Abstract
Persistent or chronic infection with the hepatitis B virus (HBV) represents one of the most common viral diseases in humans. The hepatitis B virus deploys the hepatitis B virus X protein (HBx) as a suppressor of host defenses consisting of RNAi-based silencing of viral genes. Because of its critical role in countering host defenses, HBx represents an attractive target for antiviral drugs. Here, we developed and optimized a loss-of-function screening procedure, which identified a potential pharmacophore that abrogated HBx RNAi suppression activity. In a survey of 14,400 compounds in the Maybridge Screening Collection, we prioritized candidate compounds via high-throughput screening based on reversal of green fluorescent protein (GFP)–reported, RNAi-mediated silencing in a HepG2/GFP-shRNA RNAi sensor line. The screening yielded a pharmacologically active compound, N-(2,4-difluorophenyl)-N′-[3-(1H-imidazol-1-yl) propyl] thiourea (IR415), which blocked HBx-mediated RNAi suppression indicated by the GFP reporter assay. We also found that IR415 reversed the inhibitory effect of HBx protein on activity of the Dicer endoribonuclease. We further confirmed the results of the primary screen in IR415-treated, HBV-infected HepG2 cells, which exhibited a marked depletion of HBV core protein synthesis and down-regulation of pre-genomic HBV RNA. Using a molecular interaction analysis system, we confirmed that IR415 selectively targets HBx in a concentration-dependent manner. The screening assay presented here allows rapid and improved detection of small-molecule inhibitors of HBx and related viral proteins. The assay may therefore potentiate the development of next-generation RNAi pathway-based therapeutics and promises to accelerate our search for novel and effective drugs in antiviral research.
Keywords: drug discovery; drug screening; flow cytometry; hepatitis B virus (HBV, Hep B); molecular dynamics; RNA interference (RNAi); short hairpin RNA (shRNA); surface plasmon resonance (SPR); viral protein; small molecule inhibitor; viral RNAi suppressor; HBx; antiviral agent
Introduction
Hepatitis B is a serious infectious disease of the human liver caused by the hepatitis B virus (HBV) 6. More than 2 billion people alive today have been infected with HBV at some time in their lives. Of them, 350 million remain infected chronically and become carriers of the virus. Over 1 million people die of chronic hepatitis, serum hepatitis, and serum jaundice (1). HBV infection often becomes chronic, resulting in hepatocellular carcinoma (HCC). In susceptible host cells, HBV generates a favorable environment for its replication and escapes the innate immune system including the RNAi-mediated defense of the host cell (2). Of several host immune defense mechanisms, RNA-mediated gene silencing acts as a robust inducible defense mechanism to protect the host from viral invasion and establishment (3–6). Viruses in turn evade the host RNAi degradative effects mostly by encoding suppressors of RNA silencing proteins viral suppressors of RNA silencing proteins (VSRs) (7–12). Recently, we have evaluated the role of hepatitis B virus–encoded proteins to identify HBx as a viral suppressor that confers counterdefense strategy (13). Invariably, viral suppressors are the pathogenic factors, as identified in diverse systems (9, 14–16). In light of these reports, we presumed that HBx could be a vital antiviral target.
The biochemical activity of nonstructural regulatory protein HBx is targeted toward several essential cellular functions. Hepatitis B X protein acts as a multifunctional regulator to induce HBx-responsive transcription factors like NF-κB and NF-AT (17, 18), interacts with transcriptional activator CREB/ATF (19, 20), modulates intracellular calcium signaling, and stimulates signal transduction pathways resulting in initiation of tyrosine kinases following downstream activation of mitogen-activated protein kinases (21–23). HBx can restrain cellular proliferation either by deregulating the action of cyclin-dependent kinases to affect cell cycle progression checkpoints (24) or by inducing the diverse effect on anti-apoptotic and pro-apoptotic properties linked with p53 family of proteins to determine cell fate (25, 26). Several studies have addressed the role of HBx by targeting it using either short hairpin RNAs (shRNAs) (27, 28) or siRNA for cessation of HBV replication (29). Molecular mechanisms of HBx for in vivo replication in woodchuck hepatitis virus as well as in human HBV have garnered interest, highlighting the crucial function of this protein during viral replication (30, 31). The transcriptional transactivation function of HBx stimulates the first regulatory step of synthesis of HBV 3.5 kb pre-genomic RNA transcript with parallel augmentation effect on stimulation of HBV replication, thus being indispensable for both hepatitis B transcription and replication machinery (32–34). Furthermore, hepatitis B virus is a major etiological factor of HBV-associated hepatocellular carcinoma development, and the disease prognosis conceivably contributes to potential involvement of HBx-mediated oncogenicity (35). The extending modes of action of HBx from viral replication to epigenetic modification and triggering malignancy in liver cells make HBx an attractive drug target.
Because the antiviral roles of RNAi pathways for mammalian viruses are becoming more explicit, more cogent approaches are being developed toward the discovery of RNAi-related druggable gene targets in principle. We believe no systematic approach has been reported up to now for screening small molecular ligands that can target the VSRs for inactivation of animal viruses. In the present study, we customized an assay by measuring the quantitative expression of the reporter as a readout for high-throughput screening of pharmacologically active compounds from the Maybridge library for their ability to bind HBx and inhibit its RNAi suppressor activity. Consequently, inhibition of HBx-mediated RNAi suppression was observed with putative chemical compound N-(2,4-difluorophenyl)-N′-[3-(1H-imidazol-1-yl) propyl] thiourea, termed here as IR415. The success of the primary screen was further confirmed by interference of the drug in viral replication processes in other human cell lines as well. Our results therefore provide evidence for antiviral function of host RNAi machinery in HBV replication and identify a pharmacophore that binds and blocks HBx activity.
