Abstract
Enzyme activities that improve digestion of recalcitrant plant cell wall polysaccharides may offer solutions for sustainable industries. To this end, anaerobic fungi in the rumen have been identified as a promising source of novel carbohydrate active enzymes (CAZymes) that modify plant cell wall polysaccharides and other complex glycans. Many CAZymes share insufficient sequence identity to characterized proteins from other microbial ecosystems to infer their function; thus presenting challenges to their identification. In this study, four rumen fungal genes (nf2152, nf2215, nf2523, and pr2455) were identified that encode family 39 glycoside hydrolases (GH39s), and have conserved structural features with GH51s. Two recombinant proteins, NF2152 and NF2523, were characterized using a variety of biochemical and structural techniques, and were determined to have distinct catalytic activities. NF2152 releases a single product, β1,2-arabinobiose (Ara2) from sugar beet arabinan (SBA), and β1,2-Ara2 and α-1,2-galactoarabinose (Gal-Ara) from rye arabinoxylan (RAX). NF2523 exclusively releases α-1,2-Gal-Ara from RAX, which represents the first description of a galacto-(α-1,2)-arabinosidase. Both β-1,2-Ara2 and α-1,2-Gal-Ara are disaccharides not previously described within SBA and RAX. In this regard, the enzymes studied here may represent valuable new biocatalytic tools for investigating the structures of rare arabinosyl-containing glycans, and potentially for facilitating their modification in industrial applications.
Keywords: carbohydrate, enzyme, fungi, galactose, glycoside hydrolase, arabinose, rumen
Introduction
The rumen microbiome is recognized as one of the most efficient microbial ecosystems in the degradation of plant biomass (1, 2). It contains a diverse microbial community with large numbers of bacteria, anaerobic fungi, ciliate protozoa, and bacteriophages. Of these, rumen bacteria and fungi are considered to be indispensable for plant fiber digestion. It has been estimated that at least 85% of the microbes inhabiting the rumen have not been cultured using traditional approaches (1). In recent years, our knowledge of structure and function of the rumen microbial diversity has drastically increased due to improvements in sequencing technologies (3–5).
The plant cell wall comprises cellulose, hemicellulose, and pectin (6). These polysaccharides are often interconnected, contain many diverse sugar chemistries, and display a variety of decorations and branching. Cellulose is the simplest of these three classes of polysaccharides and is composed of linear polymers of β-1,4-linked d-glucose (7). Hemicellulose is a group of branched polysaccharides that are classified according to the primary sugar within the backbone of the polymer (e.g. xylan is composed of β-1,4-linked d-xylose). Hemicelluloses contain extensive variations in their repeating structure. This can be seen in arabinoxylans, such as rye arabinoxylan (RAX). 3 RAX consists of a polymer chain of β-1,4-linked d-xylose units, many of which are 2- or 3-monosubstituted or 2,3-disubstituted by α-l-arabinose (Ara) (8). Pectin is found within the primary cell wall and the middle lamella, which punctuates the junctions between primary walls of neighboring cells, and participates in intercellular connections (9). It is a structurally complex polysaccharide that is enriched in d-galacturonic acid and divided into three distinct classes of pectic polysaccharides: homogalacturonan, rhamnogalacturonan-I (RG-I), and rhamnogalacturonan-II that display substantial variability in their structure (10, 11). For example, the side chains of RG-I can be heavily decorated with arabinans (12), such as sugar beet arabinan (SBA), which consists of an α-1,5-l-arabinofuranosyl backbone decorated with α-1,2- and α-1,3-l-arabinofuranosyl side chains.
Also present in the plant cell wall are structurally diverse cell-surface glycoproteins that are collectively referred to as arabinogalactan proteins (AGPs). This structurally diverse protein family is enriched in the amino acids: hydroxy-Pro/Pro, Ala, and Ser/Thr and heavily glycosylated (90–98% w/w). AGPs are thought to play important roles in various aspects of plant growth and development including reproduction, cell signaling, and microbial interactions; and may serve to anchor the pectic and hemicellulosic polysaccharide networks (13).
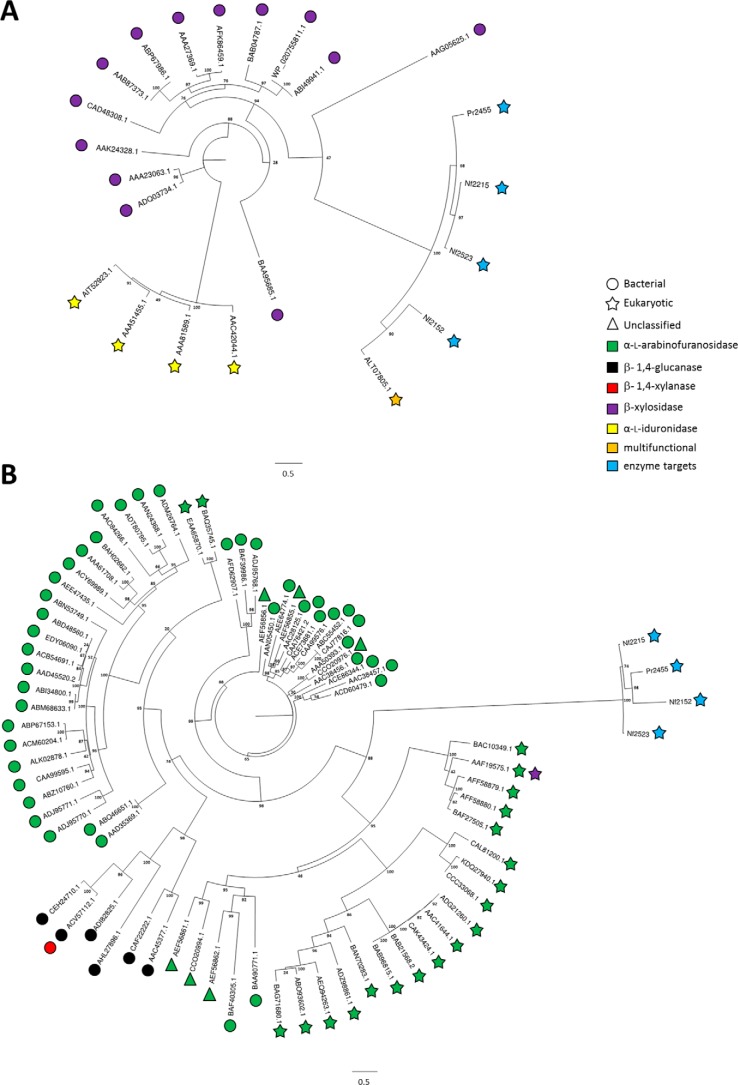
Enzymes that modify carbohydrates are referred to as carbohydrate active enzymes (CAZymes). CAZymes are categorized into “classes” based upon their catalytic function and “families” based upon sequence relatedness (14). Family relatedness does not necessarily equate to functional relatedness, however, as many CAZyme families have now been described to contain members with variations in their enzyme specificities (see Fig. 1) (15, 16), a property that is referred to as “polyspecificity.”
Figure 1.
Phylogenetic trees of four rumen fungal query sequences aligned with characterized sequences from related GH39 and GH51 families. A, GH39 and B, GH51. Each sequence leaf represents the catalytic module from the associated ascension number. Taxonomic (bacterial, eukaryotic, and unclassified) and functional (α-l-arabinofuranosidase, β-1,4-glucanase, β-1,4-xylanase, β-xylosidase, α-l-iduronidase) differences are defined within the legend. Characterized activities and organism source are depicted with different symbol colors and shapes, respectively.
The hydrolysis of a glycosidic linkage between two or more carbohydrates, or a carbohydrate and non-carbohydrate adduct, leading to the generation of a new reducing end is catalyzed by enzymes known as glycoside hydrolases (GHs) (17). Generally, hydrolysis is performed by two primary amino acid residues within the enzyme active site. Inverting GHs, which generate products that have an inverted anomeric configuration at the nascent reducing end, possess a general acid (i.e. proton donor) and a catalytic base (i.e. proton acceptor). Alternatively, retaining GHs, which generate products that retain anomeric configuration at the nascent reducing end, possess an acid/base and a nucleophile and catalyze a double displacement reaction. Often the activity of GHs is potentiated by carbohydrate-binding modules (CBMs) through “targeting” or “concentrating” effects (18). Polyspecificity is also a common feature in the binding specificities of CBM families, such as CBM13 (19). Using CBMs to identify associations with uncharacterized genes, therefore, may assist in the discovery of new enzyme activities.
Comparative analysis of fungal genomes and metatranscriptomes has revealed that rumen fungi exhibit tremendous diversity in the number and types of their CAZymes (20). Harnessing this genetic reservoir holds vast potential for biotechnological applications. For example, in vitro studies have demonstrated that rumen fluid supplemented with select combinations of CAZymes noticeably boosts the release of cellobiose, glucose, and xylose from plant cell wall structural polysaccharides (21). Anerobic fungi isolated from rumen or herbivore dung, such as phylum Neocallimastigomycota, are known to participate in the deconstruction of plant cell wall substrates by invasive rhizoidal growth, which physically disrupts recalcitrant tissues; and secrete a diverse arsenal of CAZymes (22). Many of these CAZymes share little sequence identity to characterized proteins from other microbial ecosystems (20, 23).
In this study, we identified four genes from anaerobic fungi that are predicted to encode proteins with an N-terminal GH39 followed by a CBM13 (19): nf2152, nf2523, and nf2215 from Neocallimastrix frontalis; and pr2455 from Piromyces rhizinflatus. These “fungal GH39s” comprise a new subfamily within GH39. To characterize their functions, we synthesized the genes, expressed them in Escherichia coli, and purified their gene products for biochemical characterization. The enzymes were screened against a library of plant cell wall substrates, and activity was observed on rare substrates present within SBA and RAX. Analyses of the product profiles of NF2152 and NF2523 revealed that two distinct products were released, underpinning that this subfamily is polyspecific with activities that are unique from what was reported for a related sequence, bgxg1, from Orpinomyces sp. strain C1A (24). Structural analysis of NF2152 provides insights into the molecular basis of recognition by this enzyme family.
