Abstract
Saliva of blood-feeding arthropods carries several antihemostatic compounds whose physiological role is to facilitate successful acquisition of blood. The identification of novel natural anticoagulants and the understanding of their mechanism of action may offer opportunities for designing new antithrombotics disrupting blood clotting. We report here an in-depth structural and functional analysis of the anophelin family member cE5, a salivary protein from the major African malaria vector Anopheles gambiae that specifically, tightly, and quickly binds and inhibits thrombin. Using calorimetry, functional assays, and complementary structural techniques, we show that the central region of the protein, encompassing amino acids Asp-31–Arg-62, is the region mainly responsible for α-thrombin binding and inhibition. As previously reported for the Anopheles albimanus orthologue anophelin, cE5 binds both thrombin exosite I with segment Glu-35–Asp-47 and the catalytic site with the region Pro-49–Arg-56, which includes the highly conserved DPGR tetrapeptide. Moreover, the N-terminal Ala-1–Ser-30 region of cE5 (which includes an RGD tripeptide) and the additional C-terminal serine-rich Asn-63–Glu-82 region (absent in orthologues from anophelines of the New World species A. albimanus and Anopheles darlingi) also played some functionally relevant role. Indeed, we observed decreased thrombin binding and inhibitory properties even when using the central cE5 fragment (Asp-31–Arg-62) alone. In summary, these results shed additional light on the mechanism of thrombin binding and inhibition by this family of salivary anticoagulants from anopheline mosquitoes.
Keywords: crystal structure, enzyme inhibitor, hemostasis, intrinsically disordered protein, protein-protein interaction, Anopheles, mosquito saliva
Introduction
The ability of hematophagous insects to use blood as a food source involves complex behavioral, morphological, and physiological adaptations to find suitable hosts, pierce their skin, and efficiently acquire and digest blood. As a result, the saliva of blood-feeding arthropods carries a complex mixture of compounds playing crucial roles in counteracting the three powerful and highly redundant responses of vertebrates to tissue injury: hemostasis, inflammation, and immunity (1). Blood feeding evolved independently several times in different taxa (2), and this resulted in a large variety of salivary anti-hemostatic factors targeting platelet aggregation, vasoconstriction, and blood clotting (1, 3). Salivary anticoagulants found in the mosquito family offer a good example of this convergent evolution; members of the Culicinae subfamily (i.e. Aedes and Culex species) inhibit factor Xa, whereas species belonging to the anopheline subfamily adopted a thrombin-directed anticoagulant activity (4).
The anti-thrombin of anopheline mosquitoes was first identified in the South American malaria vector Anopheles albimanus as a small cysteine-less polypeptide of 61 amino acids named anophelin (5). Synthetic anophelin was shown to be a highly specific, slow, tight-binding, reversible inhibitor of α-thrombin (6). It binds both the catalytic site and the anion binding exosite 1 (TABE1) of thrombin, a region that is known to be involved in the recognition of fibrinogen (7). A cDNA encoding the anophelin orthologue in the African malaria vector Anopheles gambiae had been previously identified by a selective cloning strategy and the encoded protein named cE5 (8). The two proteins, anophelin and cE5, are quite divergent (43% identity) with the slightly longer A. gambiae protein (82 amino acids) displaying an additional stretch of amino acids at the C terminus. Despite these differences, recombinant A. gambiae cE5 showed full preservation of anti-thrombin function, although the two proteins displayed slightly different binding kinetics. The A. albimanus anophelin is a slow-binding thrombin inhibitor (6), whereas the A. gambiae cE5 behaves as a fast-binding inhibitor (9), raising the possibility that the extended C-terminal may participate in thrombin binding.
The crystal structure of the A. albimanus anophelin in complex with human thrombin revealed its unique inhibition mechanism in comparison to other known thrombin inhibitors (10). In particular, the C-terminal segment (A(Glu-32–Pro-61)) appeared sufficient for thrombin inhibition, with residues A(Asp-33–Phe-45) blocking the exosite I and the highly conserved DPGR tetrapeptide (A(Asp-50–Arg-53)) occupying the active site cleft of the enzyme and disrupting the characteristic catalytic triad of the serine proteinase.
The release of the genomes of 16 Anopheles mosquitoes (11) allowed identifying anophelin/cE5 orthologues from several additional species, offering further insights into the evolution of this unique family of thrombin inhibitors. Multiple alignment of cE5/anophelin family members showed that, in comparison to the New World species A. albimanus and Anopheles darlingi, orthologues from the Old World Anopheles species display an extended, serine-rich, C-terminal region (10, 12). In order to investigate the possible involvement of the C-terminal region (cE5(Asn-63–Glu-82)) in thrombin-binding and inhibition as well as the role of the conserved N-terminal portion (cE5(Ala-1–Ser-30)), we performed a structural and functional analysis using both the cE5 protein and a set of cE5-derived peptides. Moreover, the three-dimensional structure of the human α-thrombin–A. gambiae cE5 complex unveiled the details of thrombin recognition and inhibition by an anophelin orthologue from an Old World Anopheles species.
Results
Structural properties of cE5 as determined by circular dichroism and NMR spectroscopy
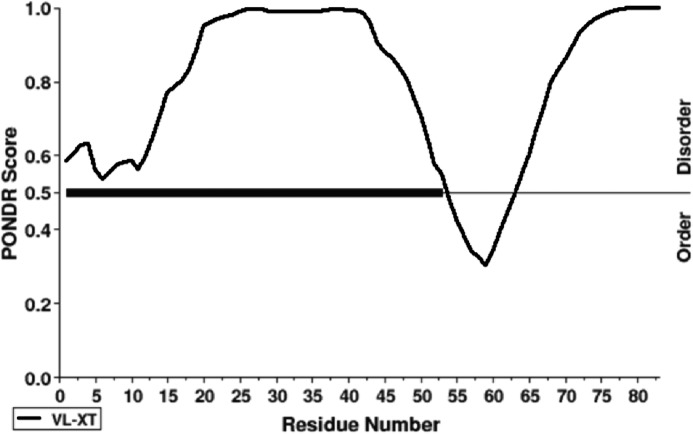
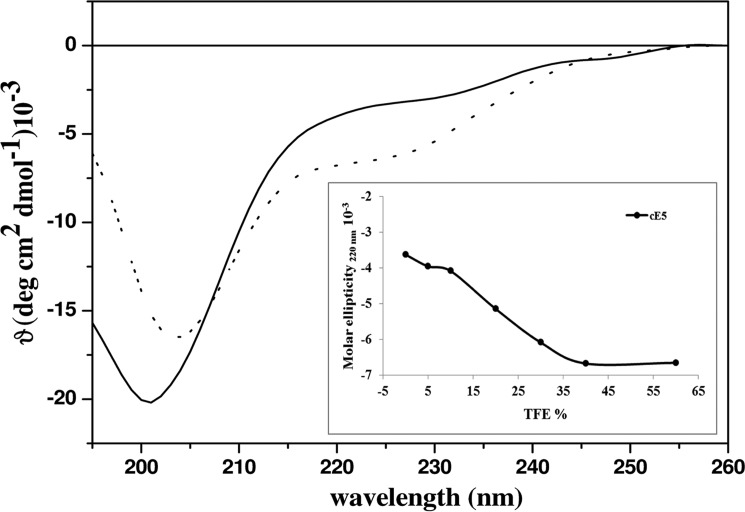
Prediction analysis carried out by DISOPRED (VL-XT predictor; www.pondr.com) 3 (50) suggested that the cE5 protein, as previously reported for the A. albimanus anophelin (10), was intrinsically disordered in solution. However, in contrast to A. albimanus anophelin, cE5 displayed a slight propensity to be structured in the region encompassing amino acids cE5(Pro-54) to cE5(Ser-64) (Fig. 1). Similar results were obtained using the database of intrinsically disordered proteins, MobiDB (not shown) (13). Furthermore, cE5 displayed a typical circular dichroism (CD) spectrum of an unstructured protein, with a minimum at 202 nm (Fig. 2). After the addition of 2,2,2-trifluoroethanol (TFE) 4 as co-solvent, which may help exploring the intrinsic tendency of a polypeptide to assume secondary structure elements (14–16), we found a very low propensity of cE5 to adopt an α-helical conformation. Indeed, even with high concentrations of TFE (60%), an increase of only ∼10% in the α-helical content was observed (inset in Fig. 2).
