Abstract
Acetylcholinesterase (AChE) hydrolyzes acetylcholine to terminate cholinergic transmission in neurons. Apart from this AChE activity, emerging evidence suggests that AChE could also function in other, non-neuronal cells. For instance, in bone, AChE exists as a proline-rich membrane anchor (PRiMA)-linked globular form in osteoblasts, in which it is proposed to play a noncholinergic role in differentiation. However, this hypothesis is untested. Here, we found that in cultured rat osteoblasts, AChE expression was increased in parallel with osteoblastic differentiation. Because several lines of evidence indicate that AChE activity in osteoblast could be triggered by Wnt/β-catenin signaling, we added recombinant human Wnt3a to cultured osteoblasts and found that this addition induced expression of the ACHE gene and protein product. This Wnt3a-induced AChE expression was blocked by the Wnt-signaling inhibitor Dickkopf protein-1 (DKK-1). We hypothesized that the Runt-related transcription factor 2 (Runx2), a downstream transcription factor in Wnt/β-catenin signaling, is involved in AChE regulation in osteoblasts, confirmed by the identification of a Runx2-binding site in the ACHE gene promoter, further corroborated by ChIP. Of note, Runx2 overexpression in osteoblasts induced AChE expression and activity of the ACHE promoter tagged with the luciferase gene. Moreover, deletion of the Runx2-binding site in the ACHE promoter reduced its activity during osteoblastic differentiation, and addition of 5-azacytidine and trichostatin A to differentiating osteoblasts affected AChE expression, suggesting epigenetic regulation of the ACHE gene. We conclude that AChE plays a role in osteoblastic differentiation and is regulated by both Wnt3a and Runx2.
Keywords: acetylcholinesterase (AChE), differentiation, DNA methylation, osteoblast, Wnt signaling, Runx2
Introduction
The active and dynamic balance between bone resorption by osteoclasts and bone formation by osteoblasts supports the lifelong bone remodeling. Imbalance of two processes results in bone disease, e.g. osteoporosis (1). Considering the bone-forming cell nature of osteoblast and the medical need for therapies based on stimulating anabolic pathways in bone, the study on possible regulation during osteoblastic differentiation therefore has an urgent need. Wnt/β-catenin signaling plays a pivotal role in bone formation and osteoblastic differentiation. Runt-related transcription factor 2 (Runx2) 2 is a downstream component of the Wnt/β-catenin-signaling pathway, and which is the master transcription factor required in determining the osteoblastic lineage (2, 3). In addition, DNA methylation during osteoblastic differentiation might have profound effects on gene expression and cell commitment, and these events could lead to bone metabolism (4).
Acetylcholinesterase (AChE) (EC 3.1.1.7) is a key enzyme needed for acetylcholine (ACh) hydrolysis in terminating cholinergic transmission in vertebrate. Three variants of transcripts are generated by alternative splicing: “read-through (AChER),” “hydrophobic (AChEH),” and “tailed (AChET)” (5). AChET exists in all vertebrates and is predominant in the brain and muscle, generating different oligomers. AChER is generated as soluble monomers (G1) (6), which is proposed to be responsible for a stress-related response (7, 8). AChEH is expressed as glycophosphatidylinositol-linked dimers (G2), and anchored on the plasma membrane of erythrocytes and lymphocytes (5, 9). AChET, well known for its cholinergic role as a neurotransmission regulator, is produced and anchored on neuronal and muscle synapses via association with anchoring proteins, e.g. proline-rich membrane anchor (PRiMA) or collagen Q (ColQ). The association forms a variety of oligomers, e.g. G4 and A12 (10, 11). The function of AChET essentially depends on a tight interaction between its C-terminal peptide with the anchoring proteins. The form of PRiMA-linked AChE exists in the mammalian brain and muscle as amphiphilic tetrameric globular forms (detergent-interacting G4 components) (12, 13).
In the last decade, the existence and function of AChE in non-neuronal tissues have been extensively focused, and emerging evidence suggests a possible non-cholinergic function of AChE in various cell types. In parallel, the non-hydrolytic action of AChE might have relevance to various diseases (14, 15). For example, the existence of AChE in bone tissue, osteoblasts, and osteoblast-like cell lines have been reported (16, 17). The possible involvement of AChE in bone development and skeleton remodeling has been proposed (18, 19). To support the possible role of AChE in bone development, we addressed a critical question here that how the enzyme being regulated during osteoblastic differentiation.