Results
Phylogenetic analysis of fungal proteins
Genes encoding NF2152, NF2215, NF2523, and PR2455 were identified from the transcriptomes of anaerobic fungal cultures previously grown on barley straw. These gene products were used as query sequences to search for entries with homology in the NCBI non-redundant database using the algorithm BLASTP. The four proteins displayed low sequence identity to GH39s and GH51s, based on pfam annotation. This was consistent when aligned to all GH39 or GH51 entries in the CAZy database (14). NF2152 had 14.9% average identity against all GH39 entries, and 12.0% average identity against all GH51 entries. Therefore, to provide insight into the potential activities of their gene products, the four fungal sequences were aligned with characterized sequences from GH39 (n = 18; Fig. 1A) and GH51 (n = 74; Fig. 1B). In both trees, the fungal target sequences partitioned as single, distantly related clades. Comparisons with the distribution of GH39 activities, which include α-l-iduronidase (EC 3.2.1.76) and β-xylosidase (EC 3.2.1.37) (14), revealed that the fungal sequences may have diverged from a common ancestor with the bacterial β-xylosidases and cluster with a “multifunctional” GH39 previously described from Orpinomyces sp. strain C1A, Bgxg1 (24). Comparison with GH51 known activities, which include endoglucanase (EC 3.2.1.4), endo-β-1,4-xylanase (EC 3.2.1.8), β-xylosidase (EC 3.2.1.37), and α-l-arabinofuranosidase (ABF; EC 3.2.1.55) (14), indicated that the clade partitions from a region of eukaryotic ABFs. Based upon these comparisons, and their high degree of relatedness to Bgxg1 (60.9–77.9%), these sequences were classified as a new subfamily of GH39s.
Functional screening of NF2152
The N-terminal domain of the four GH39 genes were synthesized and recombinant protein was produced. Based upon purification yields and protein stability, NF2152 was selected for functional screening and preliminary enzyme characterizations by thin layer chromatography (TLC) on a variety of appropriate plant cell wall substrates, including SBA, RAX, wheat arabinoxylan, β-glucan, xyloglucan, gum arabic, galactomannan, pectic galactan, arabinogalactan, RG-I, arabinofuranosyl-xylobiose, arabinofuranosyl-xylotriose, PNP-α-l-arabinofuranoside, PNP-α-l-arabinopyranoside and beech wood xylan; activity was detected primarily on SBA (Fig. 2) and RAX (Fig. 3). Based upon the multifunctional activity reported for Bgxg1 (24), NF2152 was also screened against PNP-β-d-glucopyranoside, PNP-β-d-galactopyranoside, PNP-β-d-xylopyranoside, and cellobiose under similar reaction conditions; however, no activity was observed. TLC analysis of the SBA digestion products revealed that NF2152 released a single product band that migrated slower than arabinose (Ara), xylose (Xyl), α-1,5-arabinobiose (Ara2), and α-1,5-arabinotriose (Ara3) (Fig. 2, A and B). These observations indicated that NF2152 is not a conventional ABF or xylosidase as predicted by the comparative analysis with other characterized GH39s and GH51s (Fig. 1).
Figure 2.
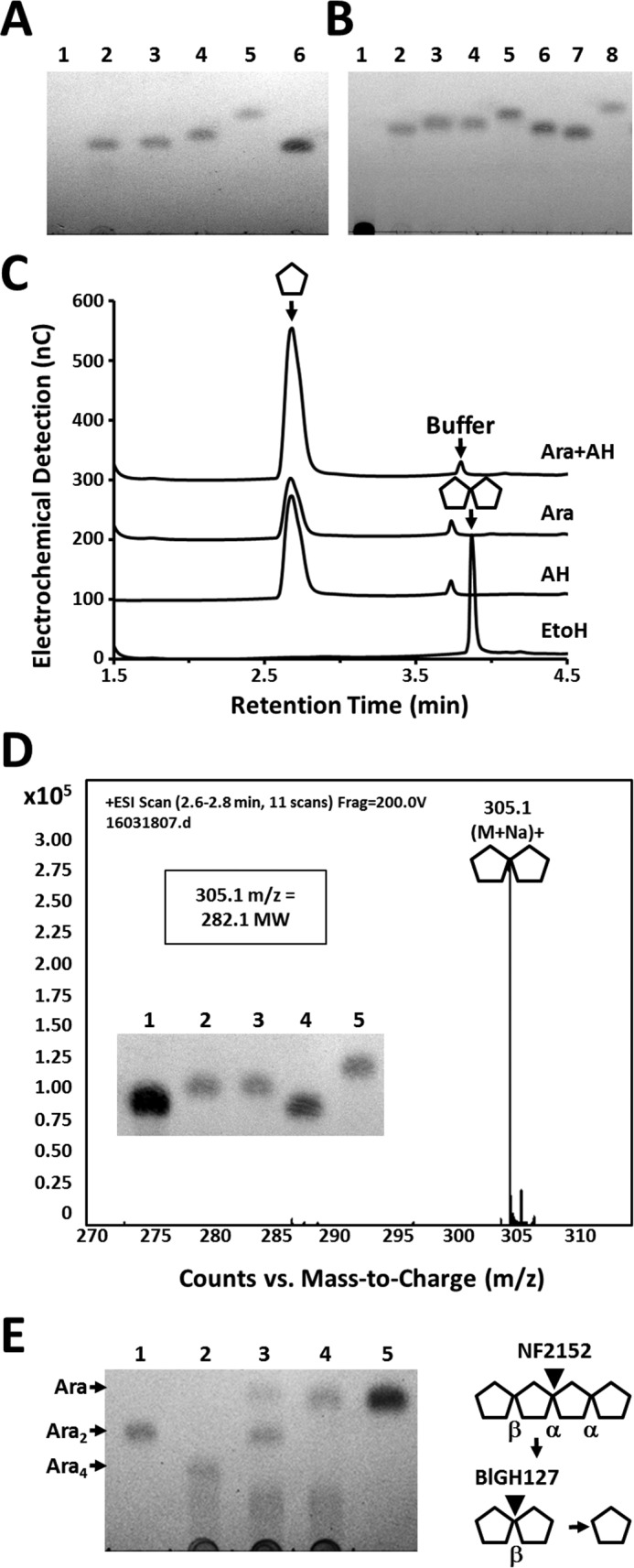
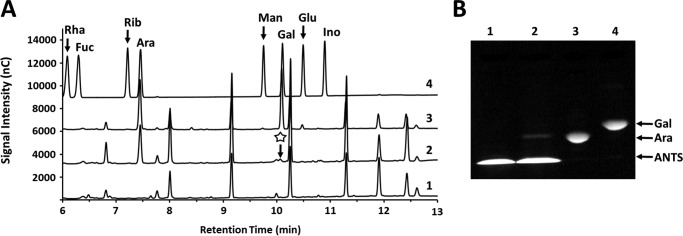
Structural analysis of the NF2152 product released from SBA. A, TLC analysis. Lane 1 = SBA; lane 2 = product before SEC purification; lane 3 = product after SEC purification; lane 4 = Ara; lane 5 = xylose; lane 6 = Gal. B, TLC of acid hydrolyzed P2 bio-gel column purified products. Lane 1 = SBA; lane 2 = product before acid hydrolysis; lane 3 = product after acid hydrolysis purification; lane 4 = Ara; lane 5 = α-1,5-Ara2; lane 6 = α-1,5-Ara3; lane 7 = Gal; and lane 8 = xylose. C, overlaid HPAEC-PAD chromatograms showing acid hydrolyzed products released from SBA. D, HR-MS spectrum displaying measured 305.1 m/z ratio and calculated mass of the Ara2 product (282.1). Inset: GH127 digestion of Ara product. Lane 1 = NF2152 product; lane 2 = GH127 digest; lane 3 = Ara; lane 4 = Gal; and lane 5 = α-1,5-Ara2. E, TLC of arabinotetraose derived from SBA (Ara4) digestion. Lane 1 = β-1,2-Ara2 product; lane 2 = Ara4 substrate; lane 3 = Ara4 digested with NF2152; lane 4 = NF2152 digestion treated with BlGH127; and lane 5 = arabinose. Right, schematic of sequential digestion.
Figure 3.
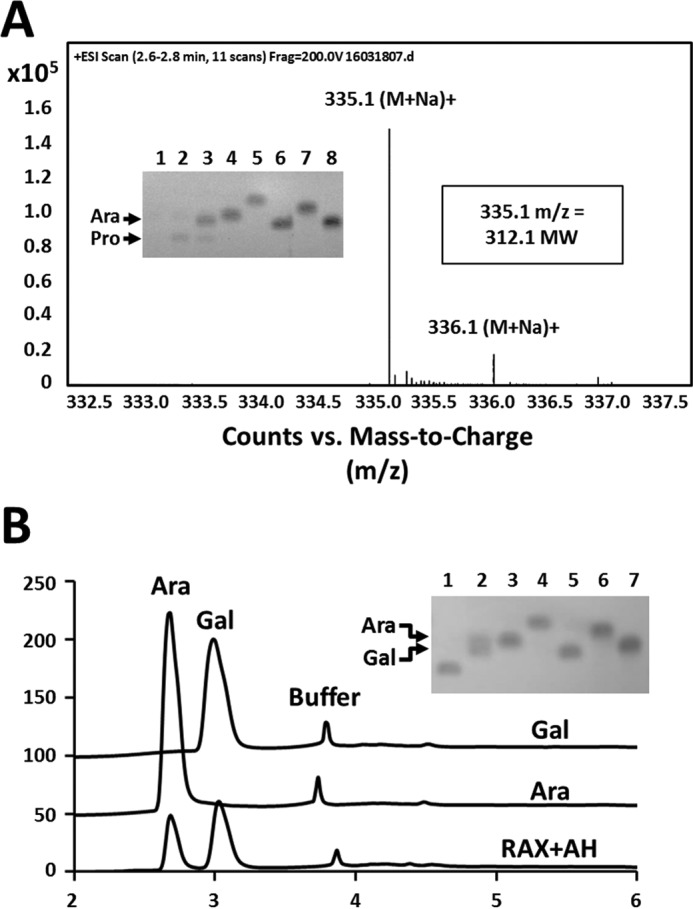
Structural analysis of the NF2523 product released from RAX. A, HR-MS spectrum of the Bio-Gel P-2 column purified NF2523 product released from RAX displaying measured 335.1 m/z ratio and calculated mass of hexose-pentose product (312.1). Inset: TLC displaying products from RAX digestions. Lane 1 = RAX; lane 2 = NF2523 digestion; lane 3 = NF2152 digestion; lane 4 = Ara; lane 5 = xylose; lane 6 = Gal; lane 7 = α-1,5-Ara2; and lane 8 = α-1,5-Ara3. Two major products are labeled. B, overlaid HPAEC-PAD chromatograms of Gal and Ara monosaccharides released by acid hydrolysis of the NF2523 product. Inset: TLC of the acid hydrolyzed NF2523 product. Lane 1 = purified product; lane 2 = acid hydrolysate; lane 3 = Ara; lane 4 = xylose; lane 5 = Gal; lane 6 = α-1,5-Ara2; lane 7 = α-1,5-Ara3.
