Abstract
RUNX1 is a member of RUNX transcription factors and plays important roles in hematopoiesis. Disruption of RUNX1 activity has been implicated in the development of hematopoietic neoplasms. Chromosomal translocations involving the RUNX1 gene are associated with several types of leukemia, including acute myeloid leukemia driven by a leukemogenic fusion protein RUNX1–RUNX1T1. Previous studies have shown that RUNX1 is an unstable protein and is subjected to proteolytic degradation mediated by the ubiquitin–proteasome pathway. However, the precise mechanisms of RUNX1 ubiquitination have not been fully understood. Furthermore, much less is known about the mechanisms to regulate the stability of RUNX1–RUNX1T1. In this study, we identified several RUNX1-interacting E3 ubiquitin ligases using a novel high-throughput binding assay. Among them, we found that STUB1 bound to RUNX1 and induced its ubiquitination and degradation mainly in the nucleus. Immunofluorescence analyses revealed that the STUB1-induced ubiquitination also promoted nuclear export of RUNX1, which probably contributes to the reduced transcriptional activity of RUNX1 in STUB1-overexpressing cells. STUB1 also induced ubiquitination of RUNX1–RUNX1T1 and down-regulated its expression. Importantly, STUB1 overexpression showed a substantial growth-inhibitory effect in myeloid leukemia cells that harbor RUNX1–RUNX1T1, whereas it showed only a marginal effect in other non-RUNX1–RUNX1T1 leukemia cells and normal human cord blood cells. Taken together, these data suggest that the E3 ubiquitin ligase STUB1 is a negative regulator of both RUNX1 and RUNX1–RUNX1T1. Activation of STUB1 could be a promising therapeutic strategy for RUNX1–RUNX1T1 leukemia.
Keywords: cancer biology, E3 ubiquitin ligase, leukemia, nuclear transport, post-translational modification (PTM), protein degradation, transcription factor, ubiquitin ligase, ubiquitin-dependent protease, ubiquitylation (ubiquitination)
Introduction
RUNX1 (also called AML1) belongs to a family of transcriptional regulators, called RUNX (1). There are three mammalian RUNX members, RUNX1, RUNX2, and RUNX3. All RUNX proteins contain a conserved Runt domain responsible for sequence-specific DNA binding, and make heterodimeric complexes with a partner protein, CBFB. 2 RUNX1 plays important roles for the generation of definitive hematopoietic stem cells and for hematopoietic differentiation to myeloid and lymphoid lineages (2). As a master regulator for hematopoiesis, RUNX1 demands precise control of its function. Ubiquitination-driven proteolytic degradation of RUNX1 is one of the mechanisms to regulate RUNX1 activity (3). Multiple lysine residues (Lys-24, Lys-43, Lys-83, Lys-90, Lys-125, Lys-144, Lys-167, Lys-182, and Lys-188), clustering within or around the Runt domain, are likely targets of ubiquitination in RUNX1 (3, 4). Previous studies have identified several candidate regulators of RUNX1 ubiquitination. CBFB and a histone methyltransferase MLL bind to the Runt domain and prevent RUNX1 degradation by reducing its polyubiquitination (4, 5). APC and SCF complex can degrade RUNX1 in a cell-cycle and phosphorylation-dependent manner (6). Other reports suggested that STUB1 (also called CHIP) is an E3 ubiquitin ligase that promotes degradation of Runx1 and Runx2 (7, 8). WWP1 and NEDD4 were also shown to promote ubiquitination of RUNX proteins (9, 10). However, E3 ubiquitin ligases that regulate stability and activity of RUNX1 have not been investigated systematically. Furthermore, the role of endogenous E3 ligases in the regulation of RUNX1 stability remains unclear because most studies described above used only cell-based assays with overexpression of those E3 ligases.
Disruption of RUNX1 activity has been implicated in the development of hematopoietic neoplasms. Chromosomal translocations involving the RUNX1 gene are associated with several types of leukemia, including acute myeloid leukemia driven by a leukemogenic fusion protein RUNX1–RUNX1T1 (also called RUNX1-ETO or AML1-MTG8). RUNX1–RUNX1T1 contains the N-terminal 177 amino acids of RUNX1 fused in-frame with almost the entire RUNX1T1 protein. RUNX1–RUNX1T1 increases self-renewal of hematopoietic stem cells and promotes leukemogenesis in cooperation with other growth-promoting mutations (11). Much less is known about the mechanisms to regulate RUNX1–RUNX1T1 stability. A study showed that RUNX1–RUNX1T1 is degraded by the combination of an E2 conjugase UbcH8 and an E3 ligase SIAH1 (12). Given that 7 of 9 ubiquitination-related lysine residues of RUNX1 are retained in RUNX1–RUNX1T1 (Fig. 1A), this fusion protein is likely subjected to clearance by the similar mechanisms that degrade RUNX1. However, this possibility needs to be demonstrated experimentally.
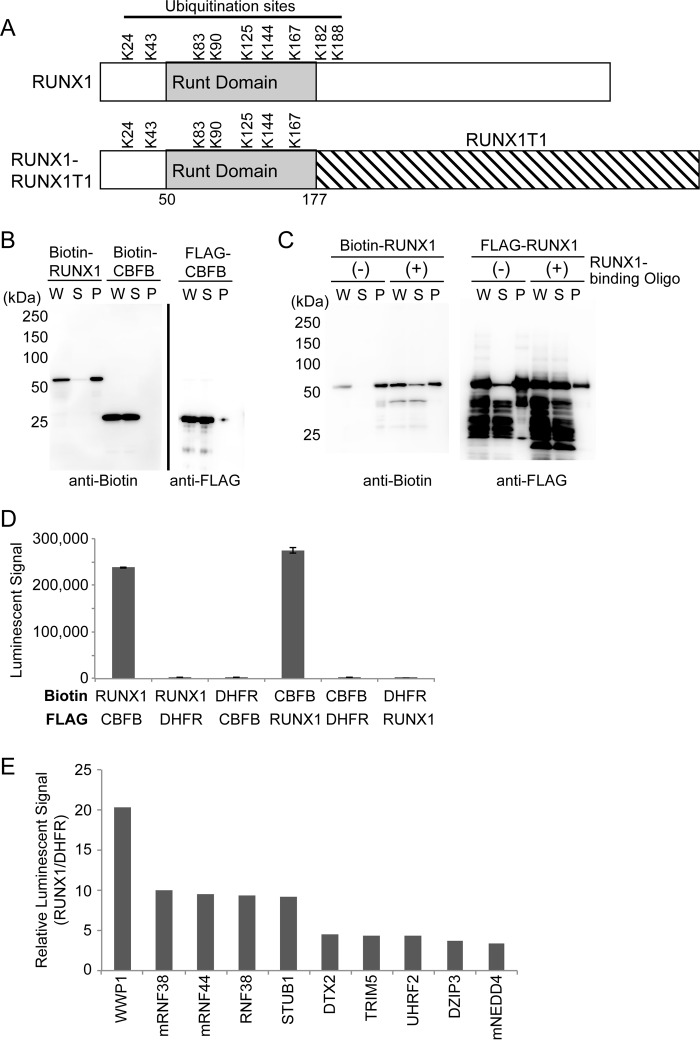
Figure 1.
AlphaScreen identified RUNX1-interacting E3 ubiquitin ligases. A, ubiquitination sites of human RUNX1 (RUNX1b, NP_001001890.1) and RUNX1–RUNX1T1. RUNX1–RUNX1T1 contains the N-terminal 177 amino acids of RUNX1 fused in-frame with almost the entire RUNX1T1 protein. Numbers indicate positions of amino acid residues from the N terminus. Runt domain is the DNA- and CBFB-binding domain. Note that 7 of 9 lysine residues of RUNX1 are retained in RUNX1–RUNX1T1. B, expression of recombinant proteins of RUNX1 and CBFB with the wheat cell-free system. RUNX1 and CBFB were synthesized as N-terminal FLAG-tagged or biotinylated forms. The total translation mixture of each protein was centrifuged, and the whole translation mixture (W), supernatant (S), and insoluble pellet (P) were subjected to SDS-PAGE followed by immunoblot analysis using anti-biotin or anti-FLAG antibodies. CBFB was detected in soluble fraction, whereas RUNX1 was observed mainly in the insoluble pellet fraction. C, RUNX1 protein could become a soluble form in the presence of oligonucleotides with RUNX1-binding sequence. D, AlphaScreen assay to detect binding between RUNX1 and CBFB. Crude translation mixtures of biotin- or FLAG–RUNX1 and biotin- or FLAG–CBFB (0.75 μl each) were used for the assay. Biotin- or FLAG-DHFR was used as a negative control of binding to RUNX1 or CBFB. E, AlphaScreen assay to detect the binding between RUNX1 and each E3 ligase. Binding of 287 E3 ligases to biotinylated RUNX1 and biotinylated DHFR was measured (supplemental Table S1), and the relative value of binding was calculated as follows: value from E3 and RUNX1/value from E3 and DHFR. The top 10 E3 ligases that showed high signals are shown.