Results
AChE expression during osteoblastic differentiation
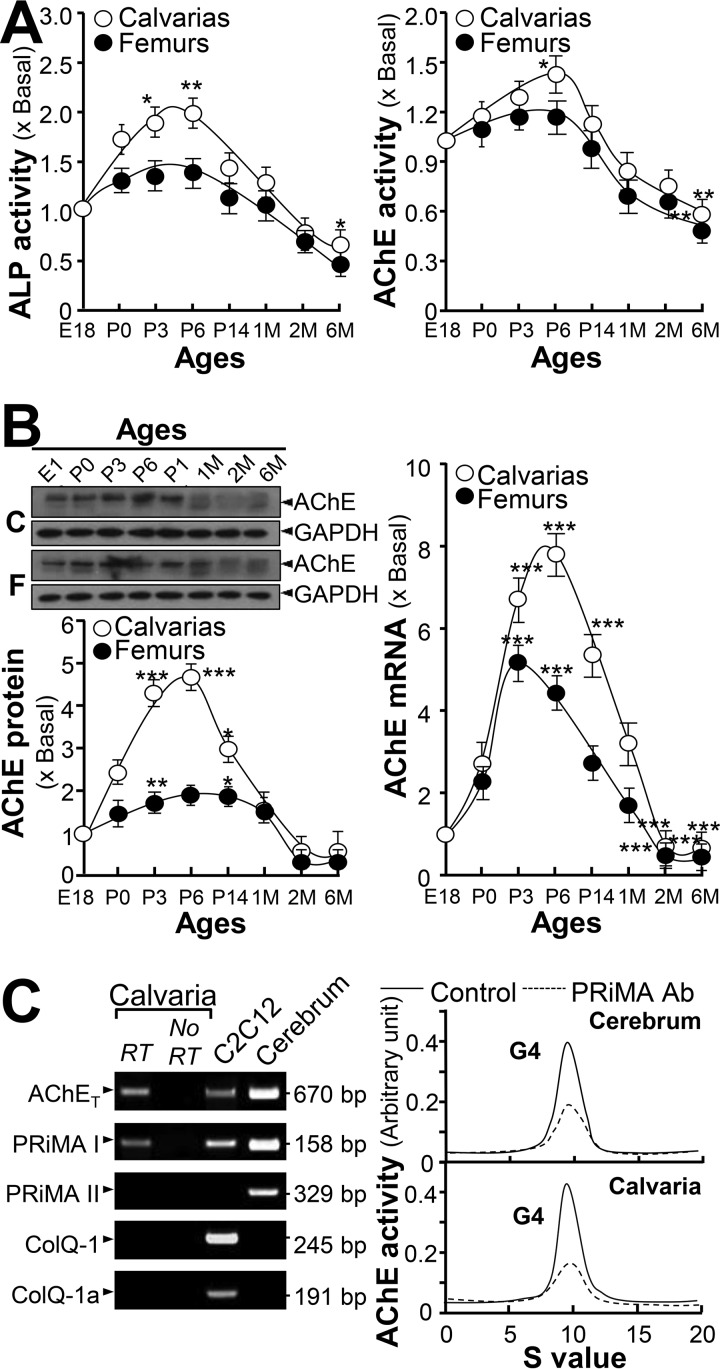
The expression profiles of AChE in calvaria and femur at different development stages, e.g. embryonic (E18), postnatal (P0, P3, P6, P14, and 1 month) and adult (2 and 6 months), were determined. A similar pattern was observed in alkaline phosphatase (ALP) activity, an indicative biochemical marker of bone differentiation, in both calvaria and femur. ALP activity was increased from E18 to P6: then this was decreased according to age (Fig. 1A). This result is consistent with a previous study (20). In line with the elevated activity of ALP, AChE activity increased from E18 to P6, and then decreased afterward (Fig. 1A). Expression of the AChE protein (∼68 kDa), as well as mRNA levels, showed a similar pattern according to developmental stages of bone, i.e. peak at earlier developmental stage (Fig. 1B). The mRNAs encoding AChE (AChET form) and PRiMA I were revealed in bone tissue, as that in the cerebrum (Fig. 1C, left panel). ColQ and PRiMA II mRNAs were below detection. The molecular form of AChE was analyzed, and the globular form (G4) of AChE was the major form being identified. Anti-PRiMA antibody precipitated the G4 form enzyme suggesting this was PRiMA-linked (Fig. 1C, right panel).
Figure 1.
Increase of AChE expression and activity during bone development. A, calvarias and femurs from rats on different stages as indicated were isolated and assayed for ALP and AChE activities. B, upper panel: protein lysates from bone tissues were analyzed by Western blotting. AChE (∼68 kDa) and GAPDH (∼35 kDa) were shown. Lower panel, quantitation of AChE protein, calibrated from the blots by densitometry. Right panel shows the real-time PCR of AChE mRNA. Values are expressed as fold-increase to basal reading. C, left panel, total RNAs were extracted from calvarias and femurs to perform PCR to determine the presence of AChET, PRiMA I, PRiMA II, ColQ-1, and ColQ-1a by using specific primers. PCR products were resolved on a 1% SYBR safe-stained agarose gel and visualized under the UV light. The identities of PCR product was confirmed by DNA sequencing. Rat cerebrum and mouse C2C12 RNAs served as positive controls. Right panel, molecular forms of AChE in calvarias and femurs were determined by sucrose density gradient analysis. PRiMA-linked G4 AChE was immunodepleted by anti-PRiMA antibody. Cerebrum served as a positive control for G4 AChE. Enzymatic activities are expressed in arbitrary units. Values are in mean ± S.E., n = 4, *, p < 0.05; **, p < 0.01; ***, p < 0.001.
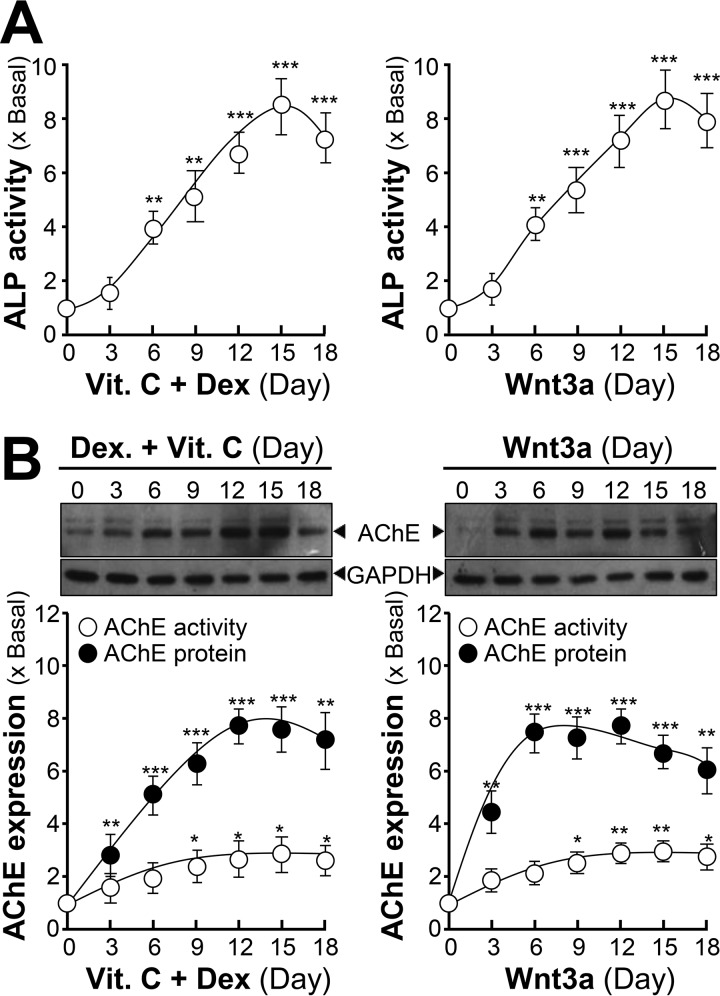
Rat osteoblast was employed to investigate the regulation of AChE during differentiation. In comparison with other osteoblastic cell lines, primary cultured osteoblasts could reflect more phenotypic properties. Under the co-treatment of dexamethasone plus vitamin C, cultured osteoblasts were induced to differentiate for 18 days. The activity of ALP was increased: the maximal induction at 8-fold was at a treatment of 15 days (Fig. 2A). In addition, recombinant human Wnt3a was applied onto the cultures, which induced the differentiation of osteoblast. The Wnt3a-induced ALP activity was similar to that of dexamethasone plus vitamin C. Under osteoblastic differentiation, the expression of AChE was induced, and the induction was similar to that of ALP (Fig. 2B). The enzymatic activity of AChE was increased to ∼2-fold, and the protein level was induced to ∼8-fold. The peak was identified at 12 days of treatment with dexamethasone plus vitamin C, or 6 days after Wnt3a treatment (Fig. 2B). An increase of AChE expression corresponding with elevated ALP activity was consistent with a previous report (21).