Chemical structure of the SBA product
To generate sufficient product for chemical characterization, a large-scale digestion of SBA was performed. Following ethanol precipitation, the soluble products were fractionated by size exclusion chromatography. Eluted fractions were screened by TLC to identify peak boundaries, and samples containing similar sized products were pooled and analyzed for purity by TLC (Fig. 2A).
To determine whether the NF2152 product had a degree of polymerization (DP) > 1, acid hydrolysis was performed. The hydrolyzed product migrated as a single band with a similar mobility to Ara on TLC (Fig. 2B) and eluted with an identical retention time as Ara when analyzed by high performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) (Fig. 2C). These results suggested that NF2152 generates an oligosaccharide that is solely composed of Ara.
Comparison with a commercially available α-1,5-Ara2 standard revealed that these arabinooligosaccharides displayed different mobility patterns in TLC plates (Fig. 2B). Additionally, a selection of GH51 and GH43 α-ABFs were unable to hydrolyze the SBA product (results not shown). This highlighted that the product has a differing chemistry, such as a modification, alternate stereochemistry (i.e. β), or positional (e.g. 1 → 2) linkage. Due to the limited availability of commercial standards, a β-1,2-arabinofuranosidase GH127 from Bifidobacterium longum (BlGH127) (25) was synthesized, purified, and used for enzymatic glycosequencing. Previously this enzyme was used to elucidate the structure of a β-1,2-arabinosyl oligosaccharide derived from an AGP glycan (25). Treatment with BlGH127 cleaved the NF2152 product completely into Ara (Fig. 2D, inset), establishing that the product is a pure oligosaccharide with a β-1,2-linkage. However, this digestion does not determine the DP of the product as the GH127 may act processively on a substrate with DP ≥ 2. Therefore, to determine the size of the arabinooligosaccharide, electrospray ionization mass spectrometry was performed (Fig. 2D). The product was determined to have a m/z ratio ((M + Na)+) of 305.1. This ratio equates to a calculated mass of 282.1, which is the absolute mass of a pentose disaccharide, and confirms that the product is a pure β-1,2-Ara2. Additionally, the product released from an arabinan-derived tetrasaccharide was characterized (Fig. 2E). Enzymatic fingerprinting showed that this arabinotetraose contains a single β-l-arabinofuranosyl linkage and three α-l-arabinofuranosyl linkages. Digestion with NF2152 releases a disaccharide product with similar mobility to the product released from SBA, and likewise, this product was cleaved to arabinose by the β-1,2-l-arabinosidase, BlGH127 (Fig. 2E). These results indicate that NF2152 must hydrolyze an α-l-arabinofuranosyl linkage and is thus is an α-l-(β-1,2)-arabinobiosidase.
Characterization of a chemically distinct product from RAX
Digestion of RAX with NF2152 generated a second band with a slower mobility, which appeared to be the primary product of a second GH39 enzyme, NF2523 (Fig. 3A). The release of two structurally distinct products by NF2152; in addition to the unique product profile of NF2523, which only released one of these two products from RAX, highlighted that there are different substrates present in RAX and that this subfamily of fungal GH39 enzymes is polyspecific with specialized variants. Notably, very little product was detected when SBA was digested with NF2523. To determine whether the second product was an arabinooligosaccharide with a DP > 2, mass spectrometry was performed. Somewhat surprisingly, the m/z ratio of the product was ((M + Na)+) of 335.1 and its calculated mass was determined to be 312.1, which is equivalent to the molecular weight of a hexose-pentose disaccharide. Therefore, acid hydrolysis was performed and the products where analyzed by TLC and HPAEC-PAD (Fig. 3B). Consistent with the MS result, two distinct monosaccharides were detected: Ara and galactose (Gal). Ara is a main compositional sugar of SBA (88%) and RAX (Ara 38%); whereas, Gal is less common (SBA = 3%; RAX = none detected). In this regard, the presence of an Ara/Gal disaccharide suggests that it is a rare structure within both polysaccharides. Alternatively, Ara and Gal are commonly linked sugars in the arabinogalactan side chains of RG-I (26). However, digests of diverse pectic substrates with GH39 enzymes did not produce any detectable products. This suggests that NF2152 and NF2523 may be targeting rare substructures within SBA or AGPs glycans that co-purify with plant cell wall polysaccharides (25).
Sequencing of the Gal/Ara product
To determine whether Ara or Gal was positioned at the reducing end of the disaccharide, two complementary techniques were used: differential gas chromatography-mass spectrometry (GC-MS) and fluorophore-assisted carbohydrate electrophoresis (FACE). First, alditol acetates were generated from intact and hydrolyzed forms of the Gal/Ara disaccharide, and then visualized by GC-MS (Fig. 4A). This technique acetylates the free reducing end; hence, the residue at the non-reducing end is protected until it is released by acid hydrolysis. In this manner, only the reducing sugar will be visible in the GC-MS chromatogram. In Fig. 4A, Ara was present before and after acid hydrolysis; however, Gal was only visible when derivatized after hydrolysis, indicating it was protected when the disaccharide is intact. This analysis revealed the product as Gal-Ara (i.e. Gal = non-reducing; Ara = reducing end), a result that was confirmed by labeling the reducing end of the disaccharide with the fluorogenic compound 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS). Following derivatization, the disaccharide was hydrolyzed into monosaccharides and analyzed by FACE (Fig. 4B). Visualization of the products revealed that only the Ara was labeled, and therefore, exposed at the reducing end of the intact disaccharide.
Figure 4.
Sequencing of the heterogeneous disaccharide released from RAX by differential labeling of the reducing end. A, GC-MS chromatograms of the RAX product. The background reaction buffer is shown in 1. The disaccharide was reduced before (2) and after (3) acid hydrolysis, and then acetylated. Only Ara is visible in the non-hydrolyzed sample (the missing Gal peak is indicated with a star); whereas, both Ara and Gal are visible when the monosaccharides are generated before reduction and acetylation. Monosaccharide standards are shown in 4 and are labeled. B, FACE analysis of the ANTS-labeled disaccharide. The NF2523 product was labeled and then acid hydrolyzed. The products were compared with various standards. Lane 1 = ANTS; lane 2 = ANTS-labeled product hydrolysate; lane 3 = ANTS-labeled Ara; lane 4 = ANTS-labeled Gal.
NMR analysis of the reducing end absolute configuration and linkage
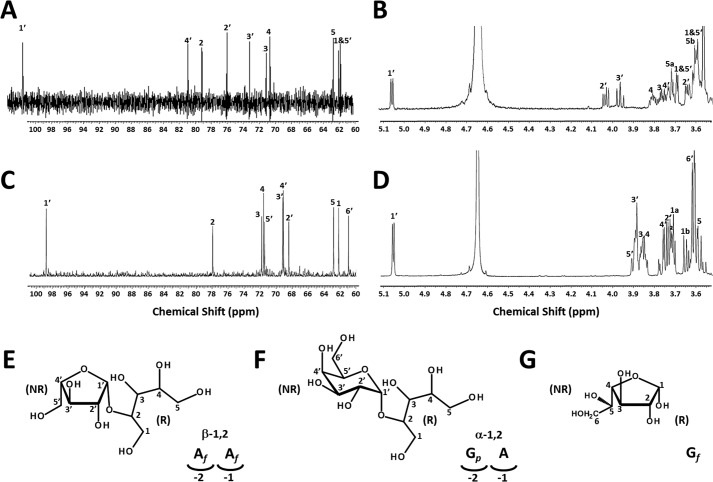
To provide further insights into the structure of the two products, both were analyzed by 1H and 13C nuclear magnetic resonance (NMR) (Fig. 5, A–D, Table 1). Both samples generated background peaks resulting from ring dynamics that obscured their interpretation; therefore, they were reduced to alditols with sodium borohydride. The product released from SBA by NF2152 was determined to be β-arabinofuranose-(1–2)-arabitol (Fig. 5E). The chemical shifts and coupling constants of its non-reducing monosaccharide suggested a β-arabinofuranose (27). The arabitol portion and the linkage were confirmed by two-dimensional NMR spectra including correlation spectroscopy (COSY), heteronuclear single quantum correlation (HSQC), and heteronuclear multiple bond correlation (HMBC). The full structure was confirmed by two-dimensional NMR.
Figure 5.
NMR analysis and structural determination of the reduced NF2152 and NF2523 products. A, 1H NMR and B, 13C NMR spectra of the purified NF2152 product. C, 1H NMR and D, 13C NMR spectra of the purified NF2523 product. E, structure of β-1,2-Ara2. Lower right: schematic representation of the β-1,2-Ara2 positioning within the active site of NF2152. F, structure of α-1,2-Gal-Ara. Lower right: schematic representation of Gal-Ara positioning within the active site of NF2523. G, structure of α-galactofuranose.
Table 1.
NMR 13C and 1H chemical shifts and coupling constants (D2O, 25 °C)
| No. | Chemical shifts |
Coupling constants | |
|---|---|---|---|
| 13C | 1H | ||
| ppm | Hz | ||
| NF2152 sample from SBA | |||
| 1 | 61.897 or 62.132 | a:3.696; b:3.589 | m |
| 2 | 79.274 | 3.630 | dd (6.5; 3.5) |
| 3 | 71.208 | 3.758 | m |
| 4 | 70.730 | 3.798 | m |
| 5 | 62.807 | a:3.596; b:3.708 | m |
| 1′ | 101.759 | 5.049 | d (5.0) |
| 2′ | 76.148 | 4.022 | dd (5.0; 8.0) |
| 3′ | 73.310 | 3.954 | t (8.0; 8.0) |
| 4′ | 81.027 | 3.732 | dt (8.0; 3.0; 3.0) |
| 5′ | 61.897 or 62.132 | a:3.696; b:3.589 | m |
| NF2523 sample from RAX | |||
| 1 | 62.200 | a: 3.706 | m |
| b: 3.649 | |||
| 2 | 77.946 | 3.714 | m |
| 3 | 71.815 | 3.846 ÷ 3.862 | m |
| 4 | 71.572 | 3.846 ÷ 3.862 | m |
| 5 | 62.830 | 3.588 ÷ 3.616 | m |
| 1′ | 98.739 | 5.051 | d (4.0) |
| 2′ | 68.430 | 3.732 | m |
| 3′ | 69.174 | 3.881 | m |
| 4′ | 69.106 | 3.744 | m |
| 5′ | 71.466 | 3.888 | m |
| 6′ | 60.971 | 3.602 ÷ 3.616 | m |
The reduced RAX product generated 1H and 13C chemical shifts and a coupling constant of the anomeric proton (4.0 Hz) of its non-reducing monosaccharide that matched those of α-galactopyranose (28, 29). The NMR data for the arabitol portion are identical with those observed for the SBA product. The reduced RAX product generated by NF2523, therefore, was identified as α-galactopyranose-(1–2)-arabitol (Fig. 5F). This ring configuration of Gal differs substantially from the furanose observed in the SBA product, which is structurally similar to galactofuranose (Fig. 5G).