Results
In silico structural modeling and virtual screening
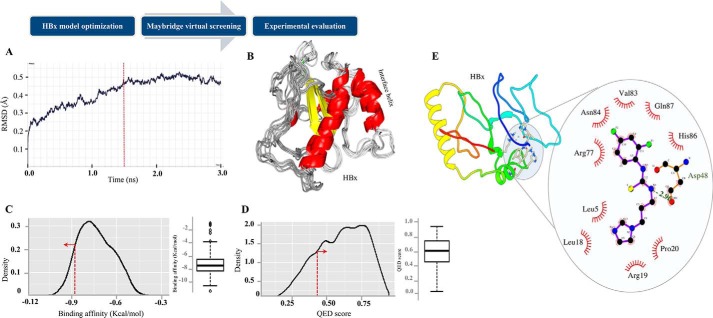
Because HBx has been described as being important for HBV replication and also functions as an RNAi suppressor protein, we aimed at identifying small molecules that bind HBx and block its activity. Although the complete structure of HBx remains to be elucidated, a small interface helix of HBx (residues 88–101) has been previously crystallized as a part of complex with another protein, Protein Data Bank code 3i7H (36). We initiated the study by modeling the 3D structure of HBx protein using threading-based I-TASSER web server (37), which computed five 3D conformation solutions. The five predicted conformational models (M1–M5) were manually scrutinized for the interface helix (residues 88–101) (36). Among the five models, two models (M1 and M2) displayed the desired helical segment and M1 model demonstrated the best theoretical conformation as revealed by the Ramachandran plot of the model structure (data not shown). The generated 3D HBx model was subjected to molecular dynamics (MD) simulation for evaluation of its conformational stability in explicit water-solvent environment (TIP3P molecules; see “Experimental Procedures”) using root mean square deviation analysis (Fig. 1A). The analysis suggested that maximum stability and minimum conformation dynamics in HBx structural coordinates was observed after 1500 picoseconds (1.5 nanoseconds) (Fig. 1B) of simulation period. Because the protein dynamics remain stable until final frame of the trajectory, we selected this structure for further analysis. For the docking studies and virtual screening, we exploited a library of 14,400 compounds from Maybridge, a preselected and prescreening library uniquely designed for lead generation. The library was evaluated for the best HBx binder by virtual screening procedure using AutoDock Vina (38). At the beginning of the virtual screening procedure, a 3D grid was designed around the active site helix of the stable ligand conformations using default parameters. The designed grid involves central coordinates (−4.27, 55.82, 22.79) with dimensions of 20 Å, 18 Å, and 38 Å in X, Y, and Z directions, respectively. AutoDock Vina generated nine different conformations for each ligand, which are sorted by binding affinity (kcal/mol). To retain the compounds with best conformation orientations on HBx surface, ligands with affinity higher than 90th percentile (−8.8 kcal/mol) were selected for subsequent analysis (Fig. 1C). In total, 1392 compounds were retained with very high affinity for HBx surface. To ensure the reliability, a more stringent filtration criterion was applied on selected compounds by computing their drug-likeliness quantitative estimation of drugs (QED) score (39). Compounds with QED score higher than the hard threshold of 0.40 among the selected compounds were shortlisted (Fig. 1D). The filtration step removed several compounds with high binding affinity predicted for HBx structure, including the compound with highest affinity score, and generated a set of 1057 drug-like compounds (supplemental data 1). Finally, a subset of 100 compounds with top affinity score from the filtered set was selected for experimental evaluation.
Figure 1.
HBx was evaluated with in silico screening. A, root mean square deviation (RMSD) of HBx model during MD simulation in explicit solvent environment. Protein tends to obtain stable conformation after 1500-picosecond simulation (red line). B, ensemble of different HBx conformations during complete simulation period. C and D, distribution analysis of QED score and binding affinity obtained for all the library compounds. For most of the compounds, the drug-likeliness is high and the largest distribution is obtained near QED score of 0.7. Red line represents the respective threshold and direction represents the values within which compounds were retained. E, the AutoDock predicted drug-binding orientation of IR415 on the modeled structure HBx. Dashed lines represent hydrogen bonds between the atoms involved, whereas hydrophobic interaction is indicated with an arc with spokes radiating toward the ligand atoms they contact.
Development and validation of drug screening platform using reversal of green fluorescent protein silencing assay in HepG2 cells
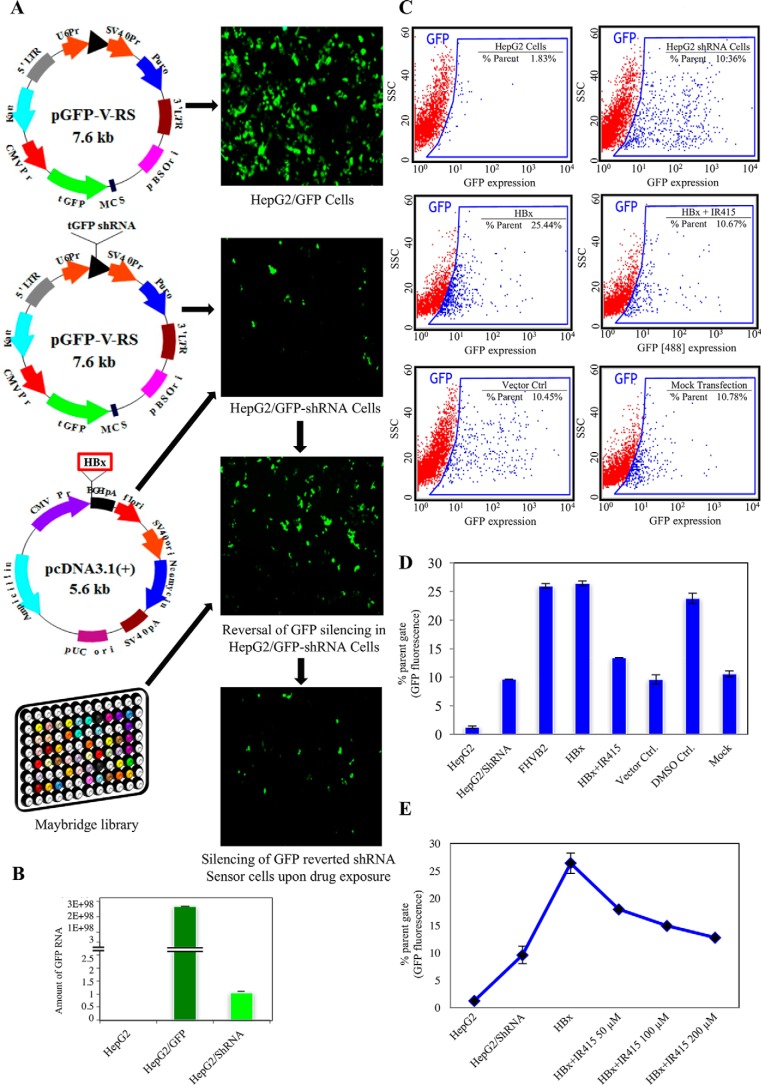
We have previously generated RNAi sensor cell lines of insect to identify RNAi silencing suppressors (RSSs) of different animal viruses. Using these lines, we identified NS4B of dengue virus and HBx of HBV as viral suppressor proteins (VSPs) (9, 12, 13). With a similar approach in the present study, we generated two stable HepG2 cell lines: (i) a reporter line, which constitutively expressed GFP and was created by integrating the turbo-GFP gene into the cellular genome using commercial vector pGFP-V-RS; (ii) second, a HepG2/GFP-shRNA RNAi sensor line where GFP expression is silenced and was generated by transfecting the HepG2 cells with a pGFP-V-RS tGFP shRNA vector (13). A schematic representation of the constructs used in the generation of the above cell lines is shown in Fig. 2A. The expression of GFP in the transgenic lines was visualized and quantitated by microscopic, quantitative RT-PCR (qRT-PCR), and FACS analyses (Fig. 2, A–C).
Figure 2.