Figure 2.
Increase of AChE expression and activity during the differentiation of cultured osteoblasts. A, primary cultured osteoblasts were treated with dexamethasone plus vitamin C (Dex, 20 nm; and Vit. C, 250 μm) or Wnt3a (200 ng/ml) for 18 days, cell lysates were collected on different days as indicated for ALP assay. B, the cell lysates from A were analyzed by Western blotting and Ellman assay. AChE (∼68 kDa) and GAPDH (∼35 kDa) are shown (upper panel). The lower panel shows quantitation of AChE activity and protein, calibrated from the blots by densitometry. Values are expressed as fold-increase to basal reading, and are in mean ± S.E., n = 4, *, p < 0.05; **, p < 0.01; ***, p < 0.001.
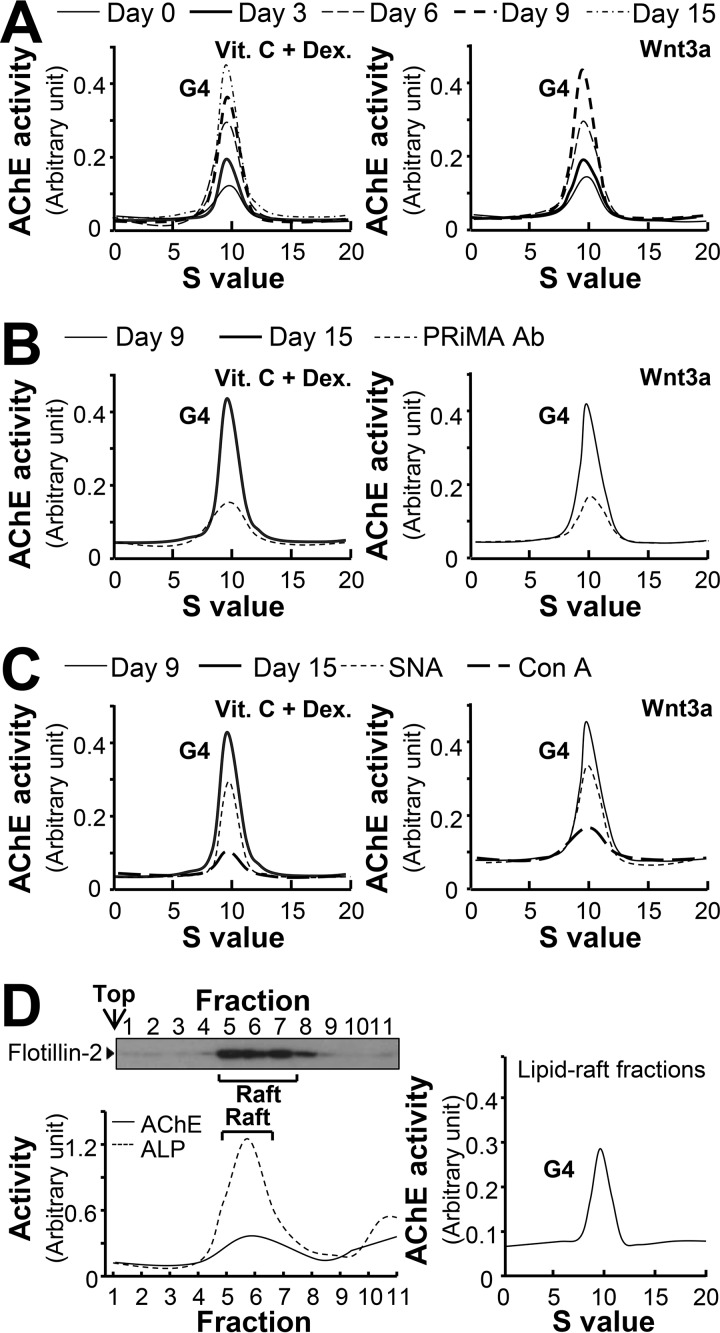
AChE is a highly glycosylated protein, and thus the N-glycosylation profile of this enzyme was determined during osteoblastic differentiation. Similar to bone tissue, cultured osteoblasts expressed AChE at the G4 form with a sedimentation index of ∼10 S (Fig. 3A). The amount of G1/G2 peak (∼4 S) was very minimal. The amount of G4 AChE was increased during the differentiation, as induced by dexamethasone plus vitamin C or Wnt3a (Fig. 3A). The specificity of G4 AChE was illustrated by precipitation with anti-PRiMA antibody (Fig. 3B), which was consistent with previous studies (11, 18).
Figure 3.