Three-dimensional structure of NF2152
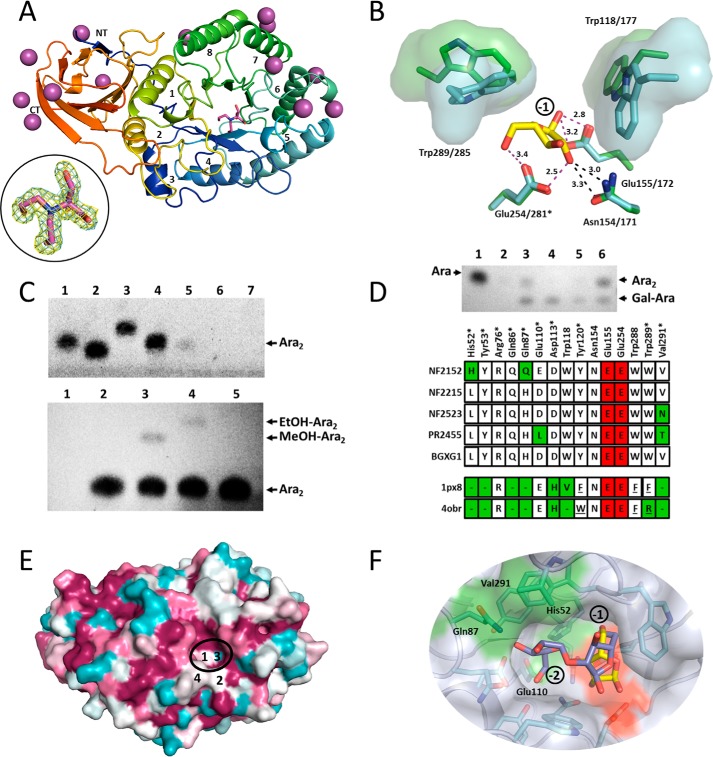
To provide further insight into the molecular basis of substrate recognition in the −1 and 2 subsites we solved the structure of NF2152. The de novo structure of NF2152 was determined to a resolution of 1.75 Å by single wavelength anomalous dispersion phasing using an iodide derivative (n = 13) (Table 2). The protein adopted the canonical GH-A clan (β/α)8 TIM-barrel-fold with a fused, hybrid β-sandwich spanning residues Met29-Asp36 and Thr321-Ala431 (Fig. 6A). The N-terminal strand formed at one end of the β-sandwich and created a “closed” bimodular domain. DaliLite v.3 (30) searches revealed that NF2152 had the highest levels of structural similarity with GH39 (PDB code 4EKJ; Zvalue = 25.1; matched Cα = 133; r.m.s. deviation = 4.2 Å2 (31)); GH51 (PDB code 2VRQ; Zvalue = 25.8; matched Cα = 154; r.m.s. deviation = 4.2 Å2 (32)); and GH30 (PDB code 4QAW, Zvalue = 25.8, matched Cα = 201; r.m.s. deviation = 4.2 Å2; (33)) families.
Table 2.
NF2152 data collection and structure statistics
| Statistics | |
|---|---|
| Data collection | |
| Beamline | Rigaku MicroMax-007HF |
| Wavelength (Å) | 1.54178 |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 36.99, 104.79, 49.45 |
| α, β, γ (°) | 90.00, 110.06, 90.00 |
| Resolution (Å)a | 28.95–1.75 (1.795–1.75) |
| Rmerge | 0.104 (0.381) |
| Wilson B-factor | 19.11 |
| 〈Ι/σΙ〉 | 12.3 (2.8) |
| Completeness (%) | 99.5 (98.8) |
| Redundancy | 4.8 (3.1) |
| No. of reflections | 170,864 |
| No. unique | 35,446 |
| Refinement | |
| Resolution (Å) | 1.75 |
| Rwork/Rfree | 0.156/0.194 |
| No. of atoms | |
| Protein | 3183 |
| Ligand | 13 (I), 12 (EDO), 13 (BTB) |
| Water | 314 |
| B-factors | |
| Protein | 18.56 |
| Ligand | 50.06 (I), 22.87 (EDO), 20.89 (BTB) |
| Water | 25.98 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.861 |
| Ramachandran (%) | |
| Preferred | 96.3 |
| Allowed | 3.7 |
| Disallowed | 0.0 |
| PDB code | 5U22 |
a Values for highest resolution shells are shown in parentheses.
Figure 6.
Structural basis of rumen fungal GH39 enzyme specificity. A, X-ray structure of NF2152 (PDB code 5U22) displayed in schematic format with a rainbow color scheme (NT = blue; CT = red). Each α/β pair are labeled 1 through 8. The 13 iodide atoms are shown as purple spheres and the bis-tris methane in magenta sticks. Inset: bound bis-tris methane represented as maximum-likelihood (69)/σa (35) weighted 2Fobs − 2Fcalc electron density map contoured at 1.0σ (0.43 e−/Å3; cyan), and Fobs − Fc map contoured at 3.0σ (0.42 e−/Å3; yellow). B, active site of NF2152 (cyan) superimposed with Ara (yellow sticks) from TmGH51 (PDB code 3UG4; green). Residues in contact with Ara in the −1 position are labeled. Distances of potential hydrogen bonds are indicated with magenta dashed lines and the distances are labeled. Steric clash of the O2 with Asn154 are indicated with black dashed lines. Tryptophans at the mouth of the active site pocket are displayed as sticks with a solvent accessible surface. Labeling system is NF2152/TmGH51. C, validation of catalytic residues and transglycosylation. Top panel, TLC of site-directed mutagenesis. All digests were performed on SBA. Lane 1 = Ara; lane 2 = Gal; lane 3 = α-1,5-Ara2; lane 4 = α-1,5-Ara3; lane 5 = NF2152; lane 6 = NF2152-E155Q; lane 7 = NF2152-E254Q. Bottom panel: TLC of primary transglycosylation reaction with NF2152 using β-1,2-arabinobiose as a substrate. Lane 1 = no product; lane 2 = water; lane 3 = MeOH; lane 4 = EtOH; lane 5 = Ara2 standard. D, polyspecificity of GH39 enzymes. Top panel, TLC of digests with GH39 enzymes. Lane 1 = Ara; lane 2 = no enzyme; lane 3 = NF2215; lane 4 = NF2523; lane 5 = PR2455; lane 6 = NF2152. All digests were performed on RAX and visualized by TLC. Bottom panel, alignment of amino acids from NF2152, NF2215, NF2523, PR2455, and Bgxg1 with GH39 structures (1px8, β-d-xylose complex; 4obr, α-l-iduronic acid complex). Residues include those that line the surface of the −1 and −2 subsites of NF2152. * indicates residues comprising the −2 subsite. Green, divergent residues; red, catalytic residues; dash, not conserved; underlined, structurally conserved, but contributed from a different loop. E, ConSurf model of GH39 fungal homologs (n = 12) mapped on the structure of NF2152. Magenta = conserved; cyan = variable. Four key residues are labeled: 1 = His52; 2 = Gln87; 3 = Glu110; and 4 = Val291. The −2 subsite in the active site cleft is highlighted with a black ellipse. F, transparent surface model of NF2152 docked with a built α-l-(β-1,2)-Ara2 ligand (purple sticks) overlaid with the superimposed Ara (yellow sticks) product from A. Residues of the active site pocket are shown as cyan sticks. The −1 and −2 subsites are labeled. The catalytic residue surfaces are shown in red, and divergent residues in the −2 subsite are labeled and shown with green surfaces.
Inspection of the putative active site of NF2152 revealed electron density consistent with the presence of a molecule of bis-tris methane (Fig. 6A, inset). This molecule was a component of the crystallization solution and proved to be essential for the reproducible generation of high quality crystals. The bis-tris methane is tightly sandwiched between Trp118 and Trp289, and forms several direct and water-mediated H-bonds. Despite multiple attempts to soak and co-crystallize NF2152 with β-1,2-Ara2 we were unable to dislodge the buffer molecule from the active site, and other conditions failed to generate diffraction quality crystals. Therefore, to investigate the potential interactions between NF2152 and Ara in the −1 subsite we performed a structural superimposition with α-l-Ara bound in the active site pocket of the GH51 ABF from Thermotoga maritima (TmGH51; PDB code 3UG4, Fig. 6B (34)). GH51s are retaining enzymes with experimentally determined catalytic residues. In TmGH51, the nucleophile is Glu281 and the acid/base is Glu172 (34), which overlay with Glu254 and Glu155, respectively, in NF2152. Both of these residues are in reasonable proximity to the C1-OH of the Ara present in the overlaid TmGH51 structure to facilitate hydrolysis. To confirm catalytic function, Glu254 and Glu155 were mutated to glutamine residues and tested for activity on SBA. Mutations to these residues resulted in the complete loss of detectable product (Fig. 6C, top panel). This suggested Glu155 and Glu254 are catalytic residues, whereas their spatial arrangement, indicated that NF2152 harnesses a retaining mechanism, consistent with other GH39s and GH51s. Previously, an unrelated retaining β-1,2-arabinobiosidase, BlGH121 was able to transglycosylate β-1,2-Ara2 in the presence of primary alcohols (27), which is indicative of retaining mechanisms. When incubated with purified β-1,2-Ara2 and MeOH or EtOH, NF2152 also generated products with shifts in mobility, which is consistent with transfer of the Ara2, through a double displacement mechanism (Fig. 6C).