Figure 1.
Disorder prediction of the A. gambiae cE5 protein. Prediction of naturally disordered regions in the primary sequence of cE5 (VL-XT predictor) (44). Residues exceeding a threshold value of 0.5 are considered disordered.
Figure 2.
Far UV CD spectra of the cE5 protein. CD spectra of cE5 (10 μm in 10 mm phosphate buffer pH 7.5) alone (solid line) and after the addition of 60% (v/v) TFE (dotted line). The inset shows the molar ellipticity at 220 nm in the presence of increasing concentrations of TFE.
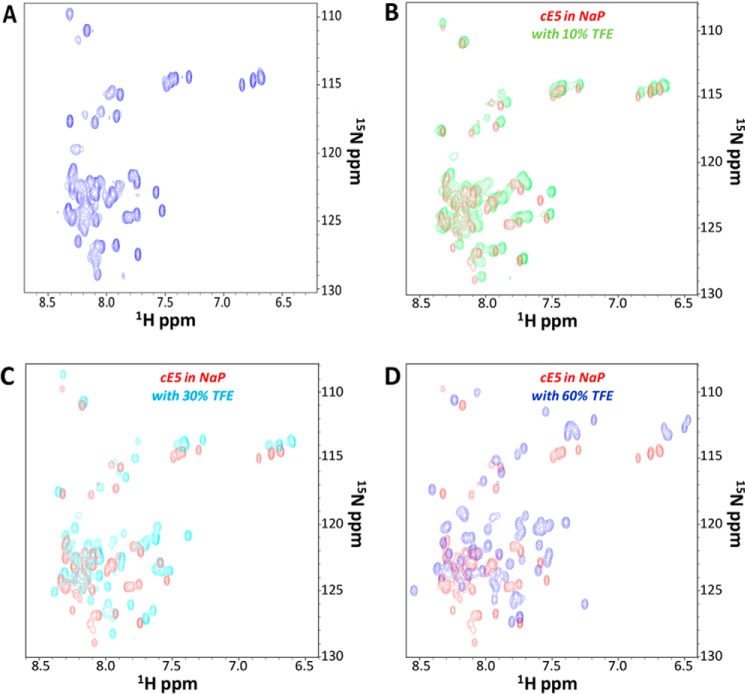
The conformational properties of cE5 were also investigated using NMR spectroscopy. The 1H,15N HSQC spectrum of 15N-labeled cE5 showed a limited chemical shift range (Fig. 3A), indicative of an unfolded molecule having few secondary structure elements, consistent with the random-coil state suggested by CD data. In addition, the 1H,15N HSQC spectra of uniformly 15N-labeled cE5 (40 μm) in the absence and the presence of different amounts of TFE (10, 30, and 60% (v/v)) (Fig. 3, B–D) were recorded in the same experimental conditions used for CD analyses. In complete agreement with the CD data, these NMR experiments indicated a continuous increase in signal dispersion (i.e. in structuration) with increasing TFE concentration. However, even at relatively high TFE concentrations, HSQC spectra still resemble those of canonical intrinsically disordered proteins (Fig. 3, B–D).
Figure 3.
NMR spectroscopy of the A. gambiae cE5 protein. A, 1H,15N HSQC spectrum of uniformly 15N-labeled cE5 (20 μm in 50 mm sodium phosphate, pH 7.8; sample total volume: 550 μl with 10% D2O). B–D, monitoring the effect of TFE on the intrinsically disordered protein cE5. Comparison of HSQC spectra of 15N-labeled cE5 in the absence (red) and in the presence of TFE at increasing concentrations: B, 10% (green); C, 30% (cyan); D, 60% (blue). NaP stands for sodium phosphate buffer.
Alignment of anophelin family members from different Anopheles species
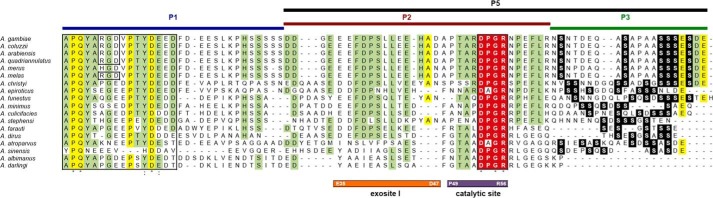
The sequencing of the genomes of 16 anopheline species (11) spanning ∼100 million years of evolution allowed identification of several additional members of the cE5/anophelin family (12). Alignment of the 18 full-length orthologues highlighted their considerable divergence: 18–77% amino acid identity among the different Anopheles species (excluding comparisons within the A. gambiae species complex). Overall, a block of 16 amino acids, including the highly conserved tetrapeptide APQY, can be recognized at the N-terminal region, and another highly conserved tetrapeptide, DPGR, is found toward the C terminus (Fig. 4). Between these two conserved blocks there is a more divergent central region enriched in acidic residues (Asp+Glu = 19–33%). A careful examination of the aligned proteins disclosed three features of potential functional relevance: (i) the N-terminal RGD tripeptide in A. gambiae and in a few other species of the complex; (ii) the conserved DPGR tetrapeptide, with proline to alanine replacement in Anopheles atroparvus and Anopheles epiroticus; (iii) the presence of a longer C-terminal in Old World anophelines.
Figure 4.
Multiple alignment of cE5/anophelin family members from 18 Anopheles species. The conserved N-terminal region and the DPGR tetrapeptide (highlighted in red) are boxed. Serine residues in the C-terminal region are shown in white lettering on a black background. Residues conserved in all or at least in 75% of the aligned sequences are highlighted in yellow and green, respectively. The peptides used in this study are represented above the aligned proteins. Regions of the cE5 protein shown by crystallographic analysis to interact with the exosite I and the catalytic site of thrombin are indicated below the alignment, as are their flanking residues. VectorBase accession numbers: A. gambiae, AGAP008004; Anopheles coluzzii, ACOM030673; Anopheles arabiensis, AARA005120; Anopheles quadriannulatus, AQUA000660; Anopheles merus, AMEM012461; Anopheles funestus, AFUN004964; Anopheles minimus, AMIN006263; Anopheles culicifacies, ACUA026308; Anopheles stephensi, ASTE006313; Anopheles dirus, ADIR011024; Anopheles sinensis, ASIS010361; A. albimanus, AALB014019. GenBankTM accession number: A. darlingi, GI:208657572. Sequences of the Anopheles melas, Anopheles christyi, A. epiroticus, Anopheles farauti, and A. atroparvus orthologues can be found in Arcà et al. (12). The sequences were aligned with Clustal Omega (43).