The G4 form of AChE remains unchanged during osteoblastic differentiation. A, primary cultured osteoblasts were treated with dexamethasone plus vitamin C (Dex. 20 nm, and Vit. C 250 μm) or Wnt3a (200 ng/ml) for a 3-day interval, cell lysates were collected on different days as indicated and subjected to sucrose density gradient analysis. B, equal amounts of proteins from differentiated cell lysates (day 9 and 15) were immune precipitated by anti-PRiMA antibody. C, equal amounts of proteins from differentiated cell lysates, the same control samples (day 9 and day 15) as in B, were incubated with or without Con A or SNA overnight, unbound and lectin-precipitated parts were separated by centrifugation. Supernatant from B and C was subjected to sucrose density gradient analysis. D, left panel: AChE activity from osteoblast membranes in detergent-resistant (Raft; fractions 5–8) and detergent-soluble (non-raft) fractions was determined after flotation in discontinuous sucrose gradients with 0.5% cold Triton X-100. Aliquots of each even fraction were analyzed by 8% SDS-PAGE, and the expression of flotillin-2 (∼55 kDa) was shown in Western blots as control (upper panel). Enzymatic activities of AChE and ALP are expressed in arbitrary units (lower panel). Right panel, sedimentation profile of AChE solubilized from the raft-enriched fractions of osteoblast was determined. Representative gradient profiles from four independent experiments are shown.
The glycosylation profile of AChE during osteoblastic differentiation was determined using two lectins, Canavalia ensiformis lectin (Con A) and Sambucus nigra lectin (SNA). Con A mainly binds to high mannose glycan chains, which is abundant in both precursor and mature AChE; whereas, SNA specifically binds to sialic acid that mainly exists in complex chains of mature AChE (22). The G4 form of AChE from differentiated osteoblasts was fully precipitated by Con A; whereas a partial enzyme was precipitated by SNA (Fig. 3C). Similar glycan compositions of AChE was also revealed in differentiated cultures treated with dexamethasone plus vitamin C or Wnt3a (Fig. 3C).
The PRiMA-linked AChE in brain is known to be associated with lipid-raft (13, 23). To determine the raft association of AChE in osteoblasts, total membrane preparations were obtained from cultured osteoblasts. The raft-enriched fractions showed the presence of a raft-associated protein, flotillin-2 (Fig. 3D). These fractions contained 60% of total membrane-bound ALP that served as a membrane raft marker here. About 50% of the total membrane-bound G4 AChE activity in osteoblasts was recovered in the raft-enriched fractions (Fig. 3D). Sedimentation analysis of the raft-enriched fractions showed that the AChE tetramer (G4) was the major form (Fig. 3D).
Wnt3a induces AChE expression
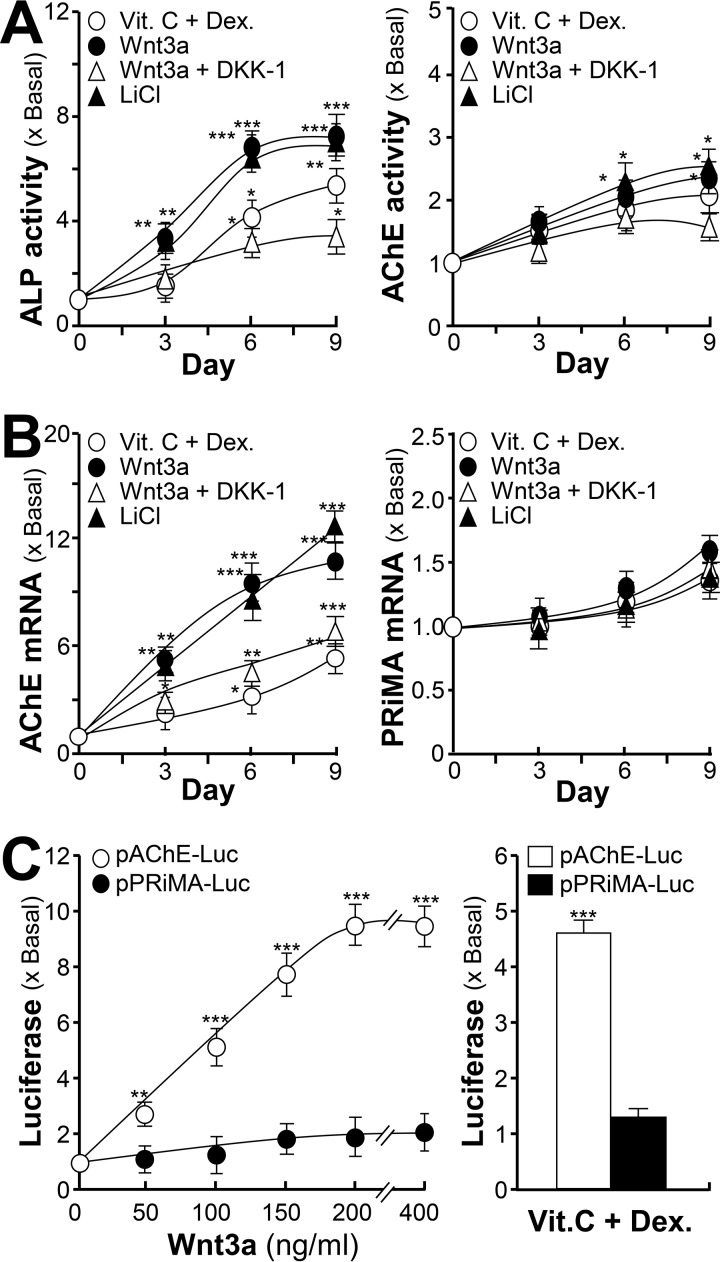
To further investigate Wnt/β-catenin signaling in AChE regulation, the validation of Wnt signaling molecules in osteoblasts was first conducted. As a crucial step in the pathway, the phosphorylation of GSK-3β was induced by treatment of Wnt3a or LiCl. LiCl is a GSK-3β inhibitor by inducing the phosphorylation of GSK-3β, used as a positive control. After 10 min of treatment, LiCl was able to induce GSK-3β phosphorylation in a transient manner (supplemental Fig. S1A). Similarly, Wnt3a was able to induce the phosphorylation of GSK-3β transiently: the maximum induction was ∼200% of increase, which was shown at 30 min, as compared with the control. DKK-1, a Wnt receptor antagonist, prevented the binding of Wnt3a onto its receptors for signal induction, was applied onto the cultures 2 h before application of LiCl, or Wnt3a. The phosphorylation of GSK-3β, induced by Wnt3a or LiCl, was inhibited after pre-treatment of DKK-1 (supplemental Fig. S1A). The nuclear translocation event of β-catenin, induced by Wnt3a, was confirmed by an increased expression of β-catenin in the nuclear fraction, which was induced by ∼3-fold (supplemental Fig. S1B).