Although interactions between RUNX1 and E3 ligases have been examined using cell-based assays, such assays have several limitations including the frequent detection of indirect interactions. Recently, we developed a novel high-throughput binding assay to identify direct interactions between E3 ligases and their target proteins. We produced a protein array containing 287 E3 ligases using a wheat cell-free protein synthesis system, and applied a luminescence-based binding assay (AlphaScreen) to screen the protein interactions (13, 14). Using this assay, we identified several E3 ligases that bind to RUNX1. Among the RUNX1-interacting E3 ligases, we found that STUB1 promotes ubiquitination of RUNX1 and RUNX1–RUNX1T1, and thereby inhibits their activities. STUB1 is a co-chaperone protein and a U-box containing E3 ligase, and has been shown to promote the ubiquitination and degradation of chaperone-bound proteins (15, 16). Importantly, STUB1 overexpression showed a growth-inhibitory effect in RUNX1–RUNX1T1 leukemia cells, indicating that activation of STUB1 can be a promising strategy to treat RUNX1–RUNX1T1-driven leukemia.
Results
AlphaScreen revealed RUNX1-interacting E3 ubiquitin ligases
We first synthesized recombinant proteins of RUNX1 and its cofactor CBFB with the wheat cell-free system in N-terminal FLAG-tagged forms and N-terminal single biotinylated forms. The recombinant CBFB was observed in the whole translation mixture (W) and the supernatant (S) after centrifugation of the former, indicating that CBFB was synthesized in a soluble form. The recombinant RUNX1 was observed mainly in the insoluble pellet (P) fraction, but addition of oligonucleotides with the RUNX1-binding sequence promoted shift of the RUNX1 protein to the soluble fraction (Fig. 1, B and C). AlphaScreen, a luminescence-based interaction assay, showed a high signal when RUNX1 and CBFB were reacted, confirming the strong interaction between these proteins (Fig. 1D). These results validated the quality of the recombinant RUNX1 protein synthesized with the wheat cell-free system, and the usefulness of the AlphaScreen assay to detect protein–protein interactions. Next, we screened RUNX1-binding ubiquitin ligases using the protein array containing 287 E3 ligases. We also assessed interaction between each E3 and DHFR to subtract the background signal of nonspecific binding. In this assay, E3 ligases that showed a relative luminescence value of more than 3 are considered good candidates for the binding partners of the substrates. Nine E3 ligases, WWP1, RNF38, RNF44, STUB1, DTX2, TRIM5, UHRF2, DZIP3, and NEDD4 showed high signals (Fig. 1E, supplemental Table S1), suggesting that these E3 ligases are likely to interact with RUNX1. Among them, STUB1, WWP1, and NEDD4 were shown to promote ubiquitination of RUNX proteins in previous reports (7–10). None of the E3 ligases showed signals as high as that of CBFB (supplemental Table S1), indicating relatively weak interaction between RUNX1 and E3 ligases. The weak interaction may be important to achieve dynamic regulation of RUNX1 stability.
STUB1 promoted ubiquitination and degradation of RUNX1
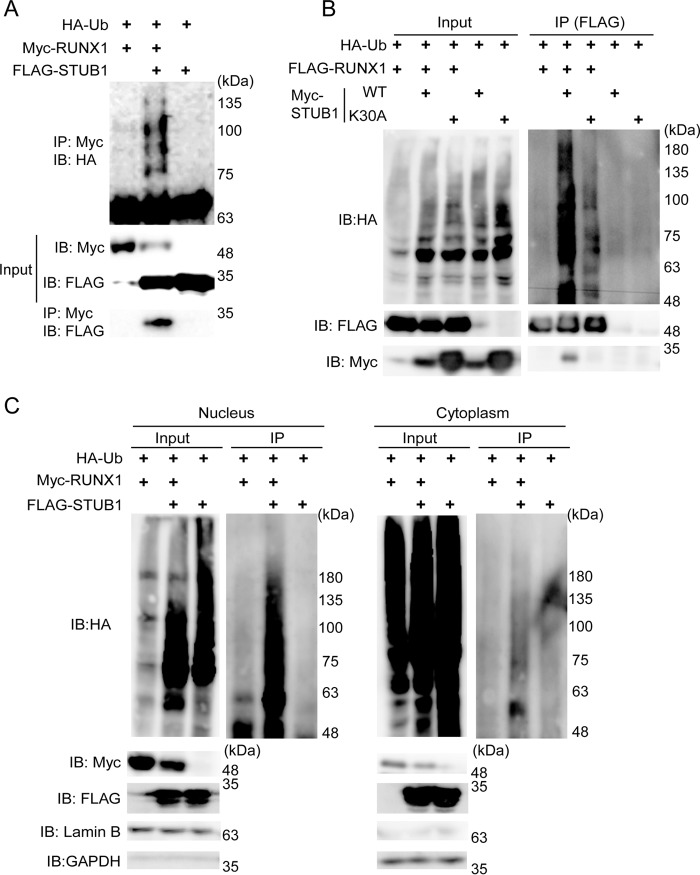
Among the candidate RUNX1-binding E3 ligases, STUB1 has been shown to interact with RUNX proteins (7, 8). We therefore first assessed whether overexpression of STUB1 promotes RUNX1 ubiquitination in a cell-based assay. We introduced Myc- or FLAG-tagged RUNX1, HA-tagged ubiquitin, and FLAG- or Myc-tagged STUB1 into 293T cells. Cell lysates were subjected to immunoprecipitation with an antibody for RUNX1 (anti-FLAG or anti-Myc), followed by immunoblotting with anti-HA to detect ubiquitinated RUNX1. We found that STUB1 bound to RUNX1, down-regulated RUNX1 expression, and strongly induced RUNX1 ubiquitination (Fig. 2A). Interestingly, STUB1-K30A, a mutant that binds neither Hsp90 nor Hsp70 chaperones (17), induced ubiquitination of RUNX1 less efficiently compared with wild-type STUB1 (Fig. 2B). The STUB1-induced ubiquitination of RUNX1 was mainly observed in the nucleus (Fig. 2C).
Figure 2.
STUB1 overexpression promoted ubiquitination and degradation of RUNX1. A, 293T cells were transfected with Myc–RUNX1, HA–ubiquitin, and FLAG–STUB1. Whole-cell extracts were immunoprecipitated with anti-Myc antibody, and ubiquitinated RUNX1 was detected with anti-HA antibody. RUNX1-bound STUB1 was detected with anti-FLAG. B, 293T cells were transfected with FLAG–RUNX1, HA–ubiquitin, Myc–STUB1, or Myc–STUB1-K30A. Whole-cell extracts were immunoprecipitated with anti-FLAG antibody, and ubiquitinated RUNX1 was detected with anti-HA antibody. RUNX1-bound STUB1 was detected with anti-Myc. STUB1-K30A showed reduced activity to induce RUNX1 ubiquitination. C, 293T cells were transfected with Myc–RUNX1, HA–ubiquitin, and FLAG–STUB1. Nuclear and cytoplasmic fractions were isolated and were immunoprecipitated with anti-Myc antibody, following the detection of ubiquitinated RUNX1 with anti-HA antibody. STUB1 induced RUNX1 ubiquitination mainly in the nucleus. IB, immunoblot.
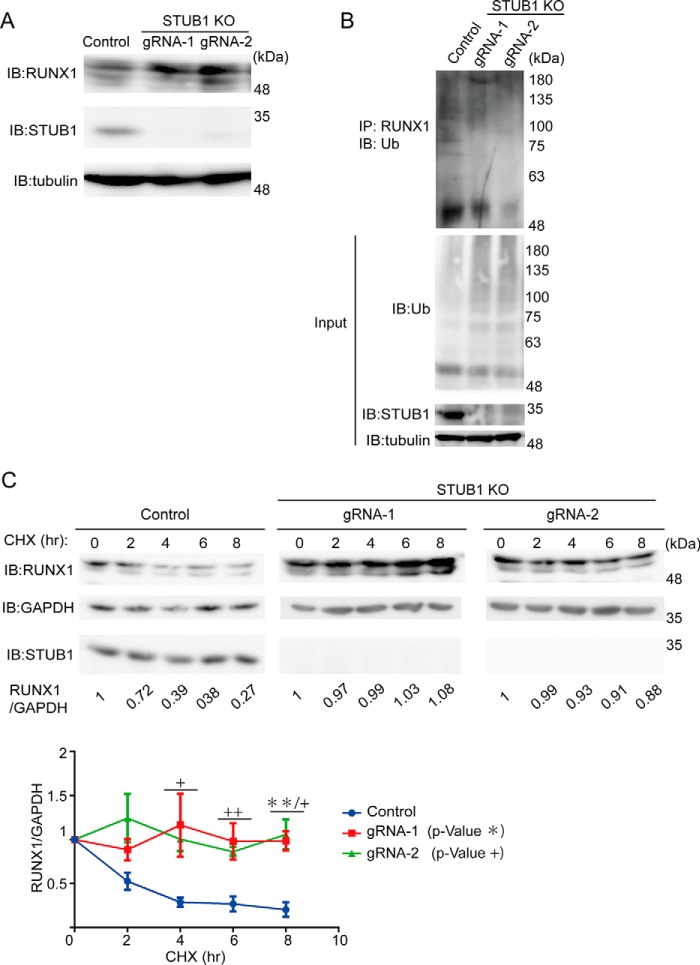
To examine the role of endogenous STUB1 in the regulation of RUNX1 stability, we depleted STUB1 in K562 cells using the CRISPR/Cas9 system. K562 cells were transduced with Cas9 and two independent guide RNAs (gRNAs) targeting STUB1. Both gRNAs induced nearly complete depletion of STUB1, and STUB1 depletion resulted in the increased protein expression and decreased ubiquitination of RUNX1 in K562 cells (Fig. 3, A and B). To compare the stability of RUNX1 protein in STUB1-intact and STUB1-depleted K562 cells, we cultured these cells in the presence of the protein synthesis inhibitor cycloheximide (CHX). Total cell extracts isolated 0, 2, 4, 6, and 8 h after the addition of CHX were subjected to immunoblotting for RUNX1. RUNX1 protein in STUB1-depleted K562 cells showed a slower rate of degradation (Fig. 3C). These data suggest that STUB1 is an E3 ubiquitin ligase that regulates RUNX1 stability.