A reversion assay in HepG2 cells was used as a drug screening platform. A, immunofluorescence analysis and schematic representation of plasmid constructs used for the generation of the GFP-expressing HepG2 line, GFP-shRNA RNAi sensor line, GFP-reverted HepG2 cells that overexpressed HBx, and re-silenced GFP reverted cell treated by small molecule (IR415). B, real-time RT-PCR analysis of GFP transcript to show relative abundance of GFP in different cell lines: HepG2 cells with no GFP, HepG2/GFP cells (dark green) and HepG2/GFP-shRNA RNAi sensor line (light green). C, dot plot analysis from FACS data shows GFP expression in different HepG2 cell lines. Plots depict forward versus side scatters (SSC) for the expression of the GFP reporter at wavelength (488) with FL1 detector (530/40). D, histogram showing levels of GFP expression in HepG2 cell lines transfected with HBx following exposure with IR415 along with suitable controls. E, graphical representation of concentration-dependent denomination of liner suppression activity of IR415 at 50 μm, 100 μm and 200 μm in HepG2/GFP-shRNA line transfected with HBx. The error bars are the standard deviation of the mean of the ordinate values.
Because we aimed at identification of inhibitors of HBx, we first re-investigated the potential RSS activity of HBx in HepG2/GFP-shRNA RNAi sensor line. The RNAi sensor line was transfected with a plasmid (pcDNATM3.1(+)) containing the HBx gene to transiently overexpress the HBx protein as described earlier (13). Analysis of GFP expression using fluorescent microscopy and FACS analysis in the HBx transfected cells revealed significantly higher levels of reversion of tGFP silencing in comparison to the RNAi sensor line (Fig. 2C). The GFP reversion parameter was compared against another VSR protein FHVB2 that served as a positive control. The expression of the HBx gene was confirmed using Western blot analysis of the cell lysates prepared from the transfected HepG2/GFP-shRNA cells using anti-HBx antibodies. The HBx protein caused at least 2-fold higher GFP reversion when compared with the mock-transfected cells under similar conditions. This change in phenotype due to HBx function was used for a high content screening assay with pharmacologically active compounds from the Maybridge library.
Because compounds from the computationally screened filtered set (n = 100) had similar HBx binding affinity scores, we randomly sampled different compounds for their experimental evaluation. The putative small molecule(s), which were able to restrain reversal of silencing of the reporter tGFP gene by blocking HBx activity, were identified as potential HBx-inhibitor(s). The fold reduction in the mean fluorescent intensity was plotted as a ratio of tGFP levels from cells treated with putative drug candidates versus control untreated cells where only HBx expression occurred. Small molecules that revealed poor reproducibility, nonspecific activity exhibiting bad dose-response profile, and nonspecific activity or that were found to disrupt the reversion assay (fluorescent artifacts or quenchers) were eliminated. After initial screening, three compounds demonstrated promising HBx inhibition potential in our preliminary assays. Motivated by the finding, we re-performed the compound evaluation at different concentrations and replicates (supplemental data 2A). To ensure the reliability of the finding, a test set of compounds for experimental re-evaluation was created. Two different criteria were considered for the choice of drugs in the test set: less binding affinity and high drug-likeliness. A total of 26 compounds from the test set were screened and none of the compounds demonstrated anti-HBx activity (supplemental data 2, B and C). IR415 was found to have a dose-dependent inhibitory effect on HBx, with a minimal effective concentration of 50 μm (Fig. 2, C–E). Data were normalized for cells growing in the presence of either IR415 alone (non-HBx transfected) or to the specified concentration of DMSO alone to observe and nullify the solvent effect. However, the effects among the replicates in reference to the controls were insignificant. IR415, therefore, with consistent effect on suppressor inhibition action, was selected for further analysis. The observation of a distinct phenotypic expression in response to drug treatment led us to investigate the effect of the drug on physiologically relevant processes such as HBx-mediated down-regulation of RNAi components.
Effect of IR415 on Dicer-mediated cleavage
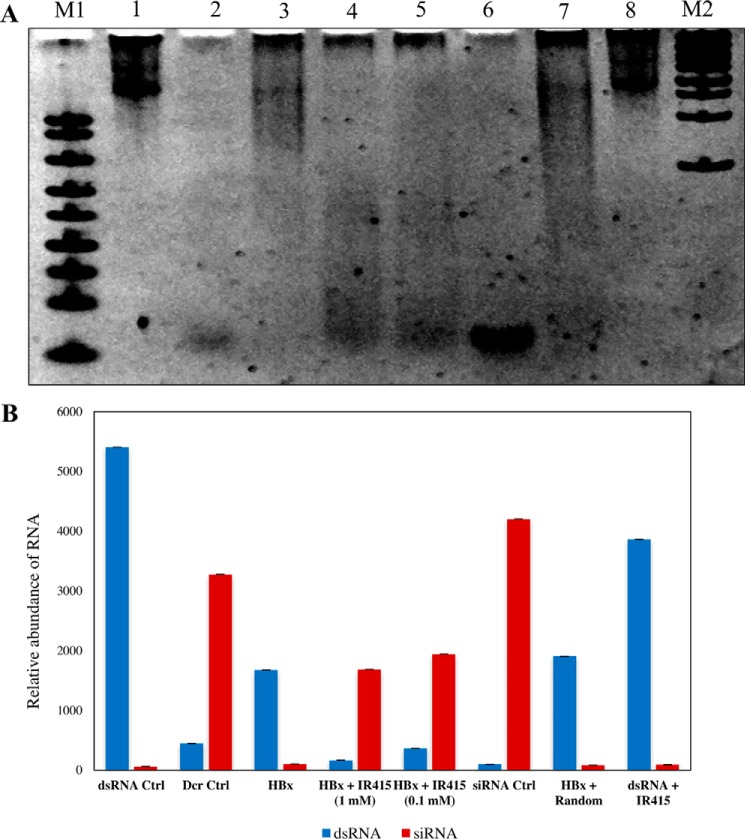
To gain insight into the mode of action of IR415, we evaluated the IR415 effect in a dicing assay in the presence of recombinant HBx protein. HBx protein was expressed in a heterologous Escherichia coli expression system and purified according to the protocol described earlier (13). In vitro dicing assay involves processing of long dsRNA substrates by commercially available human Dicer enzyme that generates siRNAs of ∼22 mers (Fig. 3A, lanes 1 and 2). The in vitro assay was performed according to the manufacturer's instructions by adding 1 unit of human Dicer enzyme to in vitro-synthesized dsRNA (∼900 nucleotides) in the presence of recombinant HBx protein and IR415 at increasing concentrations. Fig. 3A (lane 2) reveals that 1 unit of human Dicer was able to process the dsRNA substrates into siRNAs (∼22 bp), whereas 2 μg of HBx protein blocked Dicer processing, thus completely inhibiting siRNA biogenesis (lane 3). However, the digest patterns were recapitulated in the presence of IR415 in a dose-dependent manner (lanes 4 and 5). The other nonspecific/random compound from the library did not inhibit HBx-mediated restoration of dicing, thus highlighting HBx-IR415 specificity (Fig. 3, A and B). Consequences of rejuvenated siRNA generation by Dicer catalysis established the efficacy of IR415 and suggested the proficiency of IR415-HBx interactions.
Figure 3.