The tripeptide RGD is known for its involvement in binding to integrins (17). RGD-containing proteins with anti-platelet activity (disintegrins) have been found in snake venoms and in the saliva of blood-feeding arthropods (ticks and horseflies), leeches, and worms but never in mosquitoes. Disintegrins act as antagonists of integrin αIIbβ3 and inhibit fibrinogen binding to platelets and subsequent platelet cross-linking (18). Typically, integrin-binding RGD motifs are positioned in the loop of peptide hairpins formed by disulfide bonds (19), which is not the case for the cE5 RGD. We tested cE5 and the cE5-derived P1 peptide (Fig. 4) in platelet aggregation assays. We found no or very little effect on platelet aggregation (supplemental Fig. S1), which seems to rule out interaction of cE5 RGD with integrin αIIbβ3 (even though we cannot exclude binding to other integrins).
The DPGR tetrapeptide was previously found to play a crucial role in in vitro binding to α-thrombin (20) and shown to occupy the active-site cleft of the enzyme in the anophelin/thrombin crystal structure (10). Substitutions of the catalytic triad-disrupting aspartate and of the S1-targeting arginine residues in the A. albimanus protein were detrimental for the inhibitory activity (10). We do not know the effect of replacing the proline in this tetrapeptide with an alanine. However, it is conceivable that given the compact size of alanine, it may replace the pivotal proline residue without much impact in inhibitor activity.
Intriguingly, Old World anophelines (subgenera Cellia and Anopheles) carry an additional serine-enriched (29–56%) C-terminal stretch of 7–21 amino acids that is absent in the New World species A. albimanus and A. darlingi (subgenus Nyssorhynchus). It has been previously shown that the anophelin C-terminal fragment A(Glu-32–Pro-61) is mainly responsible for both binding to and inhibiting thrombin. Fragment A(Ala-1–Asp-31) did not appear to play any functional role in thrombin targeting and inhibition (10). However, A. gambiae cE5 appeared to possess higher affinity for thrombin and more potent anticoagulant activity than A. albimanus anophelin (Ki = 3.5 pm versus 34 pm; prolongation of thrombin time from 16 to 221 s for cE5 and to 84 s for anophelin) (9, 10). Moreover, anophelin and cE5 also displayed different kinetics of binding to thrombin, the former being a slow-binding thrombin inhibitor (6) and the latter behaving as a fast-binding inhibitor (9). To evaluate the possible role of the C-terminal serine-rich fragment of cE5, we designed the peptides P1 (cE5(Ala-1–Ser-30)), P2 (cE5(Asp-31–Arg-62)), P3 (cE5(Asn-63–Glu-82)), and P5 (cE5(Asp-31–Glu-82)) (Fig. 4) and assessed their interaction with thrombin by isothermal titration calorimetry (ITC) and enzymatic assays.
Binding studies
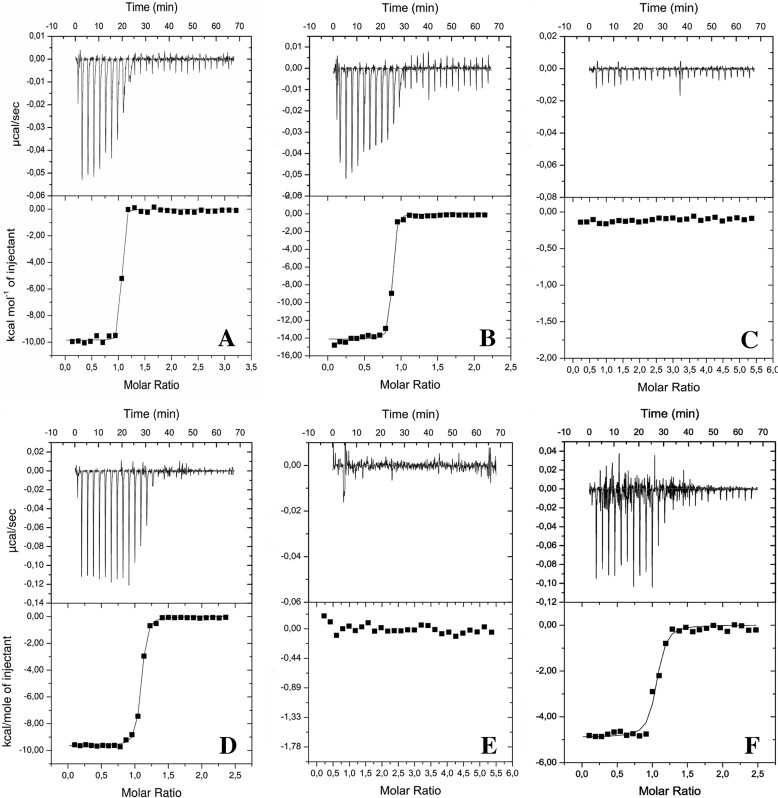
As determined by ITC, cE5 formed a very stable complex with thrombin (Kd ≤ 1 nm), with an apparent affinity slightly higher than anophelin (Kd ≤ 2 nm; Table 1, Fig. 5, A and B). In addition, binding assays with the cE5-based peptides P1, P2, P3, and P5 suggested that the region involved in the interaction with thrombin resides almost completely on the fragment cE5(Asp-31–Arg-62), corresponding to the P2 peptide (Kd ≤12 nm). This was underscored by the comparable ΔH values obtained for cE5 (−9.85 kcal/mol) and P2 (−9.66 kcal/mol) (Table 1 and Fig. 5, C–E). Surprisingly, binding experiments with a P5 yielded larger Kd (≤25 nm) and smaller ΔH (−4.89 kcal/mol) values than those obtained for P2, pointing to a negative contribution in thrombin-binding of the region corresponding to P3 in the absence of the N-terminal portion encompassed by P1 (Table 1, Fig. 5F).
Table 1.
Binding properties of cE5, anophelin, and cE5-derived peptides to thrombin as determined by ITC studies
ND, not detected.
| Ligand | Kd | ΔH | n |
|---|---|---|---|
| cal/mol | |||
| cE5 | ≤1 ± 0.2 nm | −9,848 ± 60 | 1 |
| cE5a | 0.3 nm | −8,272 | 1 |
| P1 (cE5(1–30)) | ND | ND | ND |
| P2 (cE5(31–62)) | ≤12 ± 1 nm | −9,663 ± 34 | 1 |
| P3 (cE5(63–82)) | ND | ND | ND |
| P5 (cE5(31–82)) | ≤25 ± 1 nm | −4,887 ± 75 | 1 |
| Anophelin | ≤2 ± 0.2 nm | −14,120 ± 90 | 1 |
a From displacement titration experiments.
Figure 5.
ITC analysis of thrombin binding to A. gambiae cE5, A. albimanus anophelin, and cE5-derived peptides. Isothermal titration calorimetry of thrombin interaction with A. gambiae cE5 (A), A. albimanus anophelin (B), P1 (C), P2 (D), P3 (E), and P5 (F) is shown. The top and bottom panels report raw and integrated data, respectively.
It is worth noting that the binding affinity (Ka) of cE5 to thrombin is beyond the limit of direct calorimetric determination. Under these conditions, ITC titration still provides an accurate measurement of the binding enthalpy (Table 1). However, ITC can be used to determine the complete binding thermodynamics of ligands with affinities down to the picomolar range using displacement titration (21), which was performed for this system in the presence of a weak thrombin inhibitor, the synthetic tetrapeptide d-Phe–Pro–d-Arg–Ala (22). Under these conditions, cE5 displayed a binding constant of 0.3 nm (Table 1, supplemental Fig. S2, and supplemental Equation 1), in line with the value previously reported (9, 10) and with that obtained from the thrombin amidolytic activity inhibition assays in this study (Table 2).
Table 2.