The regulatory effect of the Wnt/β-catenin signaling pathway on AChE expression was further determined. In cultured osteoblasts, application of dexamethasone plus vitamin C, or Wnt3a or LiCl, induced the expressions of ALP and AChE in a time-dependent manner, and the induction could be blocked by DKK-1 (Fig. 4A). Moreover, the mRNA level of AChE was also induced by Wnt3a or LiCl, to ∼10-fold, and induced by dexamethasone plus vitamin C to ∼6-fold. The mRNA level of PRiMA (PRiMA I) showed a minor response to the inducers (Fig. 4B), which was consistent with the un-changed protein expression of PRiMA (data not shown). DKK-1 blocked the induction of AChE mRNA expression. These lines of evidence suggested that during osteoblastic differentiation, the elevated AChE could be a result of the Wnt/β-catenin signaling cascade activation. The transcriptional activities of Wnt3a-induced genes were tested by using promoter constructs, i.e. pAChE-Luc and pPRiMA-Luc, which were being transfected in cultured osteoblasts. The induction effect of Wnt3a on pAChE-Luc was robust in a dose-dependent manner (Fig. 4C). The induction by dexamethasone plus vitamin C was used as a positive control here. The Wnt3a/dexamethasone plus vitamin C-induced pPRiMA-Luc activity was not significant (Fig. 4C).
Figure 4.
AChE expression could be enhanced by activation of Wnt/β-catenin signaling pathway. A, primary cultured osteoblasts were treated with dexamethasone plus vitamin C (Dex. 20 nm, and Vit. C 250 μm), Wnt3a (200 ng/ml), or LiCl (10 mm) with or without pre-treatment of DKK-1 (100 ng/ml) for a 3-day interval, cell lysates were collected on different days as indicated and subjected to ALP assay and AChE assay. B, as in A, total RNAs were extracted from the cultures to perform real-time PCR analysis. The mRNA expressions of AChE and PRiMA were determined by specific primers. C, left panel: cultured osteoblasts were transfected with human ACHE promoter tagged with luciferase reporter gene (pAChE-Luc) and PRiMA promoter tagged with luciferase reporter gene (pPRiMA-Luc) before the application of Wnt3a at the indicated concentrations for 3 days. Right panel, cultured osteoblasts were transfected with pAChE-Luc and pPRiMA-Luc before the application of dexamethasone plus vitamin C for 3 days as in A. Cultures were collected for luciferase assay. Values are expressed as fold-increase to basal reading, and are mean ± S.E., n = 4, each with triplicate samples, *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Runx2 is an activator for AChE expression
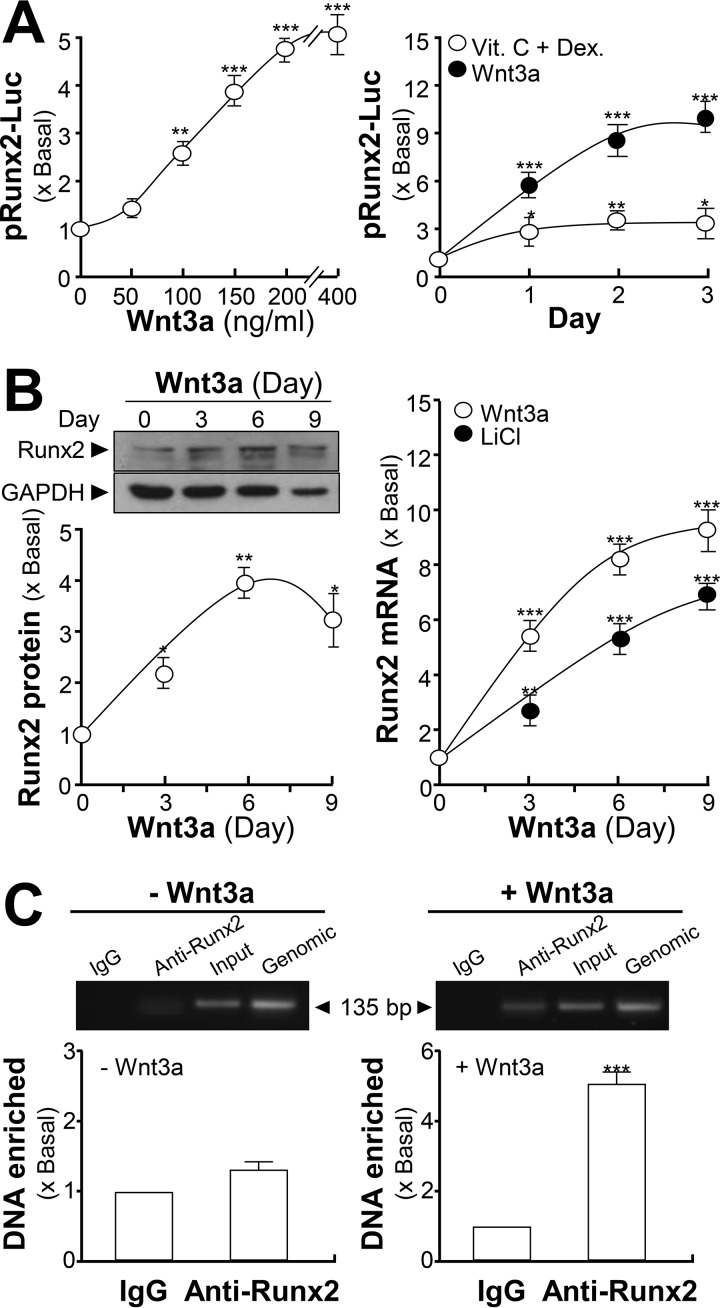
In the mammalian ACHE gene, a possible binding site for Runx2 was identified, and therefore was the target for analysis. A DNA construct pRunx2-Luc was transfected into cultured osteoblasts, and the differentiating inducers were applied. Application of Wnt3a in the transfected cells induced the promoter activity in dose- and time-dependent manners: the maximal induction was at ∼9-fold after 3 days of treatment (Fig. 5A). The treatment of dexamethasone plus vitamin C was a control having an induction of ∼3-fold. The protein expression of Runx2 (∼57 kDa) in cultured osteoblast was enhanced under Wnt3a treatment (Fig. 5B). In parallel, the mRNA level of Runx2 could be enhanced by both Wnt3a and LiCl, to ∼9- and ∼7-fold, respectively, after 9 days of treatment (Fig. 5B). The binding of Runx2 to the ACHE gene was monitored by ChIP assay. The Runx2 occupancy at ACHE promoter was barely detected in comparison to the input control. After being exposed to Wnt3a, Runx2 occupancy was induced to ∼5-fold of enrichment relative to the input control (Fig. 5C).