Although the core catalytic residues are spatially conserved between NF2152 and TmGH51, closer inspection of the C2-OH of the Ara in the −1 subsite suggested that in this orientation the Ara is poised to form an interaction with Asn154/171. This interaction provides a recognition determinant exo-acting ABFs, such as TmGH51, which only removes single Ara decorations (34). In NF2152 this interaction with Asn154 would preclude a conjugated β-1,2-Ara or α-1,2-Gal from extending deeper into the pocket. Therefore, in this subfamily of anaerobic fungal GH39 enzymes it is probable that the Ara in the −1 adopts a different orientation. In support of this, the two tryptophan residues positioned at the mouth to the active site cleft adopt strikingly different conformations in NF2152 and TmGH51 (Fig. 6B).
Blast searches of NF2152 determined that there are other related entries in the database, including bacterial entries from Clostridia spp. and Fibrobacter spp., suggesting that NF2152 and NF2523 may be representative members of a larger subfamily. To investigate the functional diversity of other fungal GH39s, digestions were performed on RAX with NF2215 and PR2455. These results demonstrated that they each have subtle differences in their product profiles when compared with NF2152 and NF2523 (Fig. 6D). These signature patterns occur despite strict conservation of residues lining the active site in the −1 and −2 subsites, which are predicted to be involved in substrate binding and catalysis (Fig. 6D). Four residue positions that displayed variations in primary structure were identified, including His52, Gln87, Glu110, and Val291. The conservation of these residues within the larger group of anaerobic fungal GH39 homologs was assessed by performing a cluster analysis with 12 sequences with Clustal Omega (36), and then mapped onto the surface of NF2152 using ConSurf (37) (Fig. 6E). Analysis of the active site confirmed that His52, Asn87, and Val291 (white) and Glu110 (cyan) represent potential hot spot sites for variation within the −2 subsite (Fig. 6E). In particular, the transition from Val291 in NF2152 to an Asn in NF2523 and Thr in PR2455 may illuminate key molecular determinants for recognition of Gal at the non-reducing end α-1,2-Gal-Ara product. An β-1,2-Ara2 with an α-l-Ara at the reducing end could be reasonably docked into the active site of NF2152, demonstrating that there is sufficient room to accommodate the product. This interaction would require a ∼20° clockwise rotation of the Ara in the −1 subsite. Importantly, His52 (3.7 Å from the O4), Gln87 (3.2 and 3.0 Å from the C2-OH and C3-OH, respectively), and Glu110 (3.8 Å from the C5-OH) are disposed in reasonable proximity to interact with the side chains of Ara in the −2 subsite (Fig. 6F). In addition, Val291 (3.8 Å from the C2-OH) may contribute to van der Waals interactions.
Discussion
Discovery of new enzyme activities that improve the digestibility of recalcitrant plant cell wall polysaccharides are promising solutions for clean, sustainable industries. Additionally, biocatalysts with novel activities that operate as surgical tools hold promise for dissecting the structures of complex glycans or generating rare or commercially valuable carbohydrates. In this regard, the rumen anaerobic fungi represent an underexplored source of potentially unique enzymes. Here we have investigated the activities of GH39 enzymes from rumen fungi that were identified by their association with CBM13s.
Most commonly, CBM binding specificity parallels the catalytic activity of the appended enzyme module, and therefore, investigating uncharacterized enzymes associated with CBMs with diverse specificities should facilitate the discovery of new CAZyme families, and potentially, activities. Using this rationale, hypothetical proteins appended to predicted CBM13s (19) were identified within fungal transcriptomes from N. frontalis and P. rhizinflatus. These sequences were trimmed to an N-terminal fragment of sufficient length to encode a functional enzyme, and embedded into extracted datasets of characterized GH39 and GH51 sequences to generate “informed” phylogenetic trees. GH39 and GH51 are well characterized polyspecific enzyme families with activity on a wide range of substrates (14). In both trees the fungal sequences partition as a single clade, distantly related to bacterial β-xylosidases and ABFs, in GH39 and GH51, respectively (Fig. 1). These phylogenetic relationships suggest that GH39 enzymes may have diverged following horizontal gene transfer from a bacterial ancestor; and based upon sequence, they may be active in the hydrolysis of either α-l-arabinofuranosyl or β-xylosyl substrates. However, with the contemporary perspective that many CAZyme families are polyspecific (15, 16), including GH39 and GH51, it has become apparent that accurate assignment of enzyme activity cannot rely solely on phylogenetic patterns and requires evidence-based biochemical characterization.
To investigate the function of these fungal GH39s, a synthetic form of NF2152 was produced and screened for activity on a variety of plant cell wall carbohydrates. NF2152 was shown to release an Ara2 product from SBA (Fig. 2) and RAX (Fig. 3A). The release of similar products from two substrates isolated from different plant sources that display different structures suggests that there is a rare substrate consistent within both sources of plant polysaccharides.
SBA is an Ara-rich glycan found within the side chains of RG-I. It is composed of a α-1,5-l-arabinofuranosyl backbone decorated with α-1,2- and α-1,3-l-arabinofuranosyl side chains. Ara is an abundant pentose in nature; and therefore, discovery of novel biocatalysts active on arabinans are promising tools for bioconversion industries. There are many enzymes known to be active on SBA. Collectively these enzymes are referred to as arabinanases (ABNs), which are found in GH43 and GH93 families (38); and ABFs, which are found in GH3, GH43, GH51, GH54, GH62, GH93, and GH127 families (25, 27, 39). ABNs catalyze the hydrolysis of the α-1,5-l-arabinofuranosyl backbone of plant cell wall arabinans, releasing arabinooligosaccharides and Ara. ABFs cleave arabinosyl decorations in an exo-fashion. Currently, several α-1,5-arabinobiosidases have been reported that processively hydrolyze the backbone of arabinan (40). Recent studies have shown that the synergistic effect of fungal enzyme mixtures supplemented with ABNs and ABFs improves the hydrolysis rate of plant biomass (41).
Arabinoxylans are an abundant polysaccharide within the plant cell wall (42). RAX possesses a d-xylosyl backbone decorated with α-1,2- and α-1,3-linked l-arabinosyl residues (42, 43). Digestion of RAX requires a combination of enzyme activities, including ABFs and xylanases, which are found mainly in GH10, GH11, and also in GH5, GH7, GH8, and GH43 (44). Multiple enzyme products that target the xylan backbone of arabinoxylan products are currently available indicating there is a market for biocatalysts that improve the digestion of RAX.
Using acid hydrolysis (Fig. 2, B and C), and diagnostic enzyme digests and HR-MS (Fig. 2, D and E), and diagnostic digestions of a purified arabinotetraose (Fig. 2E), the product released by NF2152 was determined to be β-1,2-l-Araf2. This underpinned that the target substrate is a rare, and potentially trace contaminant within SBA and RAX. Additionally, it reveals that NF2152 is not a conventional ABF or xylosidase as was suggested by phylogenetic analysis (Fig. 1). Recently, BlGH121 was reported to be the first described β-1,2-arabinobiosidase, which is active on AGP glycans that co-purify with SBA (27). Importantly, BlGH121 products are also hydrolyzed by BlGH127 (25). This suggests that GH39 enzymes, found in anaerobic fungi, and BlGH121, a family of bacterial enzymes, may have undergone functional convergence. To the best of our knowledge this small collection of enzymes represents some of the only known enzymes specific for the degradation of β-1,2-arabinofuranoside containing glycans.
The detection of a second product with lower mobility was observed when RAX was digested with NF2152 and NF2523 (Fig. 2 and 3). The composition of this heterogeneous disaccharide was determined to be Gal and Ara (Fig. 3B). To provide more insight into the product chemistry, the disaccharide was sequenced using differential alditol acetate and ANTS labeling, and analyzed by GC-MS (Fig. 4A) and FACE (Fig. 4B), respectively. Both techniques confirmed that Ara was positioned at the reducing end; and therefore, the product is a Gal-Ara disaccharide. Although NMR is the gold standard for these types of analyses, the development of differential labeling techniques can be a valuable approach for rapidly sequencing heterogeneous disaccharides if access to NMR infrastructure is lacking or product yields are limiting.
Preliminary NMR spectra of both products suffered from high backgrounds of acetate (reaction buffer) and multiple signals for the anomeric carbon, suggesting the carbohydrates were adopting several conformations. Therefore, large scale purifications of both disaccharides were performed and the products were reduced to arabitols to linearize the compounds. 1H NMR (Fig. 5, A and C) and 13C NMR (Fig. 5, B and D) analyses confirmed the order of sequence for both the Ara2 and Gal-Ara, and that both products contained β-1,2-linkages (Fig. 5, E and F). Additionally, at the non-reducing end Ara is in the furanose configuration (which is consistent with the BlGH127 digest, Fig. 2D), and Gal is in a pyranose configuration. This latter observation was somewhat surprising as we had anticipated the Gal residue may be in the α-d-galactofuranose configuration, which has an analogous ring structure to β-l-arabinofuranose and only differs by the presence of its C5-hydroxyl methyl group (Fig. 5G). This establishes that the NF2523 substrates are of plant origin, and not of fungal origin as galactofuranoses are common components of fungal cell walls (45). Furthermore, elucidation that Ara was positioned at the reducing end in both products indicates that there is a strict requirement for Ara in their −1 subsites and they display plasticity in their −2 subsites (Fig. 5, E and F).
The structure of NF2152 confirmed that it adopts the canonical TIM-barrel-fold of GH39s and share significant structural similarity with GH51s (Fig. 6A). The polyspecificity reported for these families, including ABFs, xylosidases, endoglucanases, and many others (Fig. 1) underscores the functional plasticity of this core-fold (14, 46). The position of the active site was highlighted by the presence of a bound bis-tris buffer molecule (Fig. 6A) and the superimposition of the Ara product from TmGH51 (Fig. 6B). Ara is orientated with its C1 exiting the pocket between two sequence conserved aromatics (i.e. Trp118 and Trp289), and within suitable distances to the structurally conserved catalytic residues, Glu155 and Glu254. The involvement of Glu155 and Glu254 in catalysis was confirmed by site-directed mutagenesis and their potential roles in a retaining mechanism confirmed by the transglycosylation of MeOH and EtOH (Fig. 6C). As these features are conserved in other enzymes that function as ABFs and xylosidases, the specificity of the fungal GH39 enzymes for β-1,2-linked Araf containing disaccharides is believed to result primarily from interactions within the −2 subsite (Fig. 6D). This observation is in contrast to Bgxg1, which was reported to hydrolyze monosaccharides from a variety of synthetic substrates and purified disaccharides (24). Mapping the distribution of conserved surface-exposed active site residues in fungal GH39 sequences onto the surface of NF2152 revealed four key residues, His52, Asn87, Glu110, and Val291, as being variable within the −2 subsite (Fig. 6E). Importantly, they are also in suitable proximity for potential interactions with the Araf at the reducing end of β-1,2-Araf2 (Fig. 6F). These residues, therefore, may represent genetic markers for directing the discovery of other GH39 enzymes with unique specificities.