Dicing assay was performed. A, native polyacrylamide gel (5%) image of dicing assay. Following electrophoresis, the gel was stained with ethidium bromide, photographed, and subjected to densitometric analysis. Lane 1, RNA substrate ∼900 bp as a control (in vitro transcribed); lane 2, dicing control (dicing reaction with 1 unit of Dicer protein and 1 μg of RNA); lane 3, dicing reaction with RNA substrate, Dicer protein, and 2 μg of purified HBx; lane 4, dicing reaction with RNA substrate, Dicer protein, HBx, and 1 mm IR415; lane 5, dicing reaction with RNA substrate, Dicer protein, HBx, and 0.1 mm IR415; lane 6, commercially synthesized ∼22-mer siRNA control of the dsRNA; lane 7, dicing reaction with RNA substrate, Dicer, purified HBx, and nonspecific/random compound from library; lane 8, RNA substrate with IR415. B, the relative abundance of siRNA (∼22 mers) and dsRNAs of the dicing assay. The relative abundance was plotted by using the points generated of densitometric scan of siRNA and dicing intermediates. The intensity values were obtained for both abundance of dsRNA and siRNA generation in each lane of reaction set. The x axis represents the RNA content corresponding to each specific dicing reaction as explained in A, and the y axis represents the intensity values obtained for the siRNA and dsRNA regions from the gel. ImageJ software was used for the densitometric scan.
Binding affinity and kinetics of HBx–drug interaction
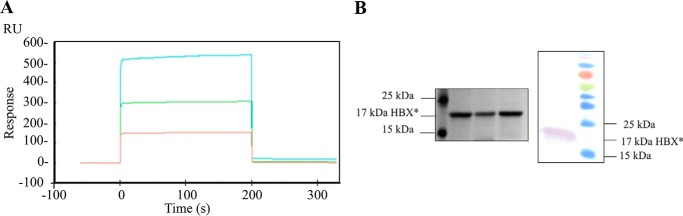
To investigate bimolecular interaction between drug and HBx, surface plasmon resonance (SPR) assays employing solubilized HBx as the immobilized ligand were performed. The stabilized HBx was covalently linked to a series S CM5 sensor chip and sensograms were recorded upon addition of different concentrations of the drug. The representative reference subtracted sensogram, characterizing the affinity of candidate drug molecule, is depicted in Fig. 4. The binding of IR415 increased linearly with time and concentration (Fig. 4). The analysis of the sensogram revealed linear increase in interaction, in response to the increasing concentration of the drug followed by complete dissociation (supplemental data 3). The rate of association (ka) or the “on rate” was 4.24 × 106 m−1 s−1 and the rate of dissociation (kd) or the “off rate” was found to be 8.475 × 10−3 s−1. Therefore, the equilibrium dissociation constant (KD), or “binding constant,” which is calculated by kd/ka was found to be 2.00 × 10−9 m or 2 nm.
Figure 4.
Analysis of molecular interaction of IR415 with HBx by SPR is shown. A, sensogram showing the binding of HBx with varying concentrations 50 μm (red), 100 μm (green), and 200 μm (cyan) of IR415. B, detection of purified HBx by means of SDS-PAGE and Western blotting.
Analysis of binding coefficient revealed significant binding as a function of concentration of IR415. Statistical analysis highlighted the linear increase in association between IR415 and the bound HBx with due course of time. The instrument proved to be extremely sensitive while detecting small molecule–protein interactions. A bulk shift in the response after reference subtraction was observed even after maintaining the same concentration of DMSO in each dilution. Nevertheless, the analysis shows proportional increase in response units to the increasing concentration of the drug. Fig. 4B shows the SDS-PAGE and Western blot of purified 17 kDa HBx protein used for dicing assay (Fig. 3A) and SPR experiments.
Transient HBV replication assays and pre-genomic RNA quantification upon drug exposure
Because the virus-encoded suppressors have an imperative role in viral genome replication, IR415 was expected to have an impact on HBV replication. Hence, we next analyzed the effect of IR415 on HBV replication in a transient HBV replication assay (40). Briefly, HepG2 cells were transfected with a plasmid containing full-length HBV viral genome of 3.2 kb (pHBV). Supernatant was harvested 48 h after the transfection and ELISA analysis was carried out to analyze the levels of hepatitis B virus surface antigen HBsAg and secretory antigen HBeAg. To know the effect of IR415 on viral replication, the HepG2 cells transfected and treated with IR415 were exposed up to 24 or 48 h. As a positive control, transfected cells were treated with tenofovir, a drug currently being used against HBV proliferation. As a negative control, a random drug from the library with lowest binding affinity for HBx or DMSO was used. On days 4 and 5 post drug treatment, HBsAg and HBeAg levels in the medium were measured respectively. Total RNA was isolated from the pHBV transfected HepG2 cells on 48 h post drug exposure and qRT-PCR was performed. HBV replication was confirmed by qRT-PCR with the detection of HBV-specific pre-genomic RNA. We also measured concentration-dependent inhibitory effect of IR415 in different time points during infection cycle and exposure period.
As shown in Fig. 5, A and B, treatment reduced the amount of both HBeAg and HBsAg in culture medium. A concentration-dependent inhibitory effect was observed in different time points during infection cycle and exposure period (Fig. 5C) and HBV replication as measured by relative expression of pre-genomic RNA (fold change) was considerably depleted (Fig. 5D) compared with the untreated and control groups in three independent experiments. Tenofovir treatment also showed similar effects as IR415 treatment. Together, our results show that putative pharmacologically relevant compound IR415 inhibits HBV replication by blocking the HBx suppressor activity, thereby implying its prospective role as an anti-HBV drug candidate.
Figure 5.
HBV antigen kinetics and pre-genomic RNA level upon drug exposure are shown. A and B, the level of HBeAg and HBsAg was measured at different time points. Antigen titers were counted for HepG2 cells transfected with full-length pHBV plasmid that contained 3.5 kb HBV genome. Tenofovir is considered as positive control and DMSO as negative control. 24/24 denotes 24-h incubation of pHBV and 24-h treatment with IR415. 48/48 h indicates 48-h incubation of pHBV for viral proliferation and 48-h exposure post drug treatment. C, concentration-dependent inhibition constrains of IR415 have been significant for both HBeAg and HBsAg production during both 24- and 48-h exposure. D, the level of pre-genomic RNA of HBV post drug exposure. Log2 value of fold change for pre-genomic RNA is in reference to HBV transfected control cell line. The mean levels (± S.D.) were calculated from three replicate transfections. IU/ml measures the antigen titer for assessment of the analytical sensitivity of HBeAg assay which indicates active viral replication and infectivity.
Discussion
With the recognition of RNAi as an important host antiviral defense mechanism, much effort is being invested in the discovery of viral proteins involved in host RNAi suppression. A number of viral encoded suppressors, such as SARS virus 7a protein and influenza A virus NS1 protein have been identified from several animal viruses that display RNAi suppression activity (10, 16, 41, 42). Our group has recently identified NS4B and HBx as viral suppressor proteins in dengue and hepatitis B viruses, respectively (9, 13). Importantly, many of these VSRs are being considered as targets for new drug candidates. Thus, RNAi suppressor proteins add to the repertoire of targets for the multidimensional antiviral therapies. We had earlier described RNAi suppressor activity of HBx that acts by blocking the host dicing activity. The present study describes the identification of a compound that blocks viral suppression activity of HBx from a library of chemical compounds.