Figure 5.
ACHE gene has a binding site of Runx2. A, cultured osteoblasts were transfected with RUNX2 promoter tagged with a luciferase reporter gene (pRunx2-Luc) before application of Wnt3a at the indicated concentrations, or different days, under the treatment of dexamethasone plus vitamin C (Dex. 20 nm, and Vit. C 250 μm) or Wnt3a (200 ng/ml). Cultures were collected for luciferase assay. B, left panel, cultured osteoblasts were treated with Wnt3a (200 ng/μl) for 3-day intervals. Cell lysates were analyzed by Western blotting. Runx2 (∼57 kDa) and GAPDH (∼35 kDa) are shown (upper panel). The lower panel shows quantitation of protein from the blots by calibrated densitometry. Right panel, cultured osteoblasts were treated with LiCl (10 mm) or Wnt3a (200 ng/ml) for 3-day intervals. Total RNAs were extracted from the cultures to perform real-time PCR analysis. C, cultured osteoblasts were treated with or without Wnt3a (200 ng/ml) for 6 days. The quantitative ChIP assay was performed. The primers flanking the Runx2-binding site of the ACHE promoter were used: sense: GAG CTG TCA GTG TGT CCT TCC GTC; antisense: GGG GCA GAC ACC CAC GTG ACA. Results were normalized with each pull-down GAPDH Ct value. Values are expressed as fold-increase to basal reading, and are in mean ± S.E., n = 4, *, p < 0.05; **, p < 0.01; ***, p < 0.001.
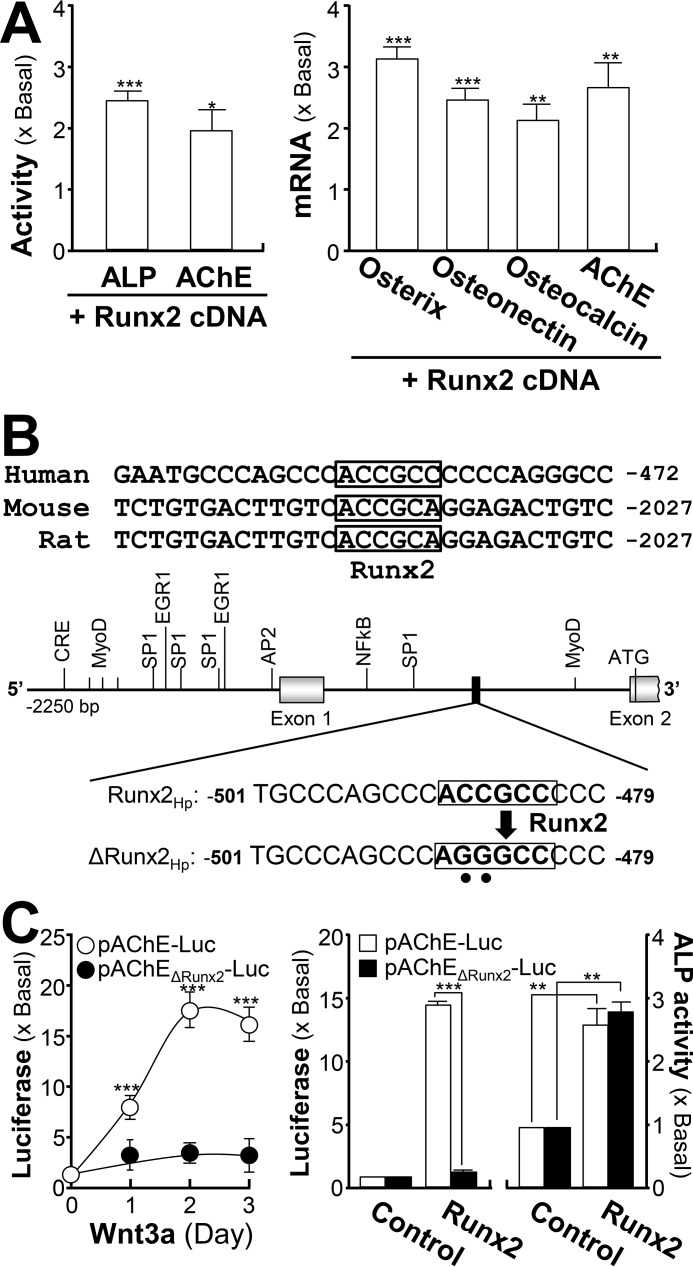
The inductive effect of Runx2 on AChE expression was further demonstrated by the overexpression system. In Runx2 cDNA-transfected osteoblasts, the enzymatic activities of ALP and AChE were induced (Fig. 6A). Similarly, the mRNA levels of bone differentiation markers, e.g. osterix, osteonectin, and osteocalcin, were induced by 2–3-fold in Runx2-overexpressed osteoblasts. In addition, the amount of AChE mRNA was increased to ∼2.5-fold (Fig. 6A). The Runx2 binding sequence within the promoters of various mammalian ACHE genes are conserved (Fig. 6B). To confirm the induction effect of Runx2 on the ACHE gene, pAChEΔRunx2-Luc, having mutation on the Runx2-binding site of the ACHE promoter, was used in transfected osteoblasts (Fig. 6B). The application of Wnt3a in the transfected osteoblasts did not show any induction on pAChEΔRunx2-Luc activity, as well as the induction by Runx2 overexpression (Fig. 6C). Being a control, the activity of pAChE-Luc was activated robustly in responding to the challenge of Wnt3a or Runx2. In contrast, the endogenous level of ALP was fully responsive to Runx2 expression in the transfected cells, i.e. serving as an internal control here (Fig. 6C).