Inhibition of human α- and γ-thrombin by A. gambiae cE5
The affinity for thrombin of the P5 fragment is comparable with that of full-length cE5. Disruption of thrombin's exosite I decreases the inhibition potency of cE5 (and P5) by 3 orders of magnitude. Ki values ± S.E. given are representative of two independent experiments.
| Inhibitor | α-Thrombin | γ-Thrombin |
|---|---|---|
| pm | nm | |
| cE5 | 5.50 ± 1.26 | 3.79 ± 0.16 |
| P5 fragment | 20.80 ± 0.99 | 10.99 ± 0.69 |
We also used NMR titration experiments and analysis of 2D 1H,15N HSQC spectra (23) to study protein-protein interactions. HSQC spectra of 15N-labeled cE5 were recorded in the absence and presence of increasing amounts of unlabeled thrombin. At cE5/thrombin ratios >1/5, numerous spectral changes could be observed, pointing to interactions taking place between the two proteins (supplemental Fig. S3). However, it was not possible to reach saturating conditions, which would have allowed gaining more detailed structural information. This is most likely due to the relatively large size of the complex, which caused severe line broadening and loss of sensitivity in the NMR spectra (supplemental Fig. S3). In addition, 2D 1H,1H NMR spectra collected for the P2 peptide showed a similar lack of relevant chemical shift dispersion and the absence of a consistent number of nuclear Overhauser effect (NOE) contacts, confirming a disordered state for the P2 peptide in aqueous solution (supplemental Fig. S4, right panel). Moreover, no significant change in either chemical shift and/or NOE intensity could be observed when comparing 2D 1H,1H NOESY (Nuclear Overhauser Enhancement Spectroscopy) experiments with the P2 peptide (470 μm) in the absence and the presence of a substoichiometric amount of thrombin (25 μm) (supplemental Fig. S4). These observations suggest that under our experimental conditions, the peptide “dynamically” binds to thrombin by preserving disorder and flexibility.
Thrombin inhibition assays
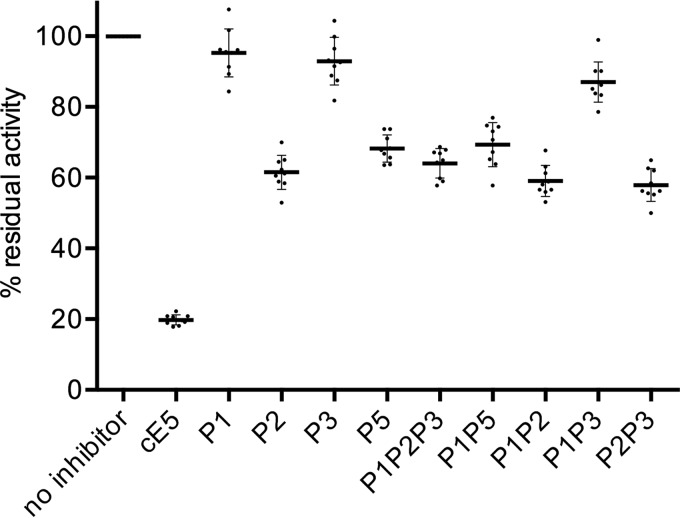
The inhibitory effect of cE5 and of peptides P1, P2, P3, and P5 on the in vitro amidolytic activity of thrombin was measured after the hydrolysis of a fluorogenic substrate. A thrombin inhibition of ∼80% was found for cE5, in line with previous measurements using a chromogenic substrate (9). The P2 peptide still retained inhibitory properties even though it was significantly less effective than cE5 (∼39% inhibition), whereas neither P1 nor P3 affected thrombin activity (Fig. 6). The inhibitory effect of P5 was not significantly different from those observed using P2 alone or any combination including either P2 or P5.
Figure 6.
Thrombin inhibition assays. Residual thrombin activity in the absence and the presence of A. gambiae cE5, or of the different peptides as indicated, was measured after the hydrolysis of the fluorogenic substrate Boc-Val-Pro-Arg-7-amido-4-methylcoumarin hydrochloride. Dots represent data from three independent experiments, each in triplicate. Thick lines show the mean values, and bars indicate S.D.
Overall, thrombin inhibition results fit quite well with ITC data, confirming that the region corresponding to the P2 peptide (cE5(Asp-31–Arg-62)) is the main segment responsible for both binding to thrombin and for the inhibitory activity. The P1 peptide (cE5(Ala-1–Ser-30)) did not bind thrombin and did not affect its activity, and these results are fully consistent with previous observations on A. albimanus anophelin (10). Moreover, our results seem to rule out any direct functional role of the serine-rich C-terminal region in thrombin inhibition. In fact, the P3 peptide alone did not bind to or inhibit thrombin, and the P5 peptide, which encompasses both P2 and P3, was not a better inhibitor or a more efficient binder than P2.
Overall structure of the A. gambiae cE5–human α-thrombin complex
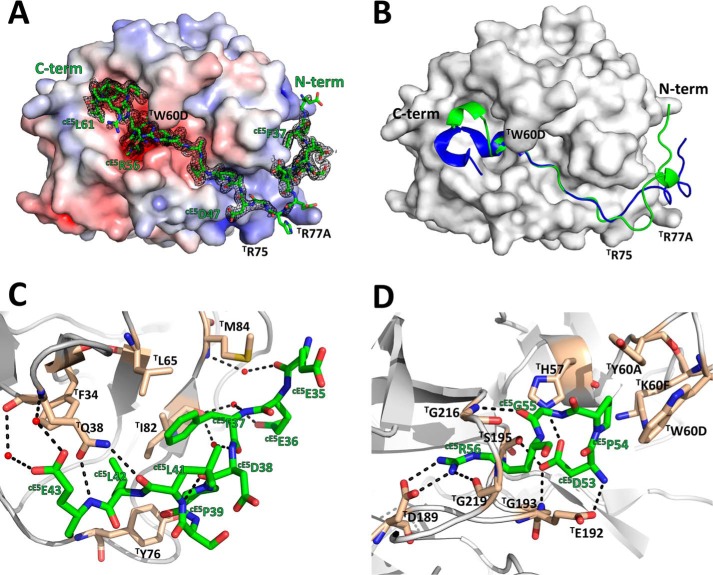
The recombinant P5 fragment (cE5(Asp-310–Glu-82)) of A. gambiae cE5 was crystallized in complex with human α-thrombin, and its structure was determined at 1.45 Å resolution (supplemental Table 1 and Fig. 7). There are three copies (A, B, and C) of the complex in the asymmetric unit: complex A comprising thrombin residues T(Ser-1E) to T(Arg-15) and T(Ile-16) to T(Gly-246) and inhibitor residues cE5(Glu-35) to cE5(Asn-63) (the chymotrypsinogen-based numbering will be used for thrombin); complex B encompasses thrombin residues T(Thr-1H) to T(Arg-15) and T(Ile-16) to T(Gly-246) and inhibitor residues cE5(Glu-36) to cE5(Leu-42) and cE5(Ala-46) to cE5(Arg-62); finally, in complex C thrombin residues T(Gly-1D) to T(Ile-14K), T(Ile-16) to T(Gln-244), and inhibitor residues cE5(Arg-52) to cE5(Leu-61) could be modeled. In all complexes, thrombin's 149 insertion loop was disordered (from T(Trp-148) to T(Lys-149E), T(Trp-148) to T(Gly-149D), and T(Thr-147) to T(Gly-149D), for complexes A to C, respectively) and could not be modeled. Although the three complexes are structurally very similar (root mean square deviations of 0.44 Å, 0.82 Å, and 0.93 Å for the superposition of the proteinase moieties of complexes A-B, A-C, and B-C, respectively), complex C displays an unusual flexibility of segments of the heavy chain of thrombin, encompassing residues T(Gln-38)–T(Lys-70), T(Arg-101)–T(Glu-127), T(Met-210)–T(Cys-220), and T(Phe-227)–T(Gln-244). Therefore, only complex A will be described herewith (Fig. 7). No significant density was present for the N-terminal tetrapeptide cE5(Asp-31) to cE5(Glu-34) or for the C-terminal 21 amino acids (cE5(Ser-64) to cE5(Glu-82) of the P5 fragment, and these segments were, therefore, excluded from the model. Superposition of the structures of free thrombin (PDB entry 3U69; Ref. 22) and the proteinase moiety of the A complex revealed that only minor rearrangements occur in the proteinase upon A. gambiae cE5 binding (root mean square deviation of 0.36 Å for 277 aligned Cα atoms).