Because the crystal structure of HBx has not been determined, we modeled the structure of HBx protein using several computational approaches such as 3D structure, relevant protonation state, and multiple tautomeric form formation. Similar approaches have been routinely performed to illustrate structures of macromolecules whose X-ray crystal structures are not available (43). Based on conformational stability in explicit solvent environments, a structure was selected for docking the small-molecular-weight compounds from Maybridge library. We chose Maybridge library because it represents a collection of small chemical entities that are pharmacologically rich and have been used by a number of studies to identify pharmacophores/inhibitors (44–47). The library has a quantifiable diversity through the application of standard chemometrics. To identify a potential HBx interacting molecule from the library, docking was performed taking into consideration various parameters such as molecular weight, calculated log P, and a number of rotatable bonds that “dock” the Maybridge small molecules into the iteratively predicted macromolecular structure of target HBx. The interaction feasibility scored for their probable complementarity based on binding affinity for hit identification and lead optimization. The most fit small molecule–HBx interactions predicted by AutoDock Vina were selected for in vivo drug profiling studies. Following the docking analysis and virtual screening, we performed biological screening of small-molecular-weight compounds through high-throughput screening platform based on reversal of silencing assay developed by us (9, 12, 13). The high-throughput screening platform employs a laboratory-developed GFP-expressing HepG2 cell line that also carries a GFP-shRNA gene. Transfection of a plasmid expressing HBx restored the GFP expression in GFP-shRNA expressing line, whereas a drug that binds HBx abrogated the expression of GFP. This shRNA-guided high-content phenotype-based assay allows rapid screening of small-molecular-weight compounds with greatest efficacy in 24- or 96-well format. Another benefit of using this system is that all steps of RNAi can be assayed in one screen, as opposed to using siRNA-induced RNAi. The phenotypic pattern thereby indicates a role of RSS-specific target site inhibition, to small RNA biogenesis to defeat the virus-mediated counterdefense. Based on binding affinities, drug-likeliness, and biological screening, we finally identified a candidate drug molecule, IR415, that effectively blocked the HBx-mediated GFP expression. Of several compounds screened, IR415 showed the predicted ability to bind to the HBx α-helix. To confirm whether the effect of IR415 was exclusive to HBx and not a consequence of its interaction with other cellular components, we performed the reversal of RNAi silencing experiment in cell types of different origin including Huh7 and HEK293 (data not shown). The effect of the drug candidate has remained the same for all in respect to the control. These results further substantiated the unlikely interaction of IR415 with other cellular components. Additionally, to determine the blocking specificity of IR415 and its exclusivity to HBx suppression activity, the compound has been used in the presence of another viral suppressor protein, the Flock House virus suppressor protein FHVB2. Negligible reversion of GFP was scored with the latter protein, proving incompetence of IR415 toward another viral suppressors (data not shown). The molecular properties of the compound IR415 are given in supplemental Table S1. We found a very limited number of experimental studies which investigate its role as an antiviral compound. In fact, none of the previous studies proposed the binding surface for the compound, and protein-ligand conformation is still not available in the Protein Data Bank (36). However, when docked, the compound demonstrated good binding affinity for the interface region of HBx surface (supplemental data 1). Fig. 1E describes a putative model of IR415 in a homology model of HBx that identifies the interactions within the drug-binding pocket. HBx helices are shown in yellow and green. For purposes of clarity, residues around IR415 at the HBx binding site are displayed.
VSRs interfere with the host RNAi pathways at different steps of RNAi activities; some inhibit the processing of dsRNA to siRNAs, whereas others bind siRNAs and sequester them, thus preventing their loading to RISC complex (48, 49). It has been suggested that HBx protein suppresses the RNAi response by directly inhibiting the dicing process. To know whether IR415 acts through this pathway, an in vitro dicing assay was performed in the presence of HBx protein and IR415 as described earlier. Significantly IR415 was able to restore the ribonuclease catalysis that was blocked by HBx as indicated by the presence of siRNAs in the reaction lane, thereby suggesting that drug IR415 blocks the suppressor activity of HBx and allows siRNA biogenesis.
The effective concentration of IR415 that was derived from cell culture experiments was further corroborated by estimating binding and kinetic constants by SPR analysis using the Biacore T200 system. The interaction of IR415 with HBx as revealed by SPR experiments substantiated its ability to directly interact with HBx and interfere with its biochemical activity. A difference in the in vivo and the in vitro effective concentrations of IR415 could be a consequence of differential rate of drug uptake in the discussed approaches. The direct interaction of drug with HBx forms a basis of specific drug development to block HBx function. It is noteworthy that IR415 can be further chemically altered for its greater access within the HepG2 cells and the altered form would then be usable in nanomolar concentrations for anti-HBV therapeutic treatment. The pharmacokinetic profile of IR415 is compatible with human applications.
HBx exhibits a plethora of activities such as in viral replication, transactivation of promoters, and activation of transcription of host genes (31–34, 50). However, complete suppression of HBV replication, e.g. depletion of HBsAg, is an imperative aim for any kind of antiviral treatment to combat against the infection. Given its role in viral replication, we tested the effect of IR415 that binds HBx during the viral replication in an in vitro culture assay and compared its efficacy with tenofovir, a potent and selective inhibitor of HBV DNA polymerase-reverse transcriptase. Anti-HBV nucleoside (tenofovir) directly works at the replication step in the HBV life cycle and thereby blocks the replication of HBV at either single strand formation or double strand formation. Decreased replication of HBV results in decreased expression of HBV antigens as well (51). Importantly, IR415 was just as effective in suppressing HBV RNA levels as tenofovir. Hence IR415 in combination with tenofovir will form an extremely potent antiviral drug as these two drugs work via independent pathways (Fig. 6). To our knowledge this is the first report that identifies a small-molecular-weight pharmacological active compound which binds a viral suppressor protein and has an antiviral property.
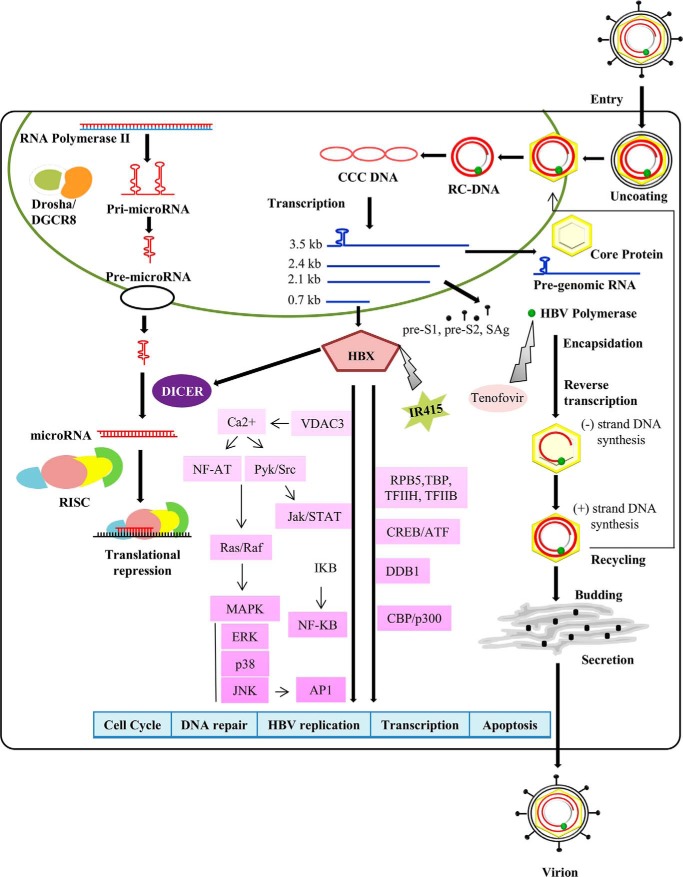
Figure 6.