Figure 3.
STUB1 depletion increased stability of RUNX1. A, K562 cells were transduced with a vector control or two independent gRNAs targeting STUB1. The gRNAs showed efficient depletion of STUB1, which resulted in the increased expression of RUNX1. B, whole-cell extracts from K562 cells described in A were immunoprecipitated with anti-RUNX1 antibody, and ubiquitinated RUNX1 was detected with anti-ubiquitin. STUB1 depletion resulted in the reduced ubiquitination of RUNX1. C, K562 cells transduced with a vector or STUB1 gRNAs were treated with 25 μg/ml of CHX for the indicated times, and cell extracts were analyzed with anti-RUNX1 antibody. Band intensities of RUNX1 and GAPDH were quantified with Multi Gauge (Science Lab), and the intensities of RUNX1 relative to GAPDH are shown. The value of RUNX1/GAPDH without CHX treatment was set to 1. Representative data (upper) and cumulative data from three independent experiments (lower, mean ± S.E., **, p < 0.01; +, p < 0.05; ++, p < 0.01) are shown. IB, immunoblot.
STUB1 altered intracellular localization of RUNX1 and inhibited its transcriptional activity
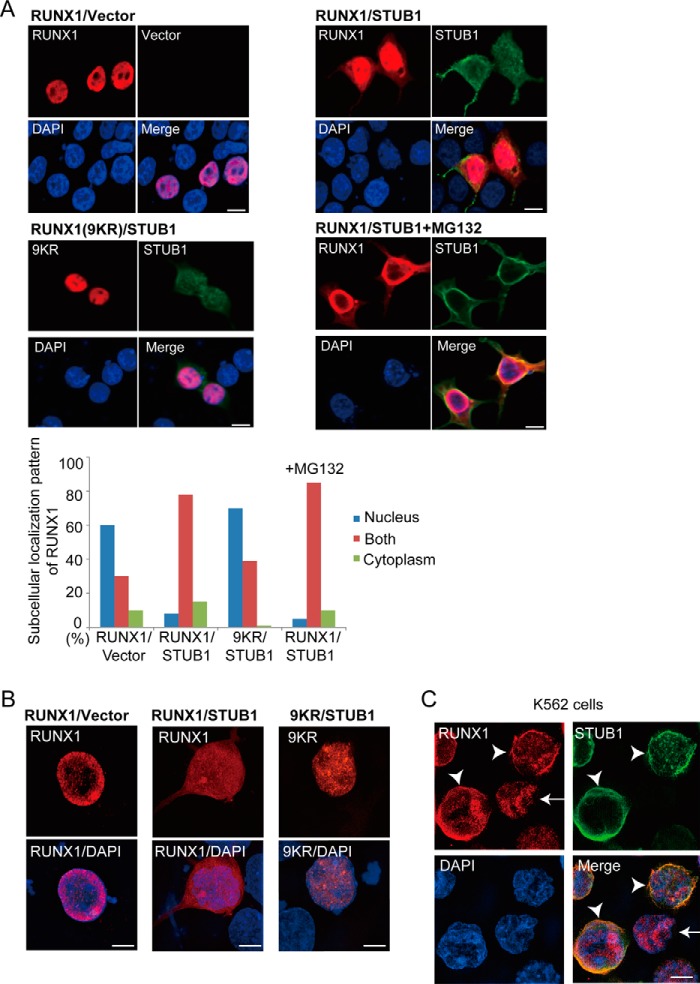
Accumulating evidence suggests that ubiquitination often alters subcellular localization of the target proteins. We therefore examined whether STUB1 overexpression affects RUNX1 localization using immunofluorescence analyses. The 293T cells were transfected with RUNX1 together with vector or FLAG-tagged STUB1, then fixed and stained with anti-RUNX1 and anti-FLAG to observe RUNX1 or STUB1, respectively. RUNX1 showed a nuclear localization pattern in most 293T cells, whereas STUB1 was mainly observed in cytoplasm. Interestingly, STUB1 overexpression sequestered RUNX1 toward the cytoplasm, which resulted in a more diffuse localization pattern of RUNX1 in both the nucleus and cytoplasm. Addition of a proteasome inhibitor, MG132, did not change the diffuse pattern of RUNX1 localization induced by STUB1 overexpression. The characteristic RUNX1 localization induced by STUB1 overexpression was not seen when we used a ubiquitination-deficient form of RUNX1 mutant (a mutant carrying arginine mutation at 9 lysine sites in RUNX1) (Fig. 4, A and B), indicating that STUB1-induced ubiquitination is a critical step for the RUNX1 redistribution. The STUB1-induced cytoplasmic sequestration of endogenous RUNX1 was also observed in K562 cells (Fig. 4C).
Figure 4.
STUB1 promoted nuclear export of RUNX1. A, 293T cells were cotransfected with RUNX1 (wild-type or the 9KR mutant) together with vector or FLAG-tagged STUB1, and stained with anti-RUNX1 (rabbit) and anti-FLAG (mouse) antibody followed by anti-rabbit Alexa 568 (red) and anti-mouse Alexa 488 (green) staining. Nuclei were visualized with DAPI (blue). Bar, 10 μm. Confocal laser scanning microscopy (Nikon A1) was used to observe localization of RUNX1 and STUB1. Note that RUNX1 displayed a more diffuse localization pattern in both nucleus and cytoplasm in many STUB1-transduced cells. This effect was not observed for the 9KR mutant. Addition of MG132 (20 μm) did not change the RUNX1 localization. Subcellular localization of RUNX1 was quantified by counting the number of cells exhibiting nuclear localization (nucleus), diffuse distribution in both nucleus and cytoplasm (both), or cytoplasmic localization (cytoplasm). B, the cells described in A were also analyzed using the super resolution microscopy (Nikon SIM), which clearly showed the diffuse localization of RUNX1 in both nucleus and cytoplasm in STUB1-transduced cells. Bar, 5 μm. C, K562 cells were transduced with FLAG-tagged STUB1 and stained with anti-RUNX1 (rabbit) and anti-FLAG (mouse) antibody followed by anti-rabbit Alexa 568 (red) and anti-mouse Alexa 633 (green) staining. Nuclei were visualized with DAPI (blue). Bar, 5 μm. The super resolution microscopy (Nikon SIM) was used to observe localization of RUNX1 and STUB1. RUNX1 was observed only in the nucleus in cells without STUB1 expression (arrow), whereas RUNX1 was observed in both nucleus and cytoplasm in STUB1-transduced cells (arrowheads).
We next investigated whether STUB1 is involved in RUNX1-mediated transcriptional regulation using the reporter plasmid pMCSFR-luc, containing the promoter region of human MCSF receptor with a RUNX1 consensus site (18). The pMCSFR-luc was cotransfected into 293T cells with RUNX1, CBFB, STUB1, or their corresponding vectors. Consistent with the previous results (18), expression of RUNX1 alone modestly, and coexpression of RUNX1 and CBFB substantially increased the reporter activity. The RUNX1- and RUNX1/CBFB-induced activations of the reporter were abrogated by STUB1 overexpression (Fig. 5). Thus, STUB1 overexpression alters intracellular localization of RUNX1 and inhibits its transcriptional activity.
Figure 5.
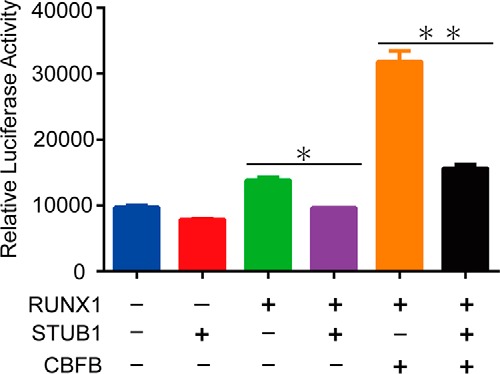
STUB1 inhibited transcriptional activity of RUNX1. 293T cells were cotransfected with pMSCFR-luc and expression vectors for RUNX1, CBFB, and STUB1. Transfection efficiency was normalized using fluorometric quantitation of enhanced green fluorescent protein in a cotransfection of pMYs-IG. All luciferase assays were performed in duplicates (data are shown as the mean ± S.E., *, p = 0.0054; **, p = 0.0055).