Figure 7.
The A. gambiae cE5 anticoagulant blocks both the active site and the exosite I regions of human α-thrombin. A, the acidic cE5(Glu-35–Asp-47) segment of cE5 (stick model with nitrogen atoms in blue, oxygen in red, and carbon in green) binds to the exosite I of thrombin (solid surface representation with positive surface electrostatic potential in blue and negative surface electrostatic potential in red), whereas the downstream segment, up to cE5(Arg-62), threads along the active site cleft of the proteinase, blocking the active site and the non-primed specificity subsites. The 2Fo − Fc electron density map (1.0-σ contour) for cE5 is represented as a black mesh. B, comparison between the A. gambiae cE5 P5 fragment (green) and A. albimanus anophelin (PDB entry 4E05; Ref. 10; blue) shown in schematic representation. Thrombin, shown as a solid white surface, is in the same orientation as in panel A. C, close-up view of the interactions established between the N-terminal segment cE5(Glu-35–Glu-43) of cE5 (colored as in panel A) and the exosite I of thrombin (gray schematic with selected residues as sticks color-coded as cE5, except for carbon atoms, colored tan). Water molecules and hydrogen bonds are represented as red spheres and dotted black lines, respectively. D, close-up view of the interaction between the strictly conserved cE5(Arg-53–Arg-56) tetrapeptide and the active-site region of thrombin (colors as in panel C). The figure was prepared with PyMOL (Schrödinger).
Unsurprisingly, A. gambiae cE5 displays the same unique mode of thrombin inhibition observed for A. albimanus anophelin (10), further confirming it as a general mechanism for this class of inhibitors (MEROPS family I77; Ref. 24; Fig. 7B). In agreement with the results of CD (Fig. 2) and NMR measurements (Fig. 3), the inhibitor adopts a mostly extended conformation, running in a reverse orientation to substrates on the surface of thrombin and interacting with both the exosite I and the active-site region of the proteinase (Fig. 7, A, C, and D). Further supporting the observed binding mode, both full-length A. gambiae cE5 and its truncated P5 fragment display 3 orders of magnitude larger inhibition constants toward the exosite I-disrupted γ-thrombin than toward α-thrombin (Table 2).
Bidentate binding of cE5 to thrombin
Similar to the A. albimanus anophelin-thrombin complex (10), the cE5 fragment spanning residues cE5(Glu-35) to cE5(Asp-47) blocks the exosite I of the proteinase (Figs. 4 and 7C). However, despite the overall resemblance, the first half of this cE5 segment runs closer to the proteinase surface, with its main-chain atoms deviating significantly from the path of the equivalent region of A. albimanus anophelin (Fig. 7B). The inhibitor's tripeptide cE5(Glu-35)–cE5(Glu-36)–cE5(Phe-37) establishes water-mediated main chain-main chain contacts with thrombin's T(Ile-82)–T(Ser-83)–T(Met-84) tripeptide. This region is also stabilized by an intramolecular hydrogen bond between the carbonyl oxygen of cE5(Asp-38) and the amide nitrogen of cE5(Leu-41), further reinforced by two interactions with the side chain of T(Gln-38), involving the main chain nitrogen of cE5(Glu-43) and the carbonyl oxygen of cE5(Leu-41). This conformation results in the placement of the side chain of cE5(Phe-37) snuggly inside the hydrophobic depression bordered by the side chains of T(Leu-65), T(Ile-82), and T(Met-84), to which it establishes van der Waals interactions. Additionally, the aromatic ring of cE5(Phe-37) also contacts the side chain of the well-conserved cE5(Leu-42), in the same position of the homologous A(Leu-41) of A. albimanus anophelin, and in turn contacting T(Phe-34) and T(Tyr-76). The side chains of cE5(Pro-39) and cE5(Leu-41) pack on top of that cE5(Phe-37), completing this strong hydrophobic cluster.
Furthermore, toward the active site cleft of thrombin, cE5(Asp-47) forms a salt bridge with T(Arg-73). Notably, the replacement of A. albimanus anophelin A(Phe-45) by the much shorter cE5(Ala-48) impairs the highly conserved stacking interaction with T(Phe-34) observed in numerous thrombin ligands, from protease-activated receptor 1 (PAR1) (25) to hirudin (26) and from boophilin (27) to anophelin (10). Instead, two polar interactions are observed between the carbonyl oxygen of cE5(Ala-48) and the side chains of T(Arg-73) and T(Gln-151). From cE5(Pro-49) all the way to cE5(Arg-56), the interactions established between thrombin and the homologous A(Thr-47–Arg-53) segment of A. albimanus anophelin are strictly conserved (10), in agreement with the remarkable amino acid sequence conservation observed in all members of the family (Fig. 4). Therefore, cE5(Arg-53) interacts with the catalytic T(His-57) and T(Ser-195), whereas cE5(Arg-56) occupies the S1 specificity pocket of the proteinase (Fig. 7D). The downstream cE5(Asn-57) interacts with the side chain of T(Glu-192), resembling the contact established by A. albimanus anophelin A(Arg-54) (10), but the main chain of the remaining ordered cE5(Pro-58)–cE5(Leu-61) tetrapeptide deviates considerably from the homologous segment of anophelin (Fig. 7B). Despite that, the position of the side chain of cE5(Phe-60) closely resembles that of A. albimanus anophelin A(Leu-55), therefore occupying the aryl binding site of thrombin.
Discussion
Overall, the cE5 central region represented by the P2 peptide appeared as the main responsible for both thrombin binding and inhibition. The fact that only the cE5(Glu-35–Asn-63) segment of peptide P5 could be modeled in the crystal structure and no significant electron density was detectable for the serine-rich C-terminal portion reinforces this main role of the region corresponding to P2. Moreover, the C-terminal region was not constrained by crystal packing, explaining its observed disorder (supplemental Fig. S5). The binding of cE5 to thrombin was to a large extent similar to that previously described for A. albimanus anophelin. Nonetheless, the cE5 N-terminal and C-terminal regions still appear to play some role in both binding and inhibition. Indeed, when these two disordered segments were missing, we observed a decrease in binding affinity (≤1 nm for cE5 versus ≤12 nm for P2) as well as in thrombin inhibition (80% for cE5 versus 39% for P2). These more efficient binding and inhibitory properties of the full-length protein are in agreement with the flanking model, already proposed for interactions involving intrinsically disordered proteins (28, 29). Unexpectedly, ITC studies indicated a negative contribution of the C-terminal region encompassed by P3 (higher Kd and lower ΔH values for P5 than for either P2 or the entire cE5 protein). This conflicts with the conservation of the C-terminal serine-rich extension among Old World anophelines, suggesting a potential functional role for this region. However, specific experimental conditions and/or the in vitro nature of our study should be taken into account. The situation may be different in vivo, where the regions encompassed by peptides P1 and P3 may provide interaction sites for other binding partners (e.g. the RGD tripeptide may be involved in binding to integrins) and/or undergo post-translational modifications (as suggested from the different mobility between recombinant and native cE5 protein in SDS-PAGE) (9). In conclusion, through structural, functional, and binding studies we shed additional light on the unique mechanism of thrombin binding and inhibition by this family of salivary anticoagulants from anopheline mosquitoes. It is anticipated here that future proteomic and structural studies on salivary proteins of hematophagous animals are expected to provide insights into novel antihemostatic molecules and novel mechanisms of disrupting blood clotting that may be of great help in the design of novel antithrombotics. This appears especially true considering (i) the fast evolutionary rate of salivary proteins (11, 12), (ii) the convergent evolutionary nature of hematophagy, and (iii) the fact that so far we acquired some knowledge on the salivary repertoires of just a few of the >14,000 arthropod species estimated to feed on blood (30).