HBx drug interaction model is shown. Pictorial representation of HBV cycle in infected cell and site of interaction of IR415 and tenofovir and their effect on virus replication is presented.
Experimental procedures
HBx MD simulation analysis
Conformational stability of the short-listed models was evaluated using molecular dynamic simulations in explicit solvent model using GROMACS (version 4.5.6) (52). To remove extremely unfavorable conformational artifact in the predicted models, we subjected the latter to restrained energy minimization. The minimized systems were allowed to undergo constant volume dynamic equilibration (NVT) for about 50,000 steps. This is followed by constant pressure dynamics (NPT) of the semi-equilibrated structure for 50,000 steps at 1 atmosphere. Finally, production simulations were carried out on the equilibrated structures under periodic boundary conditions, with a nonbonded cutoff of 10 Å. We used partial-mesh Ewald method for the treatment of long-range nonbonded interactions and SHAKE algorithm to constrain hydrogen atoms. Summarily, the MD simulations were carried out at 300 K temperature for 3000 picoseconds.
High-throughput virtual screening
AutoDock Vina (38) was used to perform molecular docking and virtual screening of Maybridge drug library. A 3D grid box was designed using PyRx interface around the interface helix on HBx surface, which was used as flexible docking site to identify compounds with strong binding affinity (kcal/mol). The docked compounds were subsequently shortlisted for scrutinizing their drug-likeliness using QED software (version 1.0.1), which is an RDkit-based implementation of Biscu-it. The IR415–HBx interaction plot has been generated using LIGPLOT (53).
Generation of HepG2 RNAi sensor line
HepG2 cells were transfected with 6 μg of plasmid construct using pGFP-V-RS series of vectors expressing tGFP (OriGene, Rockville, MD). For generation of HepG2/GFP control cell line pGFP-V-RS vector and for HepG2/GFP-shRNA RNAi sensor line, previously developed pGFP-V-RS tGFP shRNA plasmids (13) were used in the presence of jetPRIME transfection reagent (Polyplus Transfection, New York, NY). The cells were later sloughed and reseeded in a 100-mm plate in complete DMEM with puromycin concentration 1 μg/ml to 3 μg/ml for final selection pressure. Total RNA was isolated from HepG2/GFP control cell and HepG2/GFP-shRNA RNAi sensor line and GFP quantification was carried out by qRT-PCR using Verso 1-Step qRT-PCR kit, SYBR Green, ROX in PikoReal Real-Time PCR 96-Well System (Thermo Scientific). GAPDH was used as an endogenous control for RNA expression profiling. ShRNA-induced GFP down-regulation was calculated against HepG2 as negative and HepG2/GFP cell as a positive control calibrator using 2−ΔΔCt method. 20 days post selection, monoclonal colonies were observed and sorted using a BD FACSJazz™ Cell Sorter (BD Biosciences).
Compound library for high-throughput screening
Maybridge library was obtained from Thermo Fisher Scientific and dissolved in DMSO. Stock solutions were prepared at concentration 10 mm. Putative compounds were tested at concentrations between 50 and 200 μm.
Reversal of tGFP silencing assay in HepG2 cells
HepG2, HepG2/GFP, and HepG2/GFP-shRNA cells were uniformly transferred to 24-well plates containing DME complete medium with puromycin with cell density 0.8–1.0 × 105/well. HepG2/GFP-shRNA cells were individually transfected with HBx cloned in pcDNATM3.1(+) vector (Invitrogen) in the presence of jetPRIME reagent (Polyplus Transfection). The empty vector pcDNATM3.1(+), transfection reagent, and DMSO were exposed independently to serve as mock. After 24 h of incubation, fresh medium was replaced and putative compounds were exposed to transfected cells at a concentration of 200 μm in 2% DMSO. 24 h after drug exposure, cells were processed for FACS analysis. All compounds were retested in triplicate in three independent experiments.
Fluorescence microscopy and flow cytometric analysis
The level of fluorescence in the GFP reporter lines, RNAi sensor lines, and HBx transfected cells were monitored using a Nikon Eclipse TE2000-U fluorescence microscope followed by quantification using BD FACSJazz (BD Biosciences). The percentage of HBx transfected cells that reverted back silenced tGFP expression was counted by gating the region of control tGFP sensored expression in shRNA line using the BD FACS Sortware (BD Biosciences) software.
HBx protein purification
HBx was cloned into pET28a vector and expressed with His-tag fusion followed by purification as described earlier (13). Protein-containing fractions were pooled, purified to homogeneity, and concentrated by ultrafiltration at 3000 × g (centricon-30 with a cutoff of 10 kDa) followed by dicing assay. For SPR analysis protein was dialyzed in 50 mm CAPS and 150 mm NaCl (pH 11.0).
Dicing assay
Dicing assay was performed using human dicing reaction kit (Genlantis) and the reaction was carried out as described earlier (13). 2 μg of purified HBx was tested in specific dicing reactions. Dicer was preincubated briefly with different concentrations of IR415 and treated with HBx to investigate the effect of HBx protein on the dicing activity. A compound from the library with similar structure with IR415 was used as a negative control to monitor nonspecific interaction in the dicing reaction. The result was visualized using a 5% native polyacrylamide gel for the UV visualization. Densitometric scanning was performed using ImageJ software for the dsRNA and siRNA bands of dicing products.
Surface plasmon resonance
The surface plasmon resonance analysis was carried out using a Biacore T200 apparatus (GE Healthcare). HBx protein was immobilized on an S series CM5 sensor chip using 10 mm disodium tetra borate, pH 8.5, and 1 m NaCl, as suggested by the manufacturer. Following immobilization, the surface was blocked by 1 m ethanolamine, pH 8.5, and regenerated using 50 mm NaOH. Phosphate buffered saline, pH 7.4, supplemented with 2% DMSO was used as the running buffer for the experiment. The drug IR415 was dissolved in 100% DMSO to a final concentration of 10 mm and stored in glass vials. Suitable dilutions (50, 100, 200 μm) were made prior to the experiment using phosphate buffered saline, pH 7.4, while maintaining a concentration of 2% DMSO in all samples. The experiments were carried out at 25 °C, and the data were analyzed using the Biacore T200 SPR kinetics evaluation software.