Figure 6.
Runx2 regulates AChE transcription. A, cultured osteoblasts were transfected with pcDNA3, or cDNA-encoding Runx2, and cell lysates were collected 3 days after transfection. Left panel, cell lysates were subjected to ALP and AChE assays. Right panel, total RNAs were extracted from the cultures to perform real-time PCR analysis. B, deletion of the Runx2-binding site on the ACHE promoter was shown. A schematic diagram of human ACHE promoter with key transcription elements was shown. The Runx2-binding site sequence (ACC GCC) was mutated into AGG GCC on the human promoter. C, left panel, cultured osteoblasts were transiently transfected with pAChE-Luc and its mutant pAChEΔRunx2-Luc for 24 h before the application of Wnt3a (200 ng/ml) for 3 days. Right panel, cultured osteoblasts were transiently co-transfected with pAChE-Luc, or pAChEΔRunx2-Luc, with or without cDNA encoding Runx2. The cultures were collected 3 days later for luciferase assay and ALP assay. Values are expressed as fold-increase to basal reading, and are mean ± S.E., n = 5, each with triplicate samples, *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Epigenetic regulation of AChE during osteoblastic differentiation
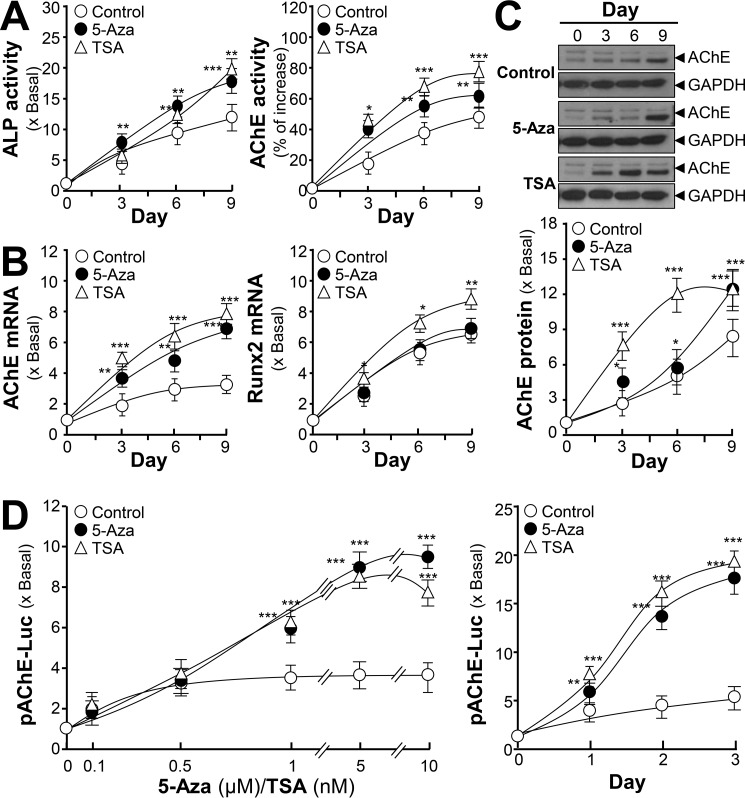
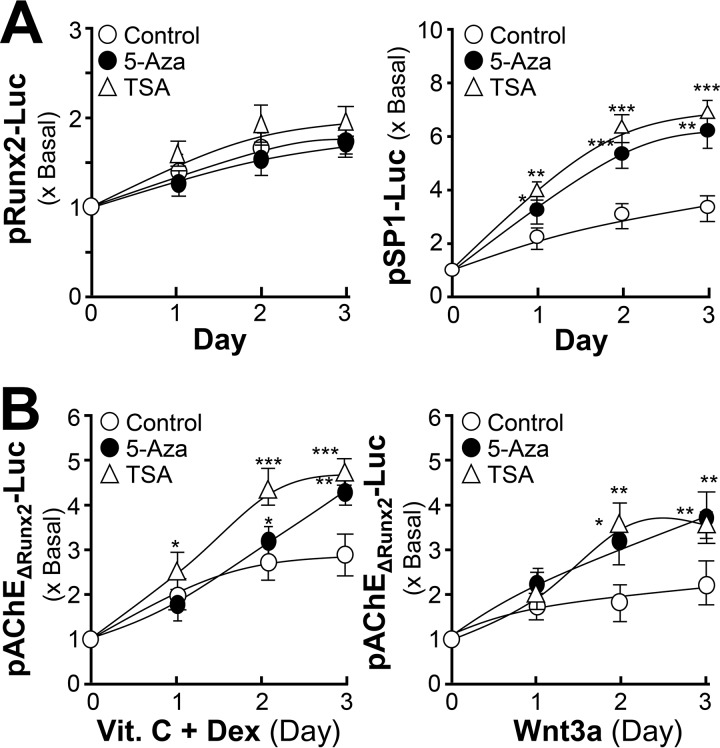
The regulation of AChE by DNA methylation was tested in cultured osteoblasts during differentiation. 5-Azacytidine (5-Aza), a DNA methyltransferases inhibitor, has been used extensively as a demethylating agent to study DNA methylation (24, 25); whereas trichostatin A (TSA) is widely used to inhibit the histone deacetylases-mediated signaling pathway (26). Two inhibitors were applied onto cultured osteoblasts. The bone differentiation markers, ALP activity and AChE activity, were induced in the present of 5-Aza and TSA during osteoblastic differentiation (Fig. 7A). In parallel, and mRNA levels of AChE and Runx2 were increased by application of 5-Aza and TSA (Fig. 7B). The amount of AChE protein was induced by 5-Aza and TSA during the differentiation (Fig. 7C). Furthermore, the pAChE-Luc activity in cultured osteoblasts was induced by 5-Aza and TSA in dose- and time-dependent manners (Fig. 7D). Thus, ACHE gene transcription should be regulated epigenetically.