Conclusion
Although many microbial ecosystems are currently being mined for beneficial enzyme activities (e.g. human distal gut) the rumen microbiota remains one of the most promising repositories of microbial enzymes active on plant cell wall polysaccharides (20). In this study, four rumen fungal genes, classified as GH39s, were selected for characterization based upon their expression and association with CBM13. CBM13s are a family with reported diversity in their binding specificities (19).
NF2152 is an α-l-(β-1,2)-arabinofuranobiosidase, and NF2523 a d-galacto-(α-1,2)-l-arabinosidase. To the best of our knowledge this is the first galacto-(α-1,2)-arabinosidase activity reported in the literature. Based upon its weak activity on the SBA oligosaccharide substrate (not shown) and the structural conservation of catalytic residues within its active site, NF2523 is most likely an α-(d-galacto-(α-1,2)-l-arabino)-sidase. Other related GH39 enzymes, NF2215 and PR2455, display subtle differences in the generation of both products but overall have similar product profiles to NF2152 and NF2523, respectively. Both β-1,2-Araf2 and α-1,2-Gal-Ara are unconventional substructures within SBA and RAX, and may also represent products released from AGP glycans that co-purify with the polysaccharides. β-1,2-Linked arabinosyl products were previously reported to be released from SBA during the characterization of the phylogenetically unrelated β-1,2-arabinobiosidase BlGH121 (27).
Very little is known about the role of AGPs in plant biology and biomass conversion, mainly due to their extensive structural complexity. Recently, it was reported that an AGP may interact directly with the pectin and hemicellulose, which challenges the traditional view that these polysaccharides are independent networks (47). In this regard, enzymes that target discreet linkages within the arabinan or AGP glycan network, such as the fungal GH39 enzymes studied here, represent tools for helping to characterize complex arabinosyl-containing glycans; and potentially, may assist in their turnover during bioconversion processes.
Experimental procedures
Phylogenetic trees
Characterized sequences from families GH51 and GH39 were extracted and then trimmed to functional modules (i.e. GH catalytic fragments) by dbCAN (48). Trimmed fragments were then merged with catalytic fragments of the fungal sequences, and aligned via multiple sequence comparison by log-expectation (49). Phylogenetic grouping was performed with FastTree 2 (50). ProtTest3 was used for the selection of the best-fit model used by FastTree (51). Finally, FigTree was used for tree creation (tree.bio.ed.ac.uk/software/figtree). 4
Gene synthesis and cloning
Sequences corresponding to the mature GH39 gene products (lacking signal peptides) of NF2152, NF2215, NF2523, and PR2455 (GenBank accession numbers: MF326649, MF326650, MF326651, and MF326652) were commercially synthesized (BioBasic Inc., Markham, Canada) codon optimized for expression in E. coli and cloned into the pET28a vector. For crystallization experiments the DNA sequence corresponding to residues 29–431 of NF2152 was PCR amplified and cloned into the NdeI and XhoI sites of pET28a to create pET28_nf2152_29–431. All constructs were confirmed by DNA sequencing (Eurofins Genomics, Louisville, KY).
Expression of synthetic genes and purification of recombinant proteins
The pET28a constructs were transformed into E. coli BL21 Star (DE3) for recombinant protein production. Cells were grown at 37 °C to an OD = 600 nm of 0.8–1.0 in LB broth containing kanamycin (50 μg ml−1). Cultures were cooled to 16 °C and gene expression was induced with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside overnight at 200 rpm. Overnight cultures were centrifuged at 6,500 × g for 10 min. Cells were resuspended in Binding Buffer (BB: 0.5 m NaCl, 20 mm Tris, pH 8.0) and lysed by sonication for 2 min of 1-s intervals of medium intensity sonic pulses at a power setting of 4.5 (Heat Systems Ultrasonics Model W-225 and probe). The cell lysate was clarified by centrifugation at 17,500 × g for 45 min and passed through a 0.45-μm filter. The filtrate was loaded onto a nickel-nitrilotriacetic acid column and purified by immobilized metal affinity chromatography. Recombinant protein was eluted via a stepwise gradient of imidazole (5, 10, 30, 100, 200, and 500 mm) in BB. Fractions containing significant amounts of protein were pooled and buffer exchanged into storage buffer (NF2152, 20 mm Tris-HCl, pH 8.0, 500 mm NaCl; NF2152_29–431, 20 mm NaPO4, pH 7.2, 100 mm NaCl; NF2215, NF2523, and PR2455, 20 mm Tris-HCl, pH 8.0, 150 mm NaCl). Following buffer exchange, samples were concentrated using a nitrogen-pressurized stirred ultrafiltration cell (Amicon) with a molecular mass cut-off of 5 kDa. Concentrated protein was filtered and passed through a HiPrep 16/60 Sephacryl S-200 HR size exclusion column (GE Healthcare) at a flow rate of 1.0 ml min−1 in storage buffer. Pure fractions were pooled and concentrated. NF2152 was further buffer exchanged into 20 mm Tris-HCl (pH 8.0), 100 mm NaCl. Protein concentration was determined using the Beer-Lambert law and extinction coefficients determined with ProtParam (52).
Enzyme characterization
The activity of NF2152 was screened on a variety of plant cell wall carbohydrates including: sugar beet arabinan (catalog number P-ARAB), wheat arabinoxylan (P-WAXYM), rye arabinoxylan (P-RAXY), β-glucan (P-BGBL), xyloglucan (P-XYGLN), galactomannan (P-GGM28), pectic galactan (P-PGAPT), arabinogalactan (P-ARGAL), rhamnogalacturonan-I (P-RHAM I), arabinofuranosyl-xylobiose (O-A3X), arabinofuranosyl-xylotriose (O-A2XX), PNP-α-l-arabinofuranoside (O-PNPAF) purchased from Megazyme International, PNP-α-l-arabinopyranoside (38018) from Glycosynth, gum arabic (G9752), and beech wood xylan (X4252) from Sigma. The enzyme concentrations, buffer, and pH were optimized for digestion through empirical studies with different substrates, enzyme concentrations, and buffers (pH values ranging from 4.0 to 8.0). The final reaction mixture contained 5 mg ml−1 of polysaccharide substrate or 1.5 mm oligosaccharide, 0.5 μm of each enzyme, and 20 mm sodium acetate buffer (pH 5.0). These reactions were incubated overnight at 37 °C with samples being removed at various time points. After incubation the samples were heat treated at 100 °C for 10 min to denature the enzyme and terminate the reaction followed by short centrifugation at 8,000 × g to pellet denatured protein from the product. Products were analyzed by TLC and HPAEC-PAD to characterize their chemistry and degree of polymerization.
Thin layer chromatography
Digested samples (total 9 μl; spotted 3 times with 3 μl each time) were spotted onto TLC plates (TLC Silica Gel 60; EMD Millipore Corp.). The samples were dried between multiple rounds of spotting. Appropriate standards (6 μl of 1 mm concentration) were also included in each run. The samples were resolved using a mobile phase of 2:1:1 butanol:water:acetic acid, dried prior to visualization with an orcinol solution (70:3, acetic acid:sulfuric acid with 1% orcinol) and heating at 100 °C for 3–5 min.
HPAEC-PAD of monosaccharide and oligosaccharide reaction products
HPAEC-PAD was performed with a Dionex ICS-3000 chromatography system (Thermo Scientific) equipped with an autosampler as well as a pulsed amperometric (PAD) detector. 10 μl of aqueous sample were injected onto an analytical (3 × 150 mm) CarboPac PA20 column (Thermo Scientific) and eluted at 0.4 ml min−1 flow rate with a sodium acetate gradient (0 to 1 min, 0 mm; 1 to 18 min, 250–850 mm; 18 to 20 min, 850 mm; 20 to 30 min, 850–0 mm) in a constant background of 100 mm NaOH. The elution was monitored with a PAD detector (standard quadratic waveform).
Ethanol precipitation
Precipitations were performed on digested products to increase their purity by separating small products (e.g. monosaccharides and disaccharides) from larger oligosaccharides and polysaccharides by their differential solubility. The digested products were dried by SpeedVac at ambient temperature or lyophilization, and then suspended in 95% ethanol (v/v, EtOH:H2O). After incubation on ice for 10 min the resuspension was mixed vigorously with a vortex to dissolve the small oligosaccharides and monosaccharides and then clarified by centrifugation at 14,000 × g for 10 min. The supernatant was removed and dried by SpeedVac. Purified carbohydrates were subsequently used directly or suspended in water at the desired concentrations for further analyses.
Acid hydrolysis
30 μg of purified dried products were incubated with 200 μl of 2.0 m trifluoroacetic acid for 4 h at 100 °C. The reaction mixture was then dried to completion in a SpeedVac followed by three washes with 100 μl of isopropyl alcohol. Released monosaccharides were analyzed by TLC and HPAEC-PAD.
Molecular weight determination by mass spectrometry
A large scale (total reaction volume: 200 ml) digest was performed to generate milligram amounts of product with NF2152 and SBA or NF2523 with RAX. The products were further purified by ethanol precipitation (95% v/v EtOH:H2O) and Bio-Gel P-2 (Bio-Rad Laboratories) size exclusion chromatography at a flow rate of 0.17 ml min−1 where distilled water was used as eluent. Extra fine (<45 μm) Bio-Gel P-2 has particle size beads with an exclusion limit of 100–1,800 Da. The elution peaks were screened for purity by TLC, pooled, and lyophilized. Mass spectrometry was carried out on the pooled fractions to determine the m/z and molecular weight of the product (Alberta Glycomics Centre, Department of Chemistry, University of Alberta, Edmonton, Canada). Electrospray ionization mass spectra were recorded on an Agilent Technologies 6220 TOF instrument. The sample was dissolved in methanol or a methanol:water mixture (1:1) and directly injected into the instrument (5 μl). The spectra were recorded in positive mode.