HBeAg and HBsAg measurement upon drug exposure
To measure the levels of antigens HBsAg and HBeAg, experiments were performed in HepG2 cells. Plasmid construct with the complete full-length HBV viral genome of 3221 bp (pHBV) was used in transient HBV replication assay (54). After 48 h transfected cells were treated with putative drug candidate. Tenofovir was used as positive control against HBV infection. To quantify the secreted HBeAg and surface HBsAg cell culture supernatant was collected and measured 48 h after drug treatment via in vitro chemiluminescent immunoassay.
HBV RNA measurement by qRT-PCR
qRT-PCR was performed using the Verso 1-Step RT-qPCR Kit, SYBR Green, ROX as per the manufacturer's instructions in PikoReal 96-well detection system (Thermo Scientific). A cycle threshold (Ct) was taken into consideration and analyzed using the 2−ΔΔCt method. The HBV replication efficiency was monitored for the pHBV transfected cells in terms of HBV pre-genomic RNA and core transcripts. The intracellular mRNA levels were normalized with GAPDH mRNA levels.
Author contributions
R. K. B., S. K. M., and P. M. designed and conceived the study. S. G. performed the experiments. A. K. and D. G. performed and analyzed the bioinformatics data for molecular modelling and docking experiment. S. K. and P. D. performed the SPR experiment. A. V. provided assistance for HBsAg and HBeAg measurement. A. K. S. and S. K. S. provided valuable suggestions for the antigen kinetics experiments. J. K. T. provided SPR instrumentation facility with experimental assistance. S. G., P. M., S. K. M., and R. K. B. analyzed the data and prepared the manuscript. All authors read and approved the final version of the paper.
Supplementary Material
Acknowledgments
We thank Pradeep Kumar (BD Biosciences) for his assistance during FACS analysis. We also appreciate the help from Dr. Sailesh Bajpai from GE Healthcare for SPR analysis.
This work was supported by Government of India, Department of Biotechnology Grant BT/PR10673/AGR/36/579/2008 and Bioinformatics Infrastructure (BIF) Grant BT/BI/25/066/2012. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental data 1–3 and Table S1.
- HBV
- hepatitis B virus
- HBx
- hepatitis B virus X protein
- MD
- molecular dynamics
- VSR
- viral suppressors of RNA
- QED
- quantitative estimation of drugs
- RSS
- RNAi silencing suppressor
- qRT-PCR
- quantitative reverse transcription-polymerase chain reaction
- SPR
- surface plasmon resonance
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid.
References
- 1. World Health Organization (2017) Global Hepatitis Report 2017. Geneva, Switzerland: World Health Organization [Google Scholar]
- 2. Skalsky R. L., and Cullen B. R. (2010) Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64, 123–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Félix M. A., Ashe A., Piffaretti J., Wu G., Nuez I., Bélicard T., Jiang Y., Zhao G., Franz C. J., Goldstein L. D., Sanroman M., Miska E. A., and Wang D. (2011) Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 9, e1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li Y., Lu J., Han Y., Fan X., and Ding S. W. (2013) RNA interference functions as an antiviral immunity mechanism in mammals. Science 342, 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maillard P. V., Ciaudo C., Marchais A., Li Y., Jay F., Ding S. W., and Voinnet O. (2013) Antiviral RNA interference in mammalian cells. Science 342, 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nicolás F. E., Torres-Martínez S., and Ruiz-Vázquez R. M. (2013) Loss and retention of RNA interference in fungi and parasites. PLoS Pathog. 9, e1003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fabozzi G., Nabel C. S., Dolan M. A., and Sullivan N. J. (2011) Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J. Virol. 85, 2512–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ji J., Glaser A., Wernli M., Berke J. M., Moradpour D., and Erb P. (2008) Suppression of short interfering RNA-mediated gene silencing by the structural proteins of hepatitis C virus. J. Gen. Virol. 89, 2761–2766 [DOI] [PubMed] [Google Scholar]
- 9. Kakumani P. K., Ponia S. S., S R. K., Sood V., Chinnappan M., Banerjea A. C., Medigeshi G. R., Malhotra P., Mukherjee S. K., and Bhatnagar R. K. (2013) Role of RNA interference (RNAi) in dengue virus replication and identification of NS4B as an RNAi suppressor. J. Virol. 87, 8870–8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karjee S., Minhas A., Sood V., Ponia S. S., Banerjea A. C., Chow V. T., Mukherjee S. K., and Lal S. K. (2010) The 7a accessory protein of severe acute respiratory syndrome coronavirus acts as an RNA silencing suppressor. J. Virol. 84, 10395–10401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scholthof H. B. (2006) The Tombusvirus-encoded P19: From irrelevance to elegance. Nat. Rev. Microbiol. 4, 405–411 [DOI] [PubMed] [Google Scholar]
- 12. Singh G., Popli S., Hari Y., Malhotra P., Mukherjee S., and Bhatnagar R. K. (2009) Suppression of RNA silencing by Flock house virus B2 protein is mediated through its interaction with the PAZ domain of Dicer. FASEB J. 23, 1845–1857 [DOI] [PubMed] [Google Scholar]
- 13. Chinnappan M., Singh A. K., Kakumani P. K., Kumar G., Rooge S. B., Kumari A., Varshney A., Rastogi A., Singh A.K., Sarin S. K., Malhotra P., Mukherjee S. K., and Bhatnagar R. K. (2014) Key elements of the RNAi pathway are regulated by hepatitis B virus replication and HBx acts as a viral suppressor of RNA silencing. Biochem. J. 462, 347–358 [DOI] [PubMed] [Google Scholar]
- 14. Bennasser Y., Le S. Y., Benkirane M., and Jeang K. T. (2005) Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22, 607–619 [DOI] [PubMed] [Google Scholar]
- 15. Haasnoot J., de Vries W., Geutjes E. J., Prins M., de Haan P., and Berkhout B. (2007) The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 3, e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li W. X., Li H., Lu R., Li F., Dus M., Atkinson P., Brydon E. W., Johnson K. L., García-Sastre A., Ball L. A., Palese P., and Ding S. W. (2004) Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. U.S.A. 101, 1350–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lara-Pezzi E., Armesilla A. L., Majano P. L., Redondo J. M., and López-Cabrera M. (1998) The hepatitis B virus X protein activates nuclear factor of activated T cells (NF-AT) by a cyclosporin A-sensitive pathway. EMBO J. 17, 7066–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lucito R., and Schneider R. J. (1992) Hepatitis B virus X protein activates transcription factor NF-κB without a requirement for protein kinase C. J. Virol. 66, 983–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maguire H. F., Hoeffler J. P., and Siddiqui A. (1991) HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science 252, 842–844 [DOI] [PubMed] [Google Scholar]
- 20. Williams J. S., and Andrisani O. M. (1995) The hepatitis B virus X protein targets the basic region-leucine zipper domain of CREB. Proc. Natl. Acad. Sci. U.S.A. 92, 3819–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benn J., and Schneider R. J. (1994) Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 91, 10350–10354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bouchard M. J., Puro R. J., Wang L., and Schneider R. J. (2003) Activation and inhibition of cellular calcium and tyrosine kinase signaling pathways identify targets of the HBx protein involved in hepatitis B virus replication. J. Virol. 77, 7713–7719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bouchard M. J., Wang L. H., and Schneider R. J. (2001) Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294, 2376–2378 [DOI] [PubMed] [Google Scholar]