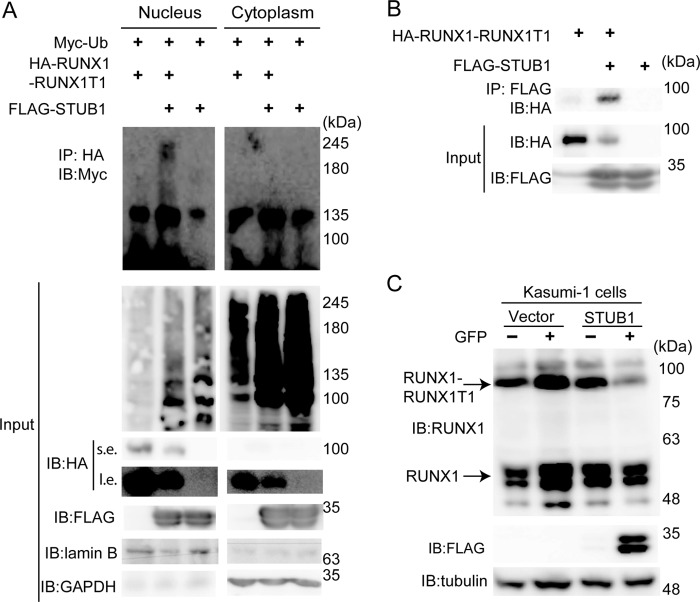
STUB1 promoted ubiquitination and degradation of RUNX1–RUNX1T1
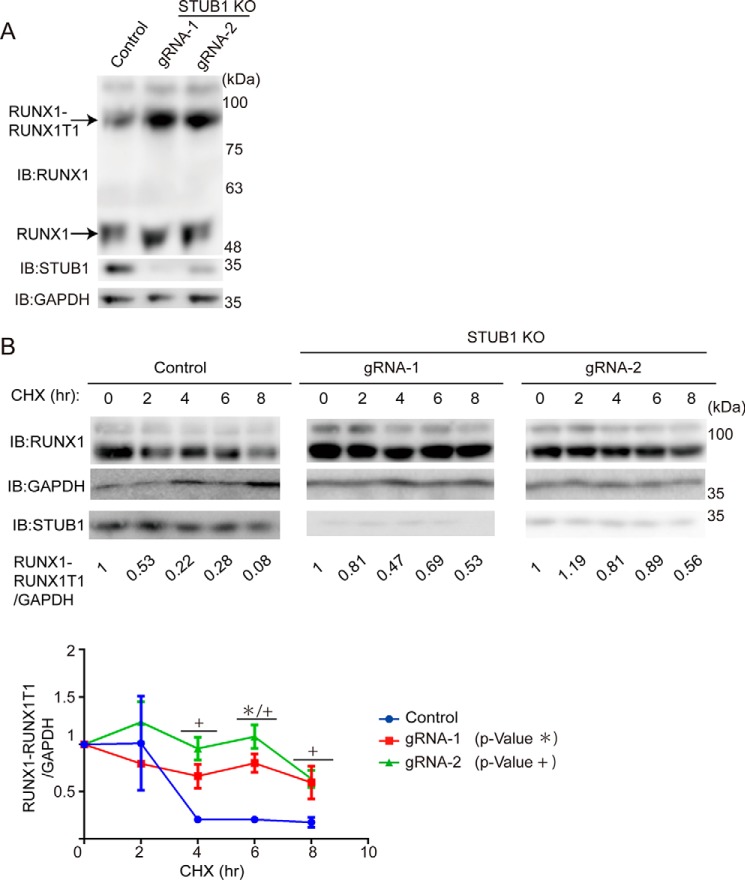
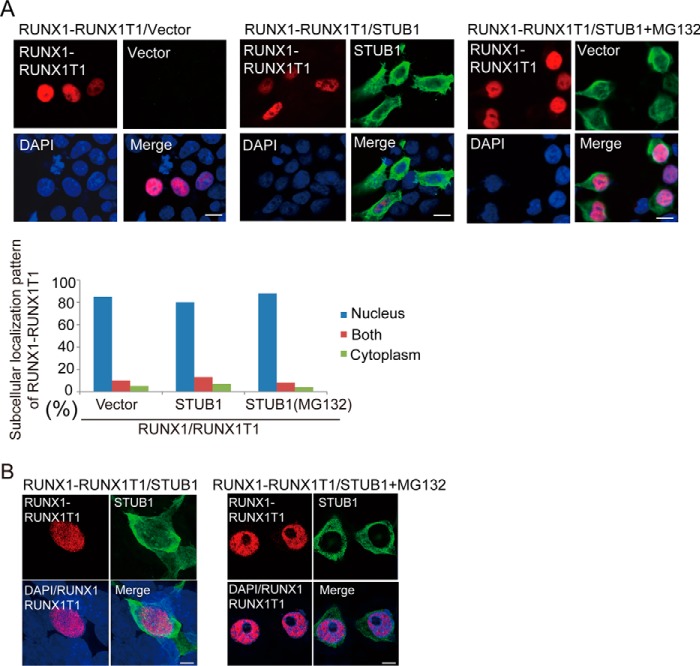
We then investigated whether STUB1 also induces ubiquitination of RUNX1–RUNX1T1. We introduced HA-tagged RUNX1–RUNX1T1, FLAG-tagged STUB1, and Myc-tagged ubiquitin into 293T cells. Cell lysates were subjected to immunoprecipitation with anti-HA, followed by immunoblotting with anti-Myc to detect ubiquitinated RUNX1–RUNX1T1. STUB1 induced ubiquitination of RUNX1–RUNX1T1 mainly in the nucleus and down-regulated its expression (Fig. 6A). Interaction between STUB1 and RUNX1–RUNX1T1 was confirmed in 293T cells transfected with both constructs (Fig. 6B). We then transduced vector or STUB1 (coexpressing GFP) into Kasumi-1, a myeloid leukemia cell line that harbors RUNX1–RUNX1T1. As expected, STUB1 overexpression induced down-regulation of both RUNX1 and RUNX1–RUNX1T1 in Kasumi-1 cells (Fig. 6C). Conversely, STUB1 depletion using the CRISPR/Cas9 system in Kasumi-1 cells led to increased stability and expression of RUNX1–RUNX1T1 (Fig. 7, A and B). Together, these data suggest that STUB1 also acts as an E3 ligase for RUNX1–RUNX1T1 to regulate its expression. However, RUNX1–RUNX1T1 was localized exclusively in the nucleus even in STUB1-overexpressing cells, indicating that intracellular localization of RUNX1–RUNX1T1 is not regulated by STUB1 (Fig. 8, A and B).
Figure 6.
STUB1 promoted ubiquitination and degradation of RUNX1–RUNX1T1. A, 293T cells were transfected with Myc–ubiquitin, HA–RUNX1–RUNX1T1, and/or FLAG–STUB1. Nuclear and cytoplasmic fractions were isolated and immunoprecipitated with anti-HA antibody, following the detection of ubiquitinated RUNX1–RUNX1T1 with anti-Myc antibody. STUB1 induced ubiquitination of RUNX1–RUNX1T1 mainly in the nucleus. s.e., short exposure; l.e., long exposure. B, 293T cells were transfected with HA–RUNX1–RUNX1T1 and/or FLAG–STUB1. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibody, following the detection of STUB1-bound RUNX1–RUNX1T1 with anti-HA antibody. C, Kasumi-1 cells were retrovirally transduced with a vector control or FLAG–STUB1 (coexpressing GFP). GFP+ cells (transduced cells) and GFP− cells (nontransduced cells) were sorted, and cell extracts were analyzed with anti-RUNX1, anti-FLAG, and anti-Tubulin antibodies. Enforced expression of STUB1 in Kasumi-1 cells resulted in down-regulation of RUNX1 and RUNX1–RUNX1T1 expression. IB, immunoblot.
Figure 7.
STUB1 depletion increased stability of RUNX1–RUNX1T1. A, Kasumi-1 cells were transduced with a vector control or two independent gRNAs targeting STUB1. The gRNAs showed efficient depletion of STUB1, which resulted in the increased expression of RUNX1 and RUNX1–RUNX1T1. B, Kasumi-1 cells transduced with a vector or STUB1 gRNAs were treated with 25 μg/ml of CHX for the indicated times, and cell extracts were analyzed with anti-RUNX1 antibody. Band intensities of RUNX1 and GAPDH were quantified with Multi Gauge (Science Lab), and the intensities of RUNX1–RUNX1T1 relative to GAPDH are shown. The value of RUNX1–RUNX1T1/GAPDH without CHX treatment was set to 1. Representative data (upper) and cumulative data from three independent experiments (lower, mean ± S.E., *, p < 0.05; +, p < 0.05) are shown. IB, immunoblot.