Carbohydrate sequencing by GC-MS
The purified product was used for gas chromatography to identify the product as well as to recognize the reducing end of the product. Gas chromatographic analysis of mono- and disaccharides requires conversion of sugars into their volatile derivatives (53). Sugars (purified product) were first converted into alditol acetates, which involved reduction of sugars with sodium borohydride (NaBH4) following conversion of polyols to polyacetate esters. Fifty micrograms of products were reduced by 200 μl of NaBH4 (10 mg/ml of NaBH4 in 1 m NH4OH). The reduced sugar was then acid hydrolyzed by 200 μl of 2 m TFA followed by an incubation at 100 °C for 4 h. The reaction mixture was then dried to completion in a SpeedVac followed by three wash steps with 100 μl of isopropyl alcohol. Released monosaccharides were acetylated by the addition of 250 μl of acetic anhydride and dried on a SpeedVac to a volume of 200 μl. The resulting solution was transferred to a GC autosampler vial containing a 250–300 micro insert and injected into gas chromatograph (Hewlett Packard 5890) where a polar capillary GC column (SP2330) and flame ionization detector was used.
Carbohydrate sequencing by FACE
FACE was performed to identify the reducing end of the digested products. Carbohydrates were fluorescently labeled at their reducing end using ANTS, and the resulting labeled sugars were separated in a high percentage (40%) polyacrylamide gel (54). 30 μg of each sample were dried and suspended by vortexing in 5 μl of fresh 0.15 m ANTS (dissolved in 15% acetic acid) and 5 μl of fresh 1 m 2-picoline borane (dissolve in 1 ml of DMSO). Samples were incubated overnight at 37 °C in a tube wrapped in foil. The labeled samples were then dried with a SpeedVac for 2–4 h or until completely dry. The labeled sugar was then acid hydrolyzed by the addition of 200 μl of 2 m TFA followed by incubation at 100 °C for 4 h. The reaction mixture was dried to completion in a SpeedVac followed by three wash steps with 100 μl of isopropyl alcohol. The dried pellet was suspended in 25 μl of FACE loading dye and run on gel immediately or stored at −20 °C wrapped in foil.
Linkage and ring configuration by NMR
Spectra were measured with a Varian VNMRS-500 MHz in D2O at 25 °C (Department of Chemistry and Biochemistry, Concordia University, Montreal, Canada). Chemical shifts and coupling constants were interpreted basing on one-dimensional NMR (1H and 13C) as well as two-dimensional NMR homonuclear correlation spectroscopy and heteronuclear single quantum correlation. The observation that the anomeric proton (proton 1′) had a coupling constant of 5.0 Hz was evidence for β-configuration of the Ara (55).
Protein crystallization NF2153 and structure solution
Purified NF2152_29–431 was buffer exchanged into 20 mm bis-tris (pH 6.0), and concentrated to 20 mg ml−1. Crystals of NF2152 were grown using the hanging drop vapor diffusion method at 18 °C in 20% (w/v) PEG 3350, 0.1 m bis-tris (pH 6.0), and 0.15 m NaI with a drop ratio of 1:2 protein solution to mother liquor. Crystals were cryoprotected with 25% (w/v) ethylene glycol and flash cooled directly in the cryostream at 100 K. Diffraction data were collected on an instrument comprising a MicroMax MM-007HF X-ray generator coupled to a Dectris Pilatus 200K detector with VariMax HF Arc Sec Confocal optics and an Oxford Cryostream Crystream 800 cryocooler. Data were processed and scaled with HKL-3000R (56). An initial model was determined by single-wavelength anomalous dispersion using SHELXC/D/E (57, 58). Phases were improved with Phaser (59), followed by successive rounds of density modification with Parrot (60) and model building with Buccaneer (61) using the CCP4 Phaser SAD pipeline (62). With an estimated solvent fraction of 35%, 13 iodide sites were identified. The resulting phases were sufficient for Arp/Warp to build a virtually complete model (63). This model was then used for iterative rounds of manual correction with COOT (64) and refinement with PHENIX (65). Water molecules were added with COOT FINDWATERS and manually checked after refinement. Throughout, refinement procedures were monitored by flagging 5% of all observation as “free” (66). Model validation was performed with MolProbity (67). Data collection and processing statistics are shown in Table 2.
Site-directed mutagenesis
Glu155 and Glu254 in NF2152 were predicted to be the acid/base and nucleophile, respectively, based upon superimposition with the GH51 from T. maritima (3UG4)(34). These residues were targeted for substitution to glutamine using site-directed mutagenesis with pET28_nf2152_29–431 for template as previously described (68). Mutations were confirmed by DNA sequencing (Eurofins Genomics). Once sequence confirmed, the mutant enzymes were tested for activity with appropriate substrates.
Transglycosylation assay
Size exclusion purified β1,2-Ara2 was incubated at a concentration of 1 mm with 1 μm NF2152 in 20 mm sodium acetate (pH 5.0) and 20% (v/v) methanol or ethanol. After overnight incubation at 37 °C the samples were heat treated at 100 °C for 10 min to terminate the reaction. Reaction products were visualized by TLC.
Docking of β-l-Araf-(1–2)-α-l-Araf with NF2153
PDB coordinates for a β-l-Araf-(1–2)-α-l-Araf molecule were generated with the GLYCAM Carbohydrate builder (www.glycam.org).4 AutoDock Vina was used to dock the Ara2 into the NF2152 active site pocket using a search area of 2,560 Å3 at an exhaustiveness setting of 8. The best fitting model had a predicted binding affinity of −6.5 kcal mol−1, with an upper and lower r.m.s. deviation bound of 2.7 and 1.3 Å, respectively.
Author contributions
D. R. J. performed enzyme characterization, product analysis, and solved the structure of NF2152; and assisted with figure generation and manuscript writing. M. S. U. performed enzyme screening, mutagenesis, and analysis of product structure; and assisted with writing and figure generation. R. J. G. generated fungal transcriptomes and gene sequences and assisted with protein crystallization. M. P. T. T. performed NMR experiments and determined structures of reduced SAB and RAX products. D. T. generated phylogenetic trees. A. B. B. and B. P. assisted with X-ray data collection and structure solution. J. B. generated the arabinotetraose. T. A. M. and R. F. helped design the study and identify target sequences. A. T. sequenced fungal transcripts, and helped in study design and NMR structure determination. L. B. S. helped in study design and interpretation of results. D. W. A. conceived of study, contributed to interpretation of the results, and wrote the initial draft of the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Professor Harry Gilbert (Newcastle University) for providing ABFs for diagnostic digestions of the SBA product that confirmed the products were α-arabinosides. In addition, we thank the Alberta Glycomics Centre for assistance with the HR-MS and Darrell Vedres (AAFC) with the GC-MS analysis.
This work was supported by Agriculture and Agri-Food Canada (AAFC) through Agriculture Innovation Program Grant AIP-P022 and Elanco Animal Health, and Natural Sciences and Engineering Research Council of Canada Discovery Grant FRN 04355 (to A. B. B.). The intellectual property surrounding the field of use for these enzymes has been protected with a United States provisional patent (No. 62/380,741 under AAFC's patent docket, PAT-13830). D. R. J., M. S. U., T. A. M., A. T., and D. W. A. are co-inventors on this patent.
The atomic coordinates and structure factors (code 5U22) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) MF326649, MF326650, MF326651, and MF326652.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- RAX
- rye arabinoxylan
- RG-I
- rhamnogalacturonan-I
- SBA
- sugar beet arabinan
- AGP
- arabinogalactan protein
- GH
- glycoside hydrolase
- CBM
- carbohydrate-binding module
- ABF
- α-l-arabinofuranosidase
- DP
- degree of polymerization
- HPAEC-PAD
- high performance anion-exchange chromatography with pulsed amperometric detection
- FACE
- fluorophore-assisted carbohydrate electrophoresis
- ANTS
- 8-aminonaphthalene-1,3,6-trisulfonic acid
- bis-tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PDB
- Protein Data Bank
- ABN
- arabinanases
- r.m.s.
- root mean square.