Experimental procedures
cE5 protein and peptides
Recombinant cE5 and 15N-labeled cE5 proteins were expressed and purified as described (9). The ORF encoding P5 (cE5(Asp-310–Glu-82)) was chemically synthesized (Life Technologies) and cloned into pETM11 (Novagen). Expression was performed as above. Protein integrity and purity was confirmed by mass spectrometry (Agilent Q-TOF LC/MS). Peptides P1 (cE5(Ala-1–Ser-30), APQYARGDVPTYDEEDFDEESLKPHSSSSS), P2 (cE5(Asp-31–Arg-62), DDGEEEFDPSLLEEHADAPTARDPGRNPEFLR), and P3 (cE5(Asn-63–Glu-82), NSNTDEQASAPAASSSESDE) were purchased from INBIOS S.r.l. (Naples, Italy). The A. albimanus anophelin was synthesized at the Peptide Synthesis Laboratory, Research Technologies Branch, NIAID, National Institutes of Health (Rockville, MD) by Dr. Jan Lukszo.
Circular dichroism analyses
CD spectra were recorded at 20 °C in the far UV (190–260 nm) on a Jasco J-710 spectropolarimeter (31). Each spectrum was obtained averaging three scans, subtracting contributions from corresponding blanks and converting the signal to mean residue ellipticity in units of degree × cm2 × dmol−1. Spectra were recorded using 10 μm cE5 in 10 mm phosphate buffer at pH 7.5 in the absence and in the presence of increasing concentrations (up to 60% (v/v)) of 2,2,2-trifluoroethanol (TFE, 99.5% isotopic purity, Sigma). Prediction analysis of secondary structure content was performed with CDPro (31, 32).
Isothermal titration calorimetry
ITC studies were performed at 22 °C with an ITC200 calorimeter (MicroCal/GE Healthcare). Anophelin and cE5 (100 μm) or the peptides P1, P2, P3, and P5 (150 μm) were titrated into a solution of human α-thrombin (20 μm; Hematologic Technologies Inc., HCT-0020). Before all titration experiments proteins and peptides were dialyzed against 20 mm HEPES, pH 7.5, 50 mm NaCl. All data were analyzed and fitted using the Microcal Origin version 7.0 software package. Binding enthalpy, dissociation constants, and stoichiometry were determined by fitting the data using a one-set-of-site-binding model. ITC runs were repeated twice to evaluate the reproducibility of the results.
Nuclear magnetic resonance spectroscopy
NMR spectra were recorded at 298 K on a Varian Unity Inova 600 MHz spectrometer provided with a cold probe. 2D 1H,15N HSQC spectra of 15N-labeled cE5 were acquired by implementing the following set of experimental conditions: 50 mm sodium phosphate buffer, pH 7.8; 550-μl sample total volume; H2O/D2O (98% deuterium, Armar Scientific, Switzerland) 90/10, 0.1%(w/v) NaN3; 5–40 μm protein concentration range. Similar experiments were conducted in phosphate buffer containing increasing amounts (10%, 30, and 60% (v/v)) of TFE. Water suppression was achieved by Excitation Sculpting (33). NMR spectra were processed with Varian software (Vnmrj version 1.1D) (34) and analyzed with the software NEASY that is included in the Cara program (http://cara.nmr.ch/doku.php)3 (35). To monitor binding to thrombin, 2D 1H,15N HSQC spectra of 15N-labeled cE5 (10 μm) were recorded for the protein in the free form and after addition of increasing amounts of unlabeled thrombin (cE5/thrombin molar ratios: 1/1.4, 1/2.7, 1/4.2, 1/7). Analysis of titration experiments and overlays of 2D spectra were performed with Sparky (36). In addition, 1H,1H NOESY 300 (37) and TOCSY 70 (total correlation spectroscopy) (38) experiments were recorded for the P2 peptide (470 μm in 50 mm sodium phosphate pH 7.8) in its unbound form and complexed to thrombin (25 μm).
Thrombin inhibition assays
Inhibition of human α-thrombin by cE5 and peptides P1, P2, P3, or P5 was evaluated after the hydrolysis of the fluorogenic substrate Boc-Val-Pro-Arg-7-amido-4-methylcoumarin hydrochloride (Boc, t-butoxycarbonyl; Sigma, B9385). Reactions were carried out in a 100-μl total volume in reaction buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl2, 20 mm CaCl2, 0.01% (v/v) Triton X-100). Human α-thrombin (0.5 nm; Sigma, T7009) was preincubated for 10 min at 37 °C with 100 nm recombinant cE5 or cE5-derived peptides. Reactions were initiated by the addition of substrate (250 μm) and hydrolysis followed at 37 °C for 60 min at 460 nm (excitation = 360 nm) on a Synergy HT microplate reader (BioTek). Data represent three independent experiments, each one in triplicate. Graphics and statistical analysis were performed using GraphPad Prism 6.0 (GraphPad Software, Inc. La Jolla, CA).
Thrombin amidolytic activity assay
The amidolytic activity of human α-thrombin or γ-thrombin (Hematologic Technologies) was followed using Tos-Gly-Pro-Arg-p-nitroanilide (Chromozym TH; Roche Applied Science) as the chromogenic substrate. Inhibition assays were performed in 50 mm Tris, pH 8.0, 50 mm NaCl, 1 mg/ml bovine serum albumin with 0.2 nm human α-thrombin or γ-thrombin, 100 μm substrate, and varying concentrations of inhibitor (0–80 nm for α-thrombin or 0–200 nm for γ-thrombin). Inhibition constants (Ki) were determined according to a tight-binding inhibitor model using the Morrison equation (39) with GraphPad Prism 6.0. All reactions were initiated by the addition of thrombin and were carried out at least in duplicate at 37 °C in 96-well microtiter plates. Reaction progress was monitored at 405 nm for 30 min on a Synergy2 multimode microplate reader (BioTek).
Platelet assays
Inhibition of collagen-mediated platelet aggregation was evaluated using human platelet-rich plasma obtained from donors at the National Institutes of Health blood bank and diluted to a density of 2 × 105 platelets/μl in Tyrode-BSA (40) (final volume 300 μl). cE5, P1, or an equal volume of Tyrode buffer was added to each sample, which was then placed in an aggregometer (40) and stirred at 1200 rpm at 37 °C for 1 min before the addition of collagen (type-1 fibrils, Chrono-log) to a concentration of 1.6 μg/ml.
