G., Nephew K. P., and Guo Z. (2014) SERPINB2 down-regulation contributes to chemoresistance in head and neck cancer. Mol. Carcinog. 53, 777–786 [DOI] [PubMed] [Google Scholar]
- 55. Yoshino H., Endo Y., Watanabe Y., and Sasaki T. (1998) Significance of plasminogen activator inhibitor 2 as a prognostic marker in primary lung cancer: association of decreased plasminogen activator inhibitor 2 with lymph node metastasis. Br. J. Cancer 78, 833–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mueller B. M., Yu Y. B., and Laug W. E. (1995) Overexpression of plasminogen activator inhibitor 2 in human melanoma cells inhibits spontaneous metastasis in scid/scid mice. Proc. Natl. Acad. Sci. U.S.A. 92, 205–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smith R., Xue A., Gill A., Scarlett C., Saxby A., Clarkson A., and Hugh T. (2007) High expression of plasminogen activator inhibitor-2 (PAI-2) is a predictor of improved survival in patients with pancreatic adenocarcinoma. World J. Surg. 31, 493–502 [DOI] [PubMed] [Google Scholar]
- 58. Jackson B., Tilli C. M., Hardman M. J., Avilion A. A., MacLeod M. C., Ashcroft G. S., and Byrne C. (2005) Late cornified envelope family in differentiating epithelia—response to calcium and ultraviolet irradiation. J. Invest. Dermatol. 124, 1062–1070 [DOI] [PubMed] [Google Scholar]
- 59. Candi E., Schmidt R., and Melino G. (2005) The cornified envelope: a model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 6, 328–340 [DOI] [PubMed] [Google Scholar]
- 60. Daley G. Q. (2008) Common themes of dedifferentiation in somatic cell reprogramming and cancer. Cold Spring Harb. Symp. Quant. Biol. 73, 171–174 [DOI] [PubMed] [Google Scholar]
- 61. Candi E., Amelio I., Agostini M., and Melino G. (2015) MicroRNAs and p63 in epithelial stemness. Cell Death Differ. 22, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bertero T., Gastaldi C., Bourget-Ponzio I., Mari B., Meneguzzi G., Barbry P., Ponzio G., and Rezzonico R. (2013) CDC25A targeting by miR-483–3p decreases CCND-CDK4/6 assembly and contributes to cell cycle arrest. Cell Death Differ. 20, 800–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Deng Z., Matsuda K., Tanikawa C., Lin J., Furukawa Y., Hamamoto R., and Nakamura Y. (2014) Late cornified envelope group I, a novel target of p53, regulates PRMT5 activity. Neoplasia 16, 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rheinwald J. G., and Green H. (1975) Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 6, 331–343 [DOI] [PubMed] [Google Scholar]
- 65. Magné N., Fischel J. L., Dubreuil A., Formento P., Poupon M.-F., Laurent-Puig P., and Milano G. (2002) Influence of epidermal growth factor receptor (EGFR), p53 and intrinsic MAP kinase pathway status of tumour cells on the antiproliferative effect of ZD1839 (“Iressa”). Br. J. Cancer 86, 1518–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rezzonico R., Loubat A., Lallemand D., Pfarr C. M., Far D. F., Proudfoot A., Rossi B., and Ponzio G. (1995) Cyclic AMP stimulates a JunD/Fra-2 AP-1 complex and inhibits the proliferation of interleukin-6-dependent cell lines. Oncogene 11, 1069–1078 [PubMed] [Google Scholar]
- 67. Le Brigand K., and Barbry P. (2007) Mediante: a web-based microarray data manager. Bioinformatics 23, 1304–1306 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.