Figure 8.
STUB1 did not promote nuclear export of RUNX1–RUNX1T1. A and B, 293T cells were cotransfected with HA-tagged RUNX1–RUNX1T1 together with vector or FLAG-tagged STUB1 in the presence or absence of MG132 (20 mm), and were stained with anti-RUNX1 (rabbit) and anti-FLAG (mouse) antibodies followed by anti-rabbit Alexa 568 (red) and anti-mouse Alexa 488 (green) staining. Nuclei were visualized with DAPI (blue). Confocal laser scanning microscopy (Nikon A1, bar, 10 μm) (A) or super resolution microscopy (Nikon SIM, Bar, 5 μm) (B) was used to observe localization of RUNX1–RUNX1T1 and STUB1. STUB1 expression did not alter the subcellular localization of RUNX1–RUNX1T1.
STUB1 inhibited the growth of RUNX1–RUNX1T1 leukemia cells
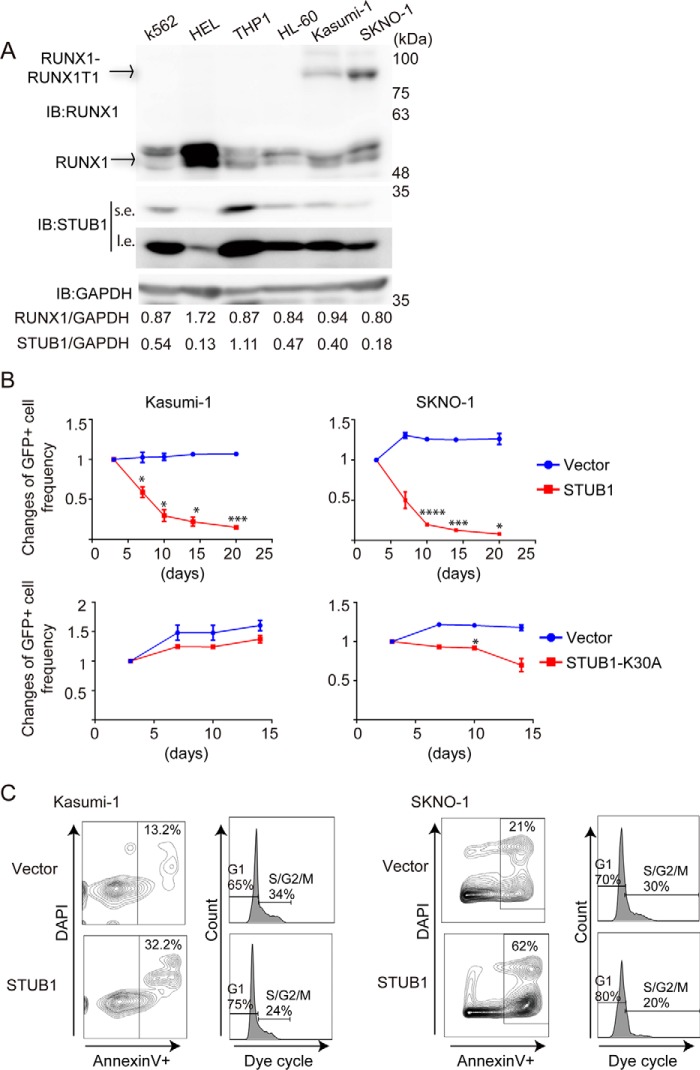
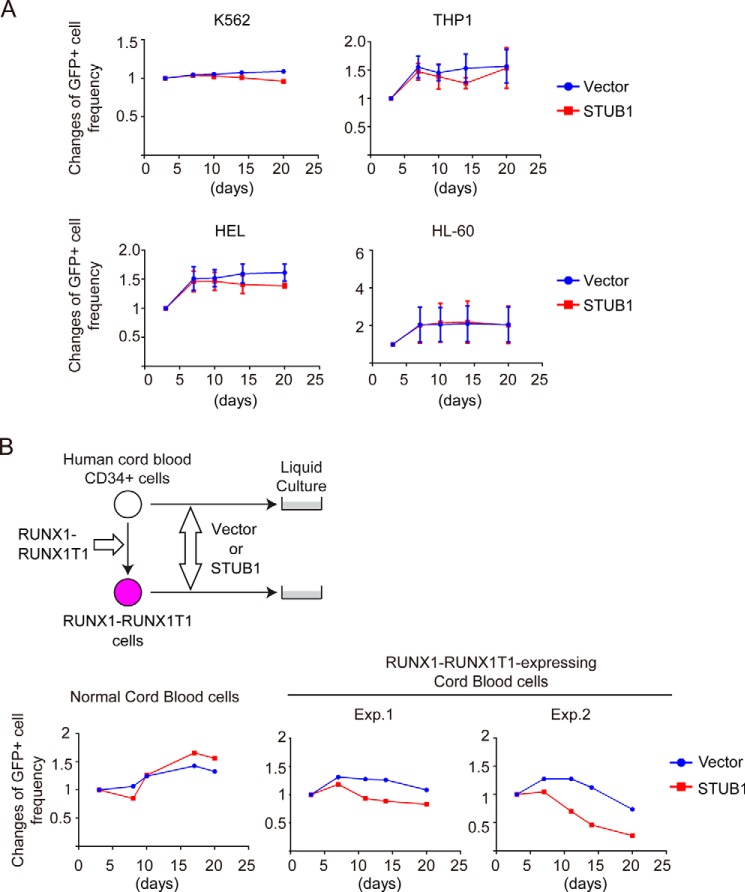
Next, we transduced vector or STUB1 (coexpressing GFP) into various myeloid leukemia cell lines and monitored the frequency of GFP-positive cells in culture. Kasumi-1 and SKNO-1 are leukemia cell lines that harbor RUNX1–RUNX1T1. K562, THP1, HEL, and HL-60 are control cell lines without RUNX1–RUNX1T1. STUB1 and RUNX1 proteins were expressed in all these cell lines. HEL cells showed the highest and the lowest expression of RUNX1 and STUB1, respectively, and STUB1 expression was relatively low in RUNX1–RUNX1T1 leukemia cells (Kasumi-1 and SKNO-1 cells) (Fig. 9A). Notably, overexpression of wild-type STUB1, but not STUB1-K30A, showed a strong growth-inhibitory effect in Kasumi-1 and SKNO-1 cells (Fig. 9B). To characterize the attenuated cell growth induced by STUB1 overexpression, we performed cell cycle and apoptosis analyses. Forced expression of STUB1 increased the frequency of annexin V+ apoptotic cells, and decreased the proportion of S/G2/M phase cells in both Kasumi-1 and SKNO-1 cells (Fig. 9C). Thus, STUB1 overexpression inhibits the growth of RUNX1–RUNX1T1 cells by inducing apoptosis and cell cycle arrest. Importantly, STUB1 overexpression showed little effect in non-RUNX1–RUNX1T1 leukemia cell lines (Fig. 10A), suggesting the specific function of STUB1 in RUNX1–RUNX1T1 leukemia. STUB1 depletion did not alter the growth of Kasumi-1 and K562 cells (data not shown).
Figure 9.
STUB1 induced apoptosis and cell cycle arrest of RUNX1–RUNX1T1 leukemia cells. A, STUB1 and RUNX1 were expressed in myeloid leukemia cell lines. RUNX1–RUNX1T1 leukemia cells (Kasumi-1 and SKNO-1) showed relatively low levels of STUB1 expression. Note that HEL cells showed the highest and the lowest expression of RUNX1 and STUB1, respectively. B, a vector control, wild-type STUB1, or STUB1-K30A (coexpressing GFP) was transduced into Kasumi-1 and SKNO-1 cells. The mixed transduction culture containing both transduced GFP+ and nontransduced GFP− cells were passaged to score the frequency of the GFP+ cell by flow cytometric analysis as a measure of the impact of the transduced gene on cellular proliferation rate. The initial frequency of GFP+ cells immediately after transduction was set as 1. Two independent experiments were performed, and data are shown as mean ± S.E. (*, p < 0.05; ***, p < 0.005; ****, p < 0.001). Overexpression of STUB1, but not STUB1-K30A, showed a strong growth-inhibitory effect in these cells. C, Kasumi-1 and SKNO-1 cells were transduced with a vector or STUB1 (coexpressing GFP), and GFP+ cells were sorted. Cell-cycle status and apoptosis were assessed in these GFP+ cells. STUB1 overexpression resulted in the increased or decreased frequency of Annexin V+ or S/G2/M phase cells, respectively. IB, immunoblot.
Figure 10.