Crystallization of A. gambiae cE5 in complex with human α-thrombin
Human α-thrombin (Hematologic Technologies) was mixed in 20 mm HEPES, pH 7.5, 125 mm NaCl with a 4-fold molar excess of P5 peptide (cE5(Asp-310–Glu-82)) and incubated on ice for 1 h. The resulting complex was concentrated by ultrafiltration using a 2-kDa cutoff centrifugal device (Sartorius). An initial crystallization screen at 20 °C on sitting drop geometry was performed at the High Throughput Crystallization Laboratory of the European Molecular Biology Laboratory (Grenoble, France). Preliminary crystallization conditions were systematically optimized in-house until single monoclinic crystals belonging to space group C2 were obtained after 4–6 days at 20 °C using the vapor diffusion sitting-drop method from drops consisting of equal volumes (1 μl) of protein complex (at 6.4 mg/ml) and precipitant solution (0.1 m PCTP, pH 5.0, 25% (w/v) PEG 1500) equilibrated against a 300-μl reservoir.
Data collection and processing
Crystals were cryoprotected by brief immersion in 0.1 m PCTP, pH 5.0, 20% (w/v) PEG1500, 20% (v/v) ethylene glycol or 0.1 m PCTP, pH 5.0, 35% (w/v) PEG1500 and flash-cooled in liquid nitrogen. Diffraction data were collected from two isomorphous crystals (2100 and 2400 images in 0.05° oscillation steps and 0.037-s exposure) on a Pilatus 6 m-F detector (Dectris) using a wavelength of 0.973 Å at beam line ID30B of the European Synchrotron Radiation Facility (Grenoble, France). Data were integrated with XDS (41), scaled with XSCALE (41), and reduced with utilities from the CCP4 program suite (42). Data collection statistics are summarized in supplemental Table 1. The raw experimental datasets for the three-dimensional structure reported in this manuscript (PDB 5NHU) are available in the SBGrid Data Bank (https://data.sbgrid.org/data/)3 (45) (Ripoll-Rozada, J., and Pereira, P. J. B. (2017) X-ray diffraction data for human thrombin-A. gambiae cE5 complex–datasets 1 and 2. Structural Biology Data Grid, V1, http://dx.doi.org/10.15785/SBGRID/430 and http://dx.doi.org/10.15785/SBGRID/431).
Structure determination and refinement
The structure of the complex was solved by molecular replacement with PHASER (46) using the coordinates of free wild-type human α-thrombin (PDB entry 3U69; Ref. 22) as the search model. Alternating cycles of model building with COOT (47) and refinement with PHENIX (48) were performed until model completion (refinement statistics are summarized in supplemental Table 1). All crystallographic software was supported by SBGrid (49). Refined coordinates and structure factors were deposited at the PDB with accession number 5NHU.
Author contributions
L. P., J. R.-R., M. L., R. R., F. L., and J. F. A. performed the research. L. P., M. L., G. F., P. J. B. P., B. A., and E. P. designed the research. L. P., J. R.-R., M. L., J. A., P. J. B. P., B. A., and E. P. analyzed the data. M. L., P. J. B. P., B. A., and E. P. wrote the paper.
Supplementary Material
Acknowledgments
We acknowledge J. M. C. Ribeiro for kindly providing the anophelin peptide, the European Synchrotron Radiation Facility (ESRF) for provision of synchrotron radiation facilities, and the ESRF staff for help with data collection. Transnational Access to the High Throughput Crystallization Laboratory of the European Molecular Biology Laboratory Grenoble Outstation was supported by the European Community-Seventh Framework Program (FP7/2007-2013) Grant Protein Production Platform (PCUBE Agreement 227764).
This work was funded by the EU FP7 Infravec project (228421) and Finanziamenti di Ateneo per la Ricerca Scientifica (C26A12SLP3) (to B. A.), in part by Portuguese funds through FCT (Fundação para a Ciência e a Tecnologia) in the form of postdoctoral fellowship SFRH/BPD/108004/2015 (to J. R.-R.), and of the project “Institute for Research and Innovation in Health Sciences” (POCI-01-0145-FEDER-007274), co-funded by the European Regional Development Fund (FEDER) through the COMPETE 2020-Operational Programme for Competitiveness and Internationalization (POCI), PORTUGAL 2020. This work was also supported by the “Structured program on bioengineered therapies for infectious diseases and tissue regeneration” (Norte-01-0145-FEDER-000012), funded by Norte Portugal Regional Operational Programme (NORTE 2020) under the PORTUGAL 2020 Partnership Agreement through FEDER. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Table S1, Figs. S1–S5, and Equation 1.
The atomic coordinates and structure factors (code 5NHU) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- TFE
- 2,2,2-trifluoroethanol
- HSQC
- heteronuclear single quantum correlation
- ITC
- isothermal titration calorimetry
- PCTP
- sodium propionate, sodium cacodylate trihydrate, bis-Tris propane
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Ribeiro J. M., and Arcà B. (2009) From sialomes to the sialoverse: an insight into salivary potion of blood-feeding insects. Adv. Insect Physiol. 37, 59–118 [Google Scholar]
- 2. Mans B. J. (2011) Evolution of vertebrate hemostatic and inflammatory control mechanisms in blood-feeding arthropods. J. Innate. Immun. 3, 41–51 [DOI] [PubMed] [Google Scholar]
- 3. Ribeiro J. M., Mans B. J., and Arcà B. (2010) An insight into the sialome of blood-feeding Nematocera. Insect. Biochem. Mol. Biol. 40, 767–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stark K. R., and James A. A. (1996) Anticoagulants in vector arthropods. Parasitol. Today 12, 430–437 [DOI] [PubMed] [Google Scholar]
- 5. Valenzuela J. G., Francischetti I. M., and Ribeiro J. M. (1999) Purification, cloning, and synthesis of a novel salivary anti-thrombin from the mosquito Anopheles albimanus. Biochemistry 38, 11209–11215 [DOI] [PubMed] [Google Scholar]
- 6. Francischetti I. M., Valenzuela J. G., and Ribeiro J. M. (1999) Anophelin: kinetics and mechanism of thrombin inhibition. Biochemistry 38, 16678–16685 [DOI] [PubMed] [Google Scholar]
- 7. De Cristofaro R., and De Candia E. (2003) Thrombin domains: structure, function and interaction with platelet receptors. J. Thromb. Thrombolysis 15, 151–163 [DOI] [PubMed] [Google Scholar]
- 8. Arcá B., Lombardo F., de Lara Capurro M., della Torre A., Dimopoulos G., James A. A., and Coluzzi M. (1999) Trapping cDNAs encoding secreted proteins from the salivary glands of the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 96, 1516–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ronca R., Kotsyfakis M., Lombardo F., Rizzo C., Currà C., Ponzi M., Fiorentino G., Ribeiro J. M., and Arcà B. (2012) The Anopheles gambiae cE5, a tight- and fast-binding thrombin inhibitor with post-transcriptionally regulated salivary-restricted expression. Insect. Biochem. Mol. Biol. 42, 610–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Figueiredo A. C., de Sanctis D., Gutiérrez-Gallego R., Cereija T. B., Macedo-Ribeiro S., Fuentes-Prior P., and Pereira P. J. (2012) Unique thrombin inhibition mechanism by anophelin, an anticoagulant from the malaria vector. Proc. Natl. Acad. Sci. U.S.A. 109, E3649–E3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neafsey D. E., Waterhouse R. M., Abai M. R., Aganezov S. S., Alekseyev M. A., Allen J. E., Amon J., Arcà B., Arensburger P., Artemov G., Assour L. A., Basseri H., Berlin A., Birren B. W., Blandin S. A., Brockman A. I., et al. (2015) Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347, 1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arcà B., Lombardo F., Struchiner C. J., and Ribeiro J. M. (2017) Anopheline salivary protein genes and gene families: an evolutionary overview after the whole genome sequence of sixteen Anopheles species. BMC Genomics 18, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Potenza E., Di Domenico T., Walsh I., and Tosatto S. C. (2015) MobiDB 2.0: an improved database of intrinsically disordered and mobile proteins. Nucleic Acids Res. 43, D315–D320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Anderson V. L., Ramlall T. F., Rospigliosi C. C., Webb W. W., and Eliezer D. (2010) Identification of a helical intermediate in trifluoroethanol-induced α-synuclein aggregation. Proc. Natl. Acad. Sci. U.S.A. 107, 18850–18855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Buck M. (1998) Trifluoroethanol and colleagues: cosolvents come of age. Recent studies with peptides and proteins. Q. Rev. Biophys. 31, 297–355 [DOI] [PubMed] [Google Scholar]