- 24. Benn J., and Schneider R. J. (1995) Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc. Natl. Acad. Sci. U.S.A. 92, 11215–11219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chirillo P., Pagano S., Natoli G., Puri P. L., Burgio V. L., Balsano C., and Levrero M. (1997) The hepatitis B virus X gene induces p53-mediated programmed cell death. Proc. Natl. Acad. Sci. U.S.A. 94, 8162–8167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Terradillos O., Pollicino T., Lecoeur H., Tripodi M., Gougeon M. L., Tiollais P., and Buendia M. A. (1998) p53-independent apoptotic effects of the hepatitis B virus HBx protein in vivo and in vitro. Oncogene 17, 2115–2123 [DOI] [PubMed] [Google Scholar]
- 27. Carmona S., Ely A., Crowther C., Moolla N., Salazar F. H., Marion P. L., Ferry N., Weinberg M. S., and Arbuthnot P. (2006) Effective inhibition of HBV replication in vivo by anti-HBx short hairpin RNAs. Mol. Ther. 13, 411–421 [DOI] [PubMed] [Google Scholar]
- 28. McCaffrey A. P., Nakai H., Pandey K., Huang Z., Salazar F. H., Xu H., Wieland S. F., Marion P. L., and Kay M. A. (2003) Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 21, 639–644 [DOI] [PubMed] [Google Scholar]
- 29. Hamasaki K., Nakao K., Matsumoto K., Ichikawa T., Ishikawa H., and Eguchi K. (2003) Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett. 543, 51–54 [DOI] [PubMed] [Google Scholar]
- 30. Chen H. S., Kaneko S., Girones R., Anderson R. W., Hornbuckle W. E., Tennant B. C., Cote P. J., Gerin J. L., Purcell R. H., and Miller R. H. (1993) The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J. Virol. 67, 1218–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zoulim F., Saputelli J., and Seeger C. (1994) Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 68, 2026–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cha M. Y., Ryu D. K., Jung H. S., Chang H. E., and Ryu W. S. (2009) Stimulation of hepatitis B virus genome replication by HBx is linked to both nuclear and cytoplasmic HBx expression. J. Gen. Virol. 90, 978–986 [DOI] [PubMed] [Google Scholar]
- 33. Keasler V. V., Hodgson A. J., Madden C. R., and Slagle B. L. (2007) Enhancement of hepatitis B virus replication by the regulatory X protein in vitro and in vivo. J. Virol. 81, 2656–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tang H., Delgermaa L., Huang F., Oishi N., Liu L., He F., Zhao L., and Murakami S. (2005) The transcriptional transactivation function of HBx protein is important for its augmentation role in hepatitis B virus replication. J. Virol. 79, 5548–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim C. M., Koike K., Saito I., Miyamura T., and Jay G. (1991) HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature 351, 317–320 [DOI] [PubMed] [Google Scholar]
- 36. Li T., Robert E. I., van Breugel P. C., Strubin M., and Zheng N. (2010) A promiscuous α-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat. Struct. Mol. Biol. 17, 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trott O., and Olson A. J. (2010) AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bickerton G. R., Paolini G. V., Besnard J., Muresan S., and Hopkins A. L. (2012) Quantifying the chemical beauty of drugs. Nat. Chem. 4, 90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Köck J., Rösler C., Zhang J.-J., Blum H. E., Nassal M., and Thoma C. (2010) Generation of covalently closed circular DNA of hepatitis B viruses via intracellular recycling is regulated in a virus specific manner. PLoS Pathog. 6, e1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bucher E., Hemmes H., de Haan P., Goldbach R., and Prins M. (2004) The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J. Gen. Virol. 85, 983–991 [DOI] [PubMed] [Google Scholar]
- 42. Li Y., Basavappa M., Lu J., Dong S., Cronkite D. A., Prior J. T., Reinecker H. C., Hertzog P., Han Y., Li W. X., Cheloufi S., Karginov F. V., Ding S. W., and Jeffrey K. L. (2016) Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat. Microbiol. 2, 16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Evers A., and Klabunde T. (2005) Structure-based drug discovery using GPCR homology modeling: successful virtual screening for antagonists of the alpha1A adrenergic receptor. J. Med. Chem. 48, 1088–1097 [DOI] [PubMed] [Google Scholar]
- 44. Dasgupta T., Chitnumsub P., Kamchonwongpaisan S., Maneeruttanarungroj C., Nichols S. E., Lyons T. M., Tirado-Rives J., Jorgensen W. L., Yuthavong Y., and Anderson K. S. (2009) Exploiting structural analysis, in silico screening, and serendipity to identify novel inhibitors of drug-resistant falciparum malaria. ACS Chem. Biol. 4, 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ekins S., Reynolds R. C., Franzblau S. G., Wan B., Freundlich J. S., and Bunin B. A. (2013) Enhancing hit identification in Mycobacterium tuberculosis drug discovery using validated dual-event Bayesian models. PLoS One 8, e63240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Major L. L., and Smith T. K. (2011) Screening the MayBridge rule of 3 fragment library for compounds that interact with the Trypanosoma brucei myo-inositol-3-phosphate synthase and/or show trypanocidal activity. Mol. Biol. Int. 2011, 389364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wahome P. G., Ahlawat S., and Mantis N. J. (2012) Identification of small molecules that suppress ricin-induced stress-activated signaling pathways. PLoS One 7, e49075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chao J. A., Lee J. H., Chapados B. R., Debler E. W., Schneemann A., and Williamson J. R. (2005) Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat. Struct. Mol. Biol. 12, 952–957 [DOI] [PubMed] [Google Scholar]
- 49. Mallory A. C., Reinhart B. J., Bartel D., Vance V. B., and Bowman L. H. (2002) A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. U.S.A. 99, 15228–15233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tang H., Oishi N., Kaneko S., and Murakami S. (2006) Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 97, 977–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zoutendijk R., Hansen B. E., van Vuuren A. J., Boucher C. A. B., and Janssen H. L. A. (2011) Serum HBsAg decline during long-term potent nucleos(t)ide analogue therapy for chronic hepatitis B and prediction of HBsAg loss. J. Infect. Dis. 204, 415–418 [DOI] [PubMed] [Google Scholar]
- 52. Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M. R., Smith J. C., Kasson P. M., van der Spoel D., Hess B., and Lindahl E. (2013) GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wallace A. C., Laskowski R. A., and Thornton J. M. (1995) LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 54. Blum H. E., Galun E., Liang T. J., von Weizsäcker F., and Wands J. R. (1991) Naturally occurring missense mutation in the polymerase gene terminating hepatitis B virus replication. J. Virol. 65, 1836–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.