STUB1 selectively inhibited the growth of RUNX1–RUNX1T1 leukemia cells. A, a vector control or STUB1 (coexpressing GFP) was transduced into K562, THP1, HEL, and HL-60 cells. The mixed transduction culture containing both transduced GFP+ and nontransduced GFP− cells were passaged to score the frequency of the GFP+ cell by flow cytometric analysis as a measure of the impact of the transduced gene on cellular proliferation rate. The initial frequency of GFP+ cells immediately after transduction was set as 1. Two independent experiments were performed, and data are shown as mean ± S.E. STUB1 overexpression showed only a marginal effect in these non-RUNX1–RUNX1T1 myeloid leukemia cells. B, human CB cells and those expressing RUNX1–RUNX1T1 were transduced with a vector control or STUB1 (coexpressing GFP). The cells were cultured in cytokine containing media to monitor the changes of GFP frequency. STUB1 inhibited the growth of RUNX1–RUNX1T1-expressing CB cells, but not that of normal CB cells.
We further assessed the effect of STUB1 overexpression in RUNX1–RUNX1T1 leukemia using a human cord blood (CB) culture assay. RUNX1–RUNX1T1 promotes self-renewal and long-term proliferation of CB cells. The CB cells expressing RUNX1–RUNX1T1 can grow over 6 months in culture retaining primitive CD34+ cells, and recapitulate features of human RUNX1–RUNX1T1 leukemia (19–21). We transduced vector or STUB1 (coexpressing GFP) into CB CD34+ cells and those expressing RUNX1–RUNX1T1. Again, STUB1 overexpression substantially inhibited the growth of RUNX1–RUNX1T1-expressing CB cells, whereas normal CB cells transduced with STUB1 grew normally (Fig. 10B). Together, these data indicate that STUB1 activation specifically inhibits the growth of RUNX1–RUNX1T1 leukemia cells.
Discussion
Although previous studies have identified several E3 ligases for RUNX1, whether physiological levels of these E3 ligases are involved in the regulation of RUNX1 ubiquitination remained unclear. Using a novel in vitro screening assay (13, 14), biochemical analyses, and the CRISPR/Cas9 system, we here demonstrated that STUB1 is a major E3 ubiquitin ligase of RUNX1. Consistent with a previous report (7), STUB1 induced ubiquitination and degradation of RUNX1 in 293T cells. A chaperon-binding deficient mutant of STUB1 (STUB1-K30A) failed to interact with RUNX1 efficiently, indicating that chaperon activity is critical for STUB1-induced RUNX1 ubiquitination. Interestingly, RUNX1–RUNX1T1 has been shown to be a client protein for the molecular chaperone Hsp90 (22, 23). Therefore, it appears that the STUB1–HSP90 chaperon complex plays a key role to regulate the stability of RUNX1 proteins. Importantly, we showed that STUB1 depletion in K562 cells led to increased stability and expression of RUNX1 protein, indicating the essential role of endogenous STUB1 to promote RUNX1 ubiquitination in vivo. Furthermore, we found that the STUB1-induced ubiquitination resulted in the reduced nuclear localization of RUNX1. Given that a nuclear localization signal is present at the end of the Runt domain in RUNX1 (24, 25), the ubiquitination sites in the nuclear localization signal region may play a role to determine RUNX1 localization. The STUB1-induced nuclear export was not seen for RUNX1–RUNX1T1, which indicates that the intracellular localization of RUNX1–RUNX1T1 is controlled by the different mechanisms. Consistent with this observation, previous reports showed that subcellular localization of RUNX1–RUNX1T1 depends on the regions in RUNX1T1, but not those in RUNX1 (26, 27). Taken together, our data suggest that STUB1 induces RUNX1 ubiquitination, promotes its degradation as well as nuclear export, and inhibits transcriptional activity of RUNX1. Whether STUB1 physiologically regulates the RUNX1 activity in hematopoiesis remains to be elucidated. Given that both RUNX1 and STUB1 play crucial roles in T-cell development and regulatory T-cells (28–31), the potential RUNX1-STUB1 interaction in the immune system may merit future studies. In addition to STUB1, our AlphaScreen assay revealed novel RUNX1-interacting ubiquitin ligases, including RNF38, DTX2, TRIM5, UHRF2, and DZIP3. The role of these E3 ligases in the regulation of RUNX1 function also warrants further investigation.
In addition to RUNX1, STUB1 also promoted ubiquitination and degradation of a fusion protein RUNX1–RUNX1T1, thereby inhibiting the growth of RUNX1–RUNX1T1 leukemia cells. The STUB1-induced down-regulation of native RUNX1 may also contribute to the impaired proliferation of RUNX1–RUNX1T1 leukemia, as suggested by several recent reports (32, 33). Thus, our data suggest that activation of STUB1 can be a therapeutic approach to treat RUNX1–RUNX1T1 leukemia. Such proteolysis-based targeting of oncoproteins has attracted considerable attention in recent years. For example, As2O3 binds to the transcription factor PML moiety of the disease-specific PML–retinoic acid receptor α (RARA) oncoprotein, and specifically induces a SUMO-dependent, ubiquitin-mediated degradation of PML–RARA (34, 35). Lenalidomide induces ubiquitination and degradation of IKZF1, IKZF3, and CSNK1A1 to show the clinical efficacy in multiple myeloma and myelodysplastic syndrome with 5q deletion (36, 37). Furthermore, small molecules promoting protein degradation, called “PROTACs,” have been recently introduced into the clinical settings (38). PROTACs are heterobifunctional small molecules that simultaneously bind a target protein and a ubiquitin ligase, enabling ubiquitination and degradation of the target. The PROTAC-based therapy, linking STUB1 and RUNX1, can be a promising therapy for RUNX1–RUNX1T1 leukemia and potentially for other leukemias with RUNX1 dysregulation.
Experimental procedures
Plasmids
For RUNX1 expression, FLAG-tagged human RUNX1b in a pMYs-IG retroviral vector or a pcDNA3 vector, Myc-tagged human RUNX1b in a pCMV2 vector, or human RUNX1b in a pcDNA3 vector were used. We introduced lysine-to-arginine mutations at the lysine residues in RUNX1 to generate RUNX1–9KR using QuikChange Lightning Site-directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). FLAG-tagged STUB1 in a pcDNA3 vector was provided by Dr. K. Kimura, and we cloned it into the pMYs-IG. Myc-tagged STUB1 and STUB1-K30A in pcDNA3 vectors were provided by Dr. J. Song, and we cloned them into the pMYs-IG. HA or Myc-tagged ubiquitin were used for ubiquitin expression. pMCSFR-luc and pCMV5-CBFβ were obtained from Addgene (18). HA-tagged RUNX1–RUNX1T1 in a pMSCV-IRES-Thy1.1 retroviral vector was used for RUNX1–RUNX1T1 expression.
Wheat cell-free protein synthesis and binding assay using AlphaScreen
The protein array containing the 287 E3s was produced by using the robotic synthesizer Gen-Decoder 1000 (CellFree Sciences, Yokohama, Japan) as described previously (13, 14). For RUNX1 and CBFB, the supernatant of the crude translation mixture after centrifugation at 22,000 × g for 10 min was subjected to SDS-PAGE followed by immunoblot analysis to determine solubility. The FLAG-tagged and biotinylated proteins on the blot were detected by using horseradish peroxidase (HRP)-conjugated anti-FLAG (M2, Sigma) or biotin (BN-34, Sigma) antibody, respectively. The following oligonucleotides, AGATGTGTGGTTAACCACAAAC and AGGTTTGTGGTTAACCACACAT, were added to obtain the soluble form of RUNX1 protein. The interactions between biotinylated RUNX1 and FLAG-tagged E3s were detected with AlphaScreen technology provided by PerkinElmer Life Sciences, as described previously (13, 14).
Cell culture
Human umbilical CB cells were obtained from Riken BRC or the Japanese Red Cross Kanto-Koshinetsu Cord Blood Bank (Tokyo, Japan). CD34+ cells were separated using the CD34 MicroBead Kit (Miltenyi Biotec). We engineered human RUNX1–RUNX1T1 cells by transducing RUNX1–RUNX1T1 into CB CD34+ cells using retrovirus, as described previously (19–21). Human CB cells and those expressing RUNX1–RUNX1T1 were cultured in Iscove's modified Dulbecco's medium containing 20% BIT9500 (StemCell Technologies, Vancouver, BC, Canada) and 10 ng/ml of human SCF, human TPO, human IL-3, human IL-6, mouse FLT3L (R & D Systems), as described previously (21). K562, Kasumi-1, THP1, HEL, and HL-60 cells were cultured in RPMI 1640 with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. SKNO-1 cells were cultured in RPMI 1640 with 10% FBS, 1% penicillin/streptomycin, and 1 ng/ml of human GM-CSF (R & D Systems).