- 16. Maestro B., Galán B., Alfonso C., Rivas G., Prieto M. A., and Sanz J. M. (2013) A new family of intrinsically disordered proteins: structural characterization of the major phasin PhaF from Pseudomonas putida KT2440. Plos ONE 8, e56904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zaccaro L., Del Gatto A., Pedone C., and Saviano M. (2009) Peptides for tumour therapy and diagnosis: current status and future directions. Curr. Med. Chem. 16, 780–795 [DOI] [PubMed] [Google Scholar]
- 18. Francischetti I. M. (2010) Platelet aggregation inhibitors from hematophagous animals. Toxicon 56, 1130–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Assumpcao T. C., Ribeiro J. M., and Francischetti I. M. (2012) Disintegrins from hematophagous sources. Toxins 4, 296–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Raffler N. A., Schneider-Mergener J., and Famulok M. (2003) A novel class of small functional peptides that bind and inhibit human α-thrombin isolated by mRNA display. Chem. Biol. 10, 69–79 [DOI] [PubMed] [Google Scholar]
- 21. Velazquez-Campoy A., and Freire E. (2006) Isothermal titration calorimetry to determine association constants for high-affinity ligands. Nat. Protoc. 1, 186–191 [DOI] [PubMed] [Google Scholar]
- 22. Figueiredo A. C., Clement C. C., Zakia S., Gingold J., Philipp M., and Pereira P. J. (2012) Rational design and characterization of d-Phe–Pro–d-Arg-derived direct thrombin inhibitors. PLoS ONE 7, e34354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pellecchia M. (2005) Solution nuclear magnetic resonance spectroscopy techniques for probing intermolecular interactions. Chem. Biol. 12, 961–971 [DOI] [PubMed] [Google Scholar]
- 24. Rawlings N. D., Barrett A. J., and Finn R. (2016) Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 44, D343–D350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gandhi P. S., Chen Z., and Di Cera E. (2010) Crystal structure of thrombin bound to the uncleaved extracellular fragment of PAR1. J. Biol. Chem. 285, 15393–15398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu C. C., Brustad E., Liu W., and Schultz P. G. (2007) Crystal structure of a biosynthetic sulfo-hirudin complexed to thrombin. J. Am. Chem. Soc. 129, 10648–10649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Macedo-Ribeiro S., Almeida C., Calisto B. M., Friedrich T., Mentele R., Stürzebecher J., Fuentes-Prior P., and Pereira P. J. (2008) Isolation, cloning and structural characterisation of boophilin, a multifunctional Kunitz-type proteinase inhibitor from the cattle tick. PLoS ONE 3, e1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sharma R., Raduly Z., Miskei M., and Fuxreiter M. (2015) Fuzzy complexes: specific binding without complete folding. FEBS Lett. 589, 2533–2542 [DOI] [PubMed] [Google Scholar]
- 29. Gruet A., Dosnon M., Blocquel D., Brunel J., Gerlier D., Das R. K., Bonetti D., Gianni S., Fuxreiter M., Longhi S., and Bignon C. (2016) Fuzzy regions in an intrinsically disordered protein impair protein-protein interactions. FEBS J. 283, 576–594 [DOI] [PubMed] [Google Scholar]
- 30. Ribeiro J. M. (1995) Blood-feeding arthropods: live syringes or invertebrate pharmacologists? Infect. Agents Dis. 4, 143–152 [PubMed] [Google Scholar]
- 31. de Paola I., Pirone L., Palmieri M., Balasco N., Esposito L., Russo L., Mazzà D., Di Marcotullio L., Di Gaetano S., Malgieri G., Vitagliano L., Pedone E., and Zaccaro L. (2015) Cullin3-BTB interface: a novel target for stapled peptides. PLoS ONE 10, e0121149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sreerama N., and Woody R. W. (1993) A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 209, 32–44 [DOI] [PubMed] [Google Scholar]
- 33. Hwang T. L., and Shaka A. J. (1995) Water suppression that works: excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. Ser. A 112, 275–279 [Google Scholar]
- 34. Contursi P., Farina B., Pirone L., Fusco S., Russo L., Bartolucci S., Fattorusso R., and Pedone E. (2014) Structural and functional studies of Stf76 from the Sulfolobus islandicus plasmid-virus pSSVx: a novel peculiar member of the winged helix-turn-helix transcription factor family. Nucleic Acids Res. 42, 5993–6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bartels C., Xia T. H., Billeter M., Güntert P., and Wüthrich K. (1995) The Program Xeasy for Computer-Supported NMR Spectral-Analysis of Biological Macromolecules. J. Biomol. NMR 6, 1–10 [DOI] [PubMed] [Google Scholar]
- 36. Goddard T. D., and Kneller D. G.. SPARKY 3, University of California, San Francisco [Google Scholar]
- 37. Kumar A., Ernst R. R., and Wüthrich K. (1980) A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 95, 1–6 [DOI] [PubMed] [Google Scholar]
- 38. Griesinger C., Otting G., Wuethrich K., and Ernst R. R. (1988) Clean TOCSY for proton spin system identification in macromolecules. J. Am. Chem. Soc. 110, 7870–7872 [Google Scholar]
- 39. Williams J. W., and Morrison J. F. (1979) The kinetics of reversible tight-binding inhibition. Methods Enzymol. 63, 437–467 [DOI] [PubMed] [Google Scholar]
- 40. Lagrue A. H., Francischetti I. M., Guimarães J. A., and Jandrot-Perrus M. (1999) Phosphatidylinositol 3′-kinase and tyrosine-phosphatase activation positively modulate convulxin-induced platelet activation: comparison with collagen. FEBS Lett. 448, 95–100 [DOI] [PubMed] [Google Scholar]
- 41. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Romero P., Obradovic Z., Li X., Garner E. C., Brown C. J., and Dunker A. K. (2001) Sequence complexity of disordered protein. Proteins 42, 38–48 [DOI] [PubMed] [Google Scholar]
- 45. Meyer P. A., Socias S., Key J., Ransey E., Tjon E. C., Buschiazzo A., Lei M., Botka C., Withrow J., Neau D., Rajashankar K., Anderson K. S., Baxter R. H., Blacklow S. C., Boggon T. J., et al. (2016) Data publication with the structural biology data grid supports live analysis. Nat. Commun. 7, 10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morin A., Eisenbraun B., Key J., Sanschagrin P. C., Timony M. A., Ottaviano M., and Sliz P. (2013) Collaboration gets the most out of software. eLife 2, e01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ward J. J., McGuffin L. J, Bryson K., Buxton B. F., and Jones D. T. (2004) The DISOPRED server for the prediction of protein disorder. Bioinformatics 20, 2138–2139 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.