Figure 7.
Epigenetic regulation of AChE during osteoblastic differentiation. A, cultured osteoblasts were treated with dexamethasone plus vitamin C (Dex. 20 nm, and Vit. C 250 μm) for 3-day intervals. During the whole process, 5-Aza (5 μm) or TSA (5 nm) were included. Cell lysates were collected on different days as indicated and subjected to ALP and AChE assays. B, as in A, total RNAs were extracted from the cultures to perform real-time PCR analysis. C, as in A, the expressions of AChE (∼68 kDa) and GAPDH (∼35 kDa) were detected by Western blot (upper panel). The lower panel shows quantitation of the AChE protein from the blots by calibrated densitometry. D, cultured osteoblasts were transiently transfected with pAChE-Luc for 24 h before treatment of dexamethasone plus vitamin C (Dex. 20 nm, and Vit. C 250 μm) for 3 days. During the whole process, 5-Aza or TSA at different concentrations as indicated were included (left panel). As in the right panel, 5-Aza (5 μm) or TSA (5 nm) were included for different days. Values are expressed as fold-increase to control reading, and are mean ± S.E., n = 4, each with triplicate samples, *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DNA methylation of ACHE gene promoter
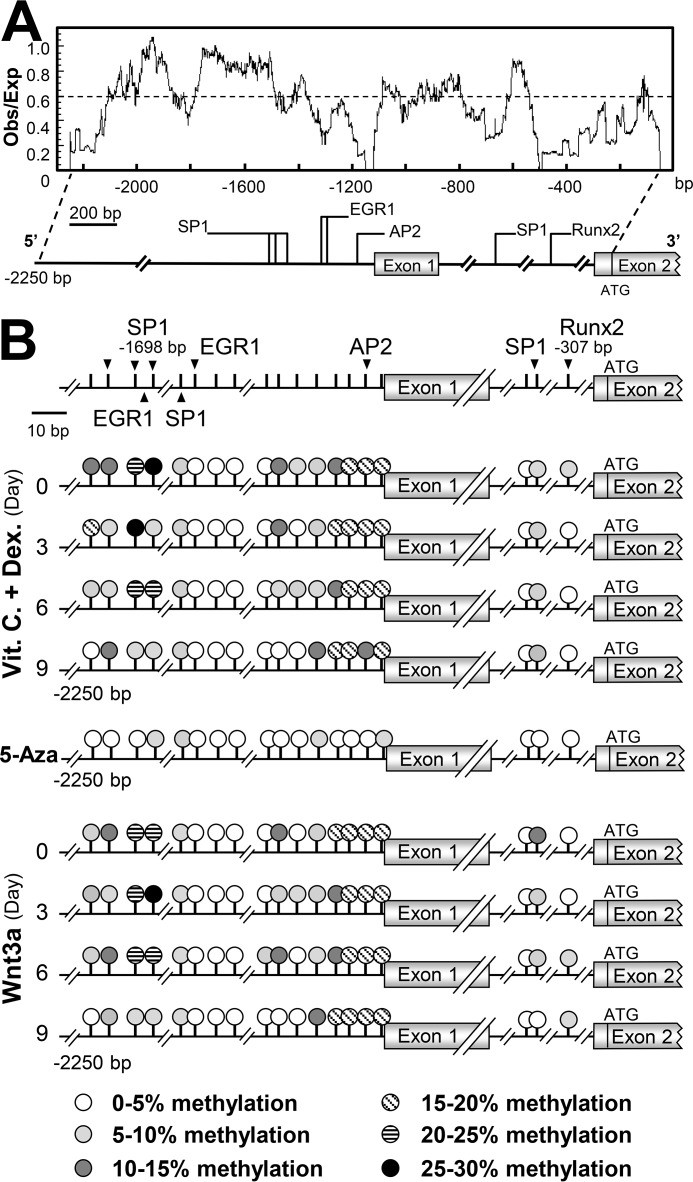
To determine DNA methylation on the ACHE promoter during osteoblastic differentiation, the CpG site-rich region in the 5′ region of the ACHE gene promoter was identified. The sequence of the rat ACHE gene was searched from the NCBI database (accession no. AF134349.1) for CpG island analysis with the CpG plot tool provided by European Bioinformatic Institute (Cambridgeshire, UK). The ratio of observed to expected (obs/exp) CpG was determined. The CpG island was defined as a sequence greater than 500 bp with the obs/exp value greater than 0.6, and the GC content was greater than 55% (25). The segment of sequence possessing a CpG obs/exp ratio greater than 0.6 was identified from −1,400 bp to −2,000 bp (Fig. 8A). This region of CpG island possesses multiple transcription factor-binding elements crucial in regulating AChE expression, e.g. specificity protein 1 (SP1), activating protein-2 (AP2), and early growth response protein 1 (EGR-1)-binding sites (Fig. 8B). The CpGs within this region and the Runx2-binding site were selected for bisulfite sequencing analysis. Pyrosequencing was performed on the genomic DNA of osteoblasts during a 9-day osteoblastic differentiation under the treatment of dexamethasone plus vitamin C or Wnt3a, and the methylation rate of each selected CpG was determined (Fig. 8B).
Figure 8.
Identification of CpG islands and methylation by bisulfite sequencing on rat ACHE promoter. A, prediction of CpG islands on the rat ACHE promoter from −2,250 bp to the start codon using EMBOSS CpG plot software was shown. Transcription elements regulating AChE expression are indicated. B, methylation status of CpGs on the rat ACHE promoter during osteoblastic differentiation induced by dexamethasone plus vitamin C (Dex. 20 nm, and Vit. C 250 μm) or Wnt3a (200 ng/ml), and under 5-Aza treatment was revealed by bisulfite pyrosequencing. The CpG site analyzed is represented