Transfection, Western blotting, and immunoprecipitation
293T cells were transiently transfected with 3 μg of vector, Myc- or FLAG-tagged RUNX1, HA-tagged RUNX1-RUNX1T1, FLAG- or Myc-tagged STUB1, and HA- or Myc-tagged ubiquitin; mixed with 30 μl of polyethylenimine. The cells were cultured 48 h after transfection and were lysed in Cell Lysis Buffer (Cell Signaling Technology, Danvers, MA; catalog number 9803). For immunoprecipitation, cell lysates were incubated with anti-Myc 9E10 (Santa Cruz Biotechnology, sc-40), anti-FLAG (Sigma, F3165), anti-HA (Roche Applied Sciences, 3F10), or anti-RUNX1 (Cell Signaling Technology, catalog number 4336) antibody for 30 min at 4 °C. Then, the samples were incubated with protein G-Sepharose (Amersham Biosciences) for 30 min at 4 °C. The precipitates were washed three times with the cell lysis buffer (Cell Signaling Technology; catalog number 9803) containing 1 mm phenylmethanesulfonyl fluoride, were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and analyzed by Western blotting. For some experiments, we isolated nuclear and cytoplasmic fractions before immunoprecipitation (see below). Western blotting analysis was performed with anti-FLAG M2-Peroxidase (Sigma, A8592), anti-Myc 9E10 (Santa Cruz Biotechnology, sc-40), anti-HA-Peroxidase (Roche Applied Science, 12CA5), anti-RUNX1 (Cell Signaling Technology, catalog number 4336), anti-STUB1 (Cell Signaling Technology, catalog number 2080), anti-Lamin (Santa Cruz Biotechnology, B1304), anti-GAPDH (Cell Signaling Technology, catalog number 5174), anti-Ubiquitin (Cell Signaling Technology, P4D1, catalog number 3936), or anti-Tubulin (Sigma, T5168). Signals were detected with SuperSignal West Pico (Pierce), and immunoreactive bands were visualized using LAS-4000 Luminescent Image Analyzer (FUJIFILM). Band intensity was measured using LabWorks Version 4.5 software (UVP, LLC).
Isolation of nuclear and cytoplasmic fractions
Cells were incubated with Hypotonic Buffer (10 mm HEPES, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DDT, 0.1% Triton X-100 with protease inhibitor mixture (Sigma)) for 20 min at 4 °C, centrifuged (8,000 rpm, 5 min), and the supernatant was collected as the cytoplasmic fraction. The insoluble pellet was lysed with Lysis buffer (0.5% Nonidet P-40 in TBS with protease inhibitor mixture (Sigma) and Benzonase (Sigma)) for 35 min at 4 °C, centrifuged (14,000 rpm, 30 min), and the supernatant was collected as the nuclear fraction.
Immunofluorescence analysis
293T cells transfected with vector, RUNX1 (wild-type or the 9KR mutant), HA-tagged RUNX1–RUNX1T, and/or FLAG-tagged STUB1 were fixed with 2% paraformaldehyde, permeabilized with 0.2% Triton X-100, blocking with 2% BSA and 5% goat serum, and then incubated with anti-RUNX1 (Abcam, ab92336), anti-FLAG (Sigma, F3165 or F7425), and anti-HA (Bio Legend, catalog number 901513) antibodies, followed by labeling with Alexa Fluor 568-conjugated anti-rabbit or -mouse antibody (Thermo Fisher, A11011 or A11030) and Alexa Fluor 488-conjugated anti-rabbit or -mouse antibody (Thermo Fisher, A11029 or A11034). K562 cells transduced with vector or FLAG-tagged STUB1 were fixed with 2% paraformaldehyde, permeabilized with 0.2% Triton X-100, blocking with 2% BSA and 5% goat serum, and then incubated with anti-RUNX1 (Abcam, ab92336), anti-FLAG (Sigma, F3165), followed by labeling with Alexa Fluor 568-conjugated anti-rabbit (Thermo Fisher, A11011) and Alexa Fluor 633-conjugated anti-mouse (Thermo Fisher, A21050). Nuclei were visualized with DAPI (Bio Legend). Fluorescent images were analyzed on a confocal microscope (Nikon A1) or super-resolution microscope (Nikon SIM). MG132 (20 μm) was added for the indicated experiments.
Flow cytometry
Cells were analyzed on a FACSVerse, and were sorted with a FACSAria (BD Biosciences, San Jose, CA).
Luciferase assay
293T cells were seeded in 12-well culture plates at a density of 100,000 per well. At 12 h after seeding, the cells were transfected with 500 ng of pMCSFR-luc (18), 100 ng of pcDNA3-RUNX1, pCMV5-CBFB, and pcDNA3-STUB1, using polyethylenimine. The cells were harvested 48 h after transfection and were assayed for the luciferase activity by means of the luciferase assay system (Promega) and a luminometer (BMG LABTECH, FLUOstar OPTIMA). Transfection efficiency was normalized using fluorometric quantitation of enhanced green fluorescent protein (39) by cotransfecting 500 ng of pMYs-IG.
Retroviral transduction
Retroviruses were generated by transient transfection of Plat-E packaging cells (40) with the retroviral constructs using the calcium–phosphate co-precipitation method. Retrovirus transduction to the cells was performed using Retronectin (Takara Bio Inc., Otsu, Shiga, Japan).
STUB1 depletion using the CRISPR/Cas9 system
To generate gRNA expression vectors targeting human STUB1, annealed oligonucleotides (CACCGGGCCGTGTATTACACCAACC and AAACGGTTGGTGTAATACACGGCCC for gRNA-1, CACCGGAAGCGCTGGAACAGCATTG and AAACCAATGCTGTTCCAGCGCTTCC for gRNA-2, obtained from FASMAC, Kanagawa, Japan) were cloned into the lentiGuide-Puro vector (41), which was obtained from Addgene (plasmid 52963). The expression vector for Cas9 (lentiCas9-Blast, 52962), and the lentiviral packaging vectors (pMD2.G, 12259 and psPAX, 12260) were also obtained from Addgene. Lentiviruses were generated by transient transfection of 293T cells with these lentiviral constructs using the calcium–phosphate co-precipitation method. K562 and Kasumi-1 cells were infected with the virus for 24 h, and were selected for stable expression of Cas9 using blasticidin (10 μg/ml) and for stable expression of gRNAs using puromycin (1 μg/ml). The cells were then harvested to examine the efficient depletion of STUB1 by immunoblotting.
Statistics
Unpaired and two-tailed t test was used to evaluate differences between groups in the luciferase assay (Fig. 5). Welch's t test were used to compared the relative stability of RUNX1 and RUNX1–RUNX1T1 in control and STUB1-depleted cells (Figs. 3C and 7B), and to compare the growth of vector- or STUB1-transduced cells (Fig. 9B).
Author contributions
T. Y. designed and performed experiments, analyzed the data, and participated in writing paper. S. S., X. L., M. T., S. A., and T. Fukushima assisted with experiments. T. Fukuyama and Y. T. analyzed the data. H. T. and T. S. performed the AlphaScreen assay. T. K. conceived the project, analyzed the data, and participated in writing the paper. S. G. conceived the project, designed and performed experiments, analyzed the data, and wrote the paper.
Supplementary Material
Acknowledgments
We thank for Keiji Kimura and Jaewhan Song for plasmids. We thank for Noriko Tokai for expert technical assistance. We also thank the Flow Cytometry Core, the Mouse Core and Microscopy Core at The Institute of Medical Science, The University of Tokyo for their help.
This work was supported by Grant-in-Aid for Research Activity 15H06162 start-up (to S. G.), a grant from Leukemia Research Fund (to S. G.), and a grant from Children's Cancer Association of Japan (to S. G.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Table S1.
- CBFB
- core-binding factor β
- gRNA
- guide RNA
- CHX
- cycloheximide
- CB
- cord blood
- PML
- promyelocytic leukemia.