References
- 1. Morgavi D. P., Kelly W. J., Janssen P. H., and Attwood G. T. (2013) Rumen microbial (meta)genomics and its application to ruminant production. Animal 7, 184–201 [DOI] [PubMed] [Google Scholar]
- 2. Ribeiro G. O., Gruninger R., Badhan A., and McAllister T. A. (2016) Mining the rumen for fibrolytic feed enzymes. Animal Front. 6, 20–26 [Google Scholar]
- 3. Hess M., Sczyrba A., Egan R., Kim T. W., Chokhawala H., Schroth G., Luo S., Clark D. S., Chen F., Zhang T., Mackie R. I., Pennacchio L. A., Tringe S. G., Visel A., Woyke T., Wang Z., and Rubin E. M. (2011) Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331, 463–467 [DOI] [PubMed] [Google Scholar]
- 4. Qi M., Wang P., O'Toole N., Barboza P. S., Ungerfeld E., Leigh M. B., Selinger L. B., Butler G., Tsang A., McAllister T. A., and Forster R. J. (2011) Snapshot of the eukaryotic gene expression in muskoxen rumen: a metatranscriptomic approach. PloS One 6, e20521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dai X., Tian Y., Li J., Luo Y., Liu D., Zheng H., Wang J., Dong Z., Hu S., and Huang L. (2015) Metatranscriptomic analyses of plant cell wall polysaccharide degradation by microorganisms in the cow rumen. Appl. Environ. Microbiol. 81, 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Houston K., Tucker M. R., Chowdhury J., Shirley N., and Little A. (2016) The plant cell wall: a complex and dynamic structure as revealed by the responses of genes under stress conditions. Front. Plant Sci. 7, 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mohnen D., Bar-Peled M., and Somerville C. (2008) Cell wall polysaccharide synthesis. Biomass Recalcitrance: Deconstructing Plant Cell Wall Bioenergy, pp. 94–187, Blackwell Publishing, John Wiley and Sons, Hoboken, NJ [Google Scholar]
- 8. Bengtsson S., Åman P., and Andersson R. (1992) Structural studies on water-soluble arabinoxylans in rye grain using enzymatic hydrolysis. Carbohydr. Polymers 17, 277–284 [Google Scholar]
- 9. Jarvis M., Briggs S., and Knox J. (2003) Intercellular adhesion and cell separation in plants. Plant Cell Environ. 26, 977–989 [Google Scholar]
- 10. Atmodjo M. A., Hao Z., and Mohnen D. (2013) Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 64, 747–779 [DOI] [PubMed] [Google Scholar]
- 11. Ndeh D., Rogowski A., Cartmell A., Luis A. S., Baslé A., Gray J., Venditto I., Briggs J., Zhang X., Labourel A., Terrapon N., Buffetto F., Nepogodiev S., Xiao Y., Field R. A., et al. (2017) Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544, 65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones L., Milne J. L., Ashford D., and McQueen-Mason S. J. (2003) Cell wall arabinan is essential for guard cell function. Proc. Natl. Acad. Sci. U.S.A. 100, 11783–11788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Showalter A. M. (2001) Arabinogalactan-proteins: structure, expression and function. Cell. Mol. Life Sci. 58, 1399–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mewis K., Lenfant N., Lombard V., and Henrissat B. (2016) Dividing the large glycoside hydrolase family 43 into subfamilies: a motivation for detailed enzyme characterization. Appl. Environ. Microbiol. 82, 1686–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aspeborg H., Coutinho P. M., Wang Y., Brumer H. 3rd, and Henrissat B. (2012) Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 12, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davies G., and Henrissat B. (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859 [DOI] [PubMed] [Google Scholar]
- 18. Boraston A. B., Bolam D. N., Gilbert H. J., and Davies G. J. (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fujimoto Z. (2013) Structure and function of carbohydrate-binding module families 13 and 42 of glycoside hydrolases, comprising a beta-trefoil fold. Biosci. Biotechnol. Biochem. 77, 1363–1371 [DOI] [PubMed] [Google Scholar]
- 20. Solomon K. V., Haitjema C. H., Henske J. K., Gilmore S. P., Borges-Rivera D., Lipzen A., Brewer H. M., Purvine S. O., Wright A. T., Theodorou M. K., Grigoriev I. V., Regev A., Thompson D. A., and O'Malley M. A. (2016) Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science 351, 1192–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Badhan A., Wang Y., Gruninger R., Patton D., Powlowski J., Tsang A., and McAllister T. (2014) Formulation of enzyme blends to maximize the hydrolysis of alkaline peroxide pretreated alfalfa hay and barley straw by rumen enzymes and commercial cellulases. BMC Biotechnol. 14, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gruninger R. J., Puniya A. K., Callaghan T. M., Edwards J. E., Youssef N., Dagar S. S., Fliegerova K., Griffith G. W., Forster R., Tsang A., McAllister T., and Elshahed M. S. (2014) Anaerobic fungi (phylum Neocallimastigomycota): advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential. FEMS Microbiol. Ecol. 90, 1–17 [DOI] [PubMed] [Google Scholar]
- 23. Couger M. B., Youssef N. H., Struchtemeyer C. G., Liggenstoffer A. S., and Elshahed M. S. (2015) Transcriptomic analysis of lignocellulosic biomass degradation by the anaerobic fungal isolate Orpinomyces sp. strain C1A. Biotechnol. Biofuels 8, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morrison J. M., Elshahed M. S., and Youssef N. (2016) A multifunctional GH39 glycoside hydrolase from the anaerobic gut fungus Orpinomyces sp. strain C1A. Peer J. 4, e2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fujita K., Takashi Y., Obuchi E., Kitahara K., and Suganuma T. (2014) Characterization of a novel β-l-arabinofuranosidase in Bifidobacterium longum funtional elucidation of a DUF1680 protein family member. J. Biol. Chem. 289, 5240–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yapo B. M. (2011) Rhamnogalacturonan-I: a structurally puzzling and functionally versatile polysaccharide from plant cell walls and mucilages. Polymer Rev. 51, 391–413 [Google Scholar]
- 27. Fujita K., Sakamoto S., Ono Y., Wakao M., Suda Y., Kitahara K., and Suganuma T. (2011) Molecular cloning and characterization of a β-l-arabinobiosidase in Bifidobacterium longum that belongs to a novel glycoside hydrolase family. J. Biol. Chem. 286, 5143–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bock K., Pedersen C., and Pedersen H. (1984) Carbon-13 nuclear magnetic resonance data for oligosaccharides. Adv. Carbohydr. Chem. Biochem. 42, 193–225 [Google Scholar]
- 29. Duus J., Gotfredsen C. H., and Bock K. (2000) Carbohydrate structural determination by NMR spectroscopy: modern methods and limitations. Chem. Rev. 100, 4589–4614 [DOI] [PubMed] [Google Scholar]
- 30. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Santos C. R., Polo C. C., Corrêa J. M., Simão Rde C., Seixas F. A., and Murakami M. T. (2012) The accessory domain changes the accessibility and molecular topography of the catalytic interface in monomeric GH39 beta-xylosidases. Acta Crystallogr. D Biol. Crystallogr. 68, 1339–1345 [DOI] [PubMed] [Google Scholar]
- 32. Paës G., Skov L. K., O'Donohue M. J., Rémond C., Kastrup J. S., Gajhede M., and Mirza O. (2008) The structure of the complex between a branched pentasaccharide and Thermobacillus xylanilyticus GH-51 arabinofuranosidase reveals xylan-binding determinants and induced fit. Biochemistry 47, 7441–7451 [DOI] [PubMed] [Google Scholar]
- 33. Sainz-Polo M. A., Valenzuela S. V., González B., Pastor F. I., and Sanz-Aparicio J. (2014) Structural analysis of glucuronoxylan-specific Xyn30D and its attached CBM35 domain gives insights into the role of modularity in specificity. J. Biol. Chem. 289, 31088–31101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Im D. H., Kimura K., Hayasaka F., Tanaka T., Noguchi M., Kobayashi A., Shoda S., Miyazaki K., Wakagi T., and Fushinobu S. (2012) Crystal structures of glycoside hydrolase family 51 α-l-arabinofuranosidase from Thermotoga maritima. Biosci. Biotechnol. Biochem. 76, 423–428 [DOI] [PubMed] [Google Scholar]
- 35. Read R. J. (1986) Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallog. Sect. A 42, 140–149 [Google Scholar]
- 36. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Systems Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goldenberg O., Erez E., Nimrod G., and Ben-Tal N. (2009) The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 37, D323–D327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., and Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shi H., Ding H., Huang Y., Wang L., Zhang Y., Li X., and Wang F. (2014) Expression and characterization of a GH43 endo-arabinanase from Thermotoga thermarum. BMC Biotechnol. 14, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sakamoto T., and Thibault J.-F. (2001) Exo-arabinanase of Penicillium chrysogenum able to release arabinobiose from α-1,5-l-arabinan. Appl. Environ. Microbiol. 67, 3319–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Santos C. R., Polo C. C., Costa M. C., Nascimento A. F., Meza A. N., Cota J., Hoffmam Z. B., Honorato R. V., Oliveira P. S., Goldman G. H., et al. (2014) Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases. J. Biol. Chem. 289, 7362–7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vinkx C., and Delcour J. (1996) Rye (Secale cereale L.) Arabinoxylans: a critical review. J. Cereal Sci. 24, 1–14 [Google Scholar]
- 43. Vinkx C., Reynaert H., Grobet P., and Delcour J. (1993) Physicochemical and functional properties of rye nonstarch polysaccharides: V. variability in the structure of water-soluble arabinoxylans. Cereal Chem. 70, 311–311 [Google Scholar]
- 44. Collins T., Gerday C., and Feller G. (2005) Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 29, 3–23 [DOI] [PubMed] [Google Scholar]
- 45. Tefsen B., Ram A. F., van Die I., and Routier F. H. (2012) Galactofuranose in eukaryotes: aspects of biosynthesis and functional impact. Glycobiology 22, 456–469 [DOI] [PubMed] [Google Scholar]
- 46. Tóth-Petróczy A., and Tawfik D. S. (2014) The robustness and innovability of protein folds. Curr. Opin. Struct. Biol. 26, 131–138 [DOI] [PubMed] [Google Scholar]
- 47. Tan L., Eberhard S., Pattathil S., Warder C., Glushka J., Yuan C., Hao Z., Zhu X., Avci U., Miller J. S., Baldwin D., Pham C., Orlando R., Darvill A., Hahn M. G., Kieliszewski M. J., and Mohnen D. (2013) An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25, 270–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yin Y., Mao X., Yang J., Chen X., Mao F., and Xu Y. (2012) dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, W445–W451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Price M. N., Dehal P. S., and Arkin A. P. (2010) FastTree 2: approximately maximum-likelihood trees for large alignments. PloS One 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Darriba D., Taboada G. L., Doallo R., and Posada D. (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gasteiger E., Hoogland C., Gattiker A., Duvaud S. e., Wilkins M. R., Appel R. D., and Bairoch A. (2005) Protein identification and analysis tools on the ExPASy server. Methods Mol. Biol. 112, 531–552 [DOI] [PubMed] [Google Scholar]
- 53. Blakeney A. B., Harris P. J., Henry R. J., and Stone B. A. (1983) A simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr. Res. 113, 291–299 [Google Scholar]
- 54. Gao N. (2005) Fluorophore-assisted carbohydrate electrophoresis: a sensitive and accurate method for the direct analysis of dolichol pyrophosphate-linked oligosaccharides in cell cultures and tissues. Methods 35, 323–327 [DOI] [PubMed] [Google Scholar]
- 55. Joseleau J.-P., Chambat G., Vignon M., and Barnoud F. (1977) Chemical and 13C NMR studies of two arabinans from the inner bark of young stems of Rosa Glauca. Carbohydr. Res. 58, 165–175 [Google Scholar]
- 56. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 57. Sheldrick G. M. (2010) Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schneider T. R., and Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 59. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cowtan K. (2010) Recent developments in classical density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 62. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 63. Langer G., Cohen S. X., Lamzin V. S., and Perrakis A. (2008) Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nature Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 65. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brünger A. T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 67. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. McLean R., Hobbs J. K., Suits M. D., Tuomivaara S. T., Jones D. R., Boraston A. B., and Abbott D. W. (2015) Functional analyses of resurrected and contemporary enzymes illuminate an evolutionary path for the emergence of exolysis in polysaccharide lyase family 2. J. Biol. Chem. 290, 21231–21243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 70. Rambaut A. (2014) FigTree – Tree Figure Drawing Tool, Version 1.4.2, Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, United Kingdom [Google Scholar]
- 71. Woods Group (2017) Carbohydrate Builder – GLYCAM Web, Complex Carbohydrate Research Center, University of Georgia, Athens, GA [Google Scholar]