References
- 1. Ito Y., Bae S. C., and Chuang L. S. (2015) The RUNX family: developmental regulators in cancer. Nat. Rev. Cancer 15, 81–95 [DOI] [PubMed] [Google Scholar]
- 2. Link K. A., Chou F. S., and Mulloy J. C. (2010) Core binding factor at the crossroads: determining the fate of the HSC. J. Cell. Physiol. 222, 50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goyama S., Huang G., Kurokawa M., and Mulloy J. C. (2015) Posttranslational modifications of RUNX1 as potential anticancer targets. Oncogene 34, 3483–3492 [DOI] [PubMed] [Google Scholar]
- 4. Huang G., Shigesada K., Ito K., Wee H. J., Yokomizo T., and Ito Y. (2001) Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 20, 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang G., Zhao X., Wang L., Elf S., Xu H., Zhao X., Sashida G., Zhang Y., Liu Y., Lee J., Menendez S., Yang Y., Yan X., Zhang P., Tenen D. G., et al. (2011) The ability of MLL to bind RUNX1 and methylate H3K4 at PU.1 regulatory regions is impaired by MDS/AML-associated RUNX1/AML1 mutations. Blood 118, 6544–6552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Biggs J. R., Peterson L. F., Zhang Y., Kraft A. S., and Zhang D. E. (2006) AML1/RUNX1 phosphorylation by cyclin-dependent kinases regulates the degradation of AML1/RUNX1 by the anaphase-promoting complex. Mol. Cell. Biol. 26, 7420–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shang Y., Zhao X., Xu X., Xin H., Li X., Zhai Y., He D., Jia B., Chen W., and Chang Z. (2009) CHIP functions an E3 ubiquitin ligase of Runx1. Biochem. Biophys. Res. Commun. 386, 242–246 [DOI] [PubMed] [Google Scholar]
- 8. Li X., Huang M., Zheng H., Wang Y., Ren F., Shang Y., Zhai Y., Irwin D. M., Shi Y., Chen D., and Chang Z. (2008) CHIP promotes Runx2 degradation and negatively regulates osteoblast differentiation. J. Cell Biol. 181, 959–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jones D. C., Wein M. N., Oukka M., Hofstaetter J. G., Glimcher M. J., and Glimcher L. H. (2006) Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science 312, 1223–1227 [DOI] [PubMed] [Google Scholar]
- 10. Chen C., and Matesic L. E. (2007) The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. 26, 587–604 [DOI] [PubMed] [Google Scholar]
- 11. Goyama S., and Mulloy J. C. (2011) Molecular pathogenesis of core binding factor leukemia: current knowledge and future prospects. Int. J. Hematol 94, 126–133 [DOI] [PubMed] [Google Scholar]
- 12. Krämer O. H., Müller S., Buchwald M., Reichardt S., and Heinzel T. (2008) Mechanism for ubiquitylation of the leukemia fusion proteins AML1-ETO and PML-RARα. FASEB J. 22, 1369–1379 [DOI] [PubMed] [Google Scholar]
- 13. Takahashi H., Uematsu A., Yamanaka S., Imamura M., Nakajima T., Doi K., Yasuoka S., Takahashi C., Takeda H., and Sawasaki T. (2016) Establishment of a wheat cell-free synthesized protein array containing 250 human and mouse E3 ubiquitin ligases to identify novel interaction between E3 ligases and substrate proteins. PLoS ONE 11, e0156718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takahashi H., Nozawa A., Seki M., Shinozaki K., Endo Y., and Sawasaki T. (2009) A simple and high-sensitivity method for analysis of ubiquitination and polyubiquitination based on wheat cell-free protein synthesis. BMC Plant Biol. 9, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang J., Ballinger C. A., Wu Y., Dai Q., Cyr D. M., Höhfeld J., and Patterson C. (2001) CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 276, 42938–42944 [DOI] [PubMed] [Google Scholar]
- 16. Ballinger C. A., Connell P., Wu Y., Hu Z., Thompson L. J., Yin L. Y., and Patterson C. (1999) Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19, 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seo J., Lee E. W., Sung H., Seong D., Dondelinger Y., Shin J., Jeong M., Lee H. K., Kim J. H., Han S. Y., Lee C., Seong J. K., Vandenabeele P., and Song J. (2016) CHIP controls necroptosis through ubiquitylation- and lysosome-dependent degradation of RIPK3. Nat. Cell. Biol. 18, 291–302 [DOI] [PubMed] [Google Scholar]
- 18. Zhang D. E., Hetherington C. J., Meyers S., Rhoades K. L., Larson C. J., Chen H. M., Hiebert S. W., and Tenen D. G. (1996) CCAAT enhancer-binding protein (C/EBP) and AML1 (CBF α2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol. Cell. Biol. 16, 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mulloy J. C., Cammenga J., Berguido F. J., Wu K., Zhou P., Comenzo R. L., Jhanwar S., Moore M. A., and Nimer S. D. (2003) Maintaining the self-renewal and differentiation potential of human CD34+ hematopoietic cells using a single genetic element. Blood 102, 4369–4376 [DOI] [PubMed] [Google Scholar]
- 20. Mulloy J. C., Cammenga J., MacKenzie K. L., Berguido F. J., Moore M. A., and Nimer S. D. (2002) The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood 99, 15–23 [DOI] [PubMed] [Google Scholar]
- 21. Goyama S., Schibler J., Gasilina A., Shrestha M., Lin S., Link K. A., Chen J., Whitman S. P., Bloomfield C. D., Nicolet D., Assi S. A., Ptasinska A., Heidenreich O., Bonifer C., Kitamura T., Nassar N. N., and Mulloy J. C. (2016) UBASH3B/Sts-1-CBL axis regulates myeloid proliferation in human preleukemia induced by AML1-ETO. Leukemia 30, 728–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bots M., Verbrugge I., Martin B. P., Salmon J. M., Ghisi M., Baker A., Stanley K., Shortt J., Ossenkoppele G. J., Zuber J., Rappaport A. R., Atadja P., Lowe S. W., and Johnstone R. W. (2014) Differentiation therapy for the treatment of t(8;21) acute myeloid leukemia using histone deacetylase inhibitors. Blood 123, 1341–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang G., Thompson M. A., Brandt S. J., and Hiebert S. W. (2007) Histone deacetylase inhibitors induce the degradation of the t(8;21) fusion oncoprotein. Oncogene 26, 91–101 [DOI] [PubMed] [Google Scholar]
- 24. Lu J., Maruyama M., Satake M., Bae S. C., Ogawa E., Kagoshima H., Shigesada K., and Ito Y. (1995) Subcellular localization of the α and β subunits of the acute myeloid leukemia-linked transcription factor PEBP2/CBF. Mol. Cell. Biol. 15, 1651–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanno T., Kanno Y., Chen L. F., Ogawa E., Kim W. Y., and Ito Y. (1998) Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the β subunit. Mol. Cell. Biol. 18, 2444–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barseguian K., Lutterbach B., Hiebert S. W., Nickerson J., Lian J. B., Stein J. L., van Wijnen A. J., and Stein G. S. (2002) Multiple subnuclear targeting signals of the leukemia-related AML1/ETO and ETO repressor proteins. Proc. Natl. Acad. Sci. U.S.A. 99, 15434–15439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nagel S., Hambach L., Krauter J., Venturini L., Heidenreich O., Ganser A., and Heil G. (2002) Analysis of the nuclear distribution of the translocation t(8;21)-derived fusion protein AML1/ETO by confocal laser scanning microscopy. J. Hematother. Stem Cell Res. 11, 401–408 [DOI] [PubMed] [Google Scholar]
- 28. Taniuchi I., Osato M., Egawa T., Sunshine M. J., Bae S. C., Komori T., Ito Y., and Littman D. R. (2002) Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 [DOI] [PubMed] [Google Scholar]
- 29. Ono M., Yaguchi H., Ohkura N., Kitabayashi I., Nagamura Y., Nomura T., Miyachi Y., Tsukada T., and Sakaguchi S. (2007) Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature 446, 685–689 [DOI] [PubMed] [Google Scholar]
- 30. Chen Z., Barbi J., Bu S., Yang H. Y., Li Z., Gao Y., Jinasena D., Fu J., Lin F., Chen C., Zhang J., Yu N., Li X., Shan Z., Nie J., et al. (2013) The ubiquitin ligase Stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor Foxp3. Immunity 39, 272–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang S., Li Y., Hu Y. H., Song R., Gao Y., Liu H. Y., Shu H. B., and Liu Y. (2013) STUB1 is essential for T-cell activation by ubiquitinating CARMA1. Eur. J. Immunol. 43, 1034–1041 [DOI] [PubMed] [Google Scholar]
- 32. Goyama S., Schibler J., Cunningham L., Zhang Y., Rao Y., Nishimoto N., Nakagawa M., Olsson A., Wunderlich M., Link K. A., Mizukawa B., Grimes H. L., Kurokawa M., Liu P. P., Huang G., and Mulloy J. C. (2013) Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J. Clin. Invest. 123, 3876–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ben-Ami O., Friedman D., Leshkowitz D., Goldenberg D., Orlovsky K., Pencovich N., Lotem J., Tanay A., and Groner Y. (2013) Addiction of t(8;21) and inv(16) acute myeloid leukemia to native RUNX1. Cell Rep. 4, 1131–1143 [DOI] [PubMed] [Google Scholar]
- 34. Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., and Hay R. T. (2008) RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell. Biol. 10, 538–546 [DOI] [PubMed] [Google Scholar]
- 35. Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., and de Thé H. (2008) Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell. Biol. 10, 547–555 [DOI] [PubMed] [Google Scholar]
- 36. Krönke J., Udeshi N. D., Narla A., Grauman P., Hurst S. N., McConkey M., Svinkina T., Heckl D., Comer E., Li X., Ciarlo C., Hartman E., Munshi N., Schenone M., et al. (2014) Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krönke J., Fink E. C., Hollenbach P. W., MacBeth K. J., Hurst S. N., Udeshi N. D., Chamberlain P. P., Mani D. R., Man H. W., Gandhi A. K., Svinkina T., Schneider R. K., McConkey M., Järås M., Griffiths E., et al. (2015) Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature 523, 183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deshaies R. J. (2015) Protein degradation: prime time for PROTACs. Nat. Chem. Biol. 11, 634–635 [DOI] [PubMed] [Google Scholar]
- 39. Dandekar D. H., Kumar M., Ladha J. S., Ganesh K. N., and Mitra D. (2005) A quantitative method for normalization of transfection efficiency using enhanced green fluorescent protein. Anal. Biochem. 342, 341–344 [DOI] [PubMed] [Google Scholar]
- 40. Morita S., Kojima T., and Kitamura T. (2000) Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 41. Sanjana N. E., Shalem O., and Zhang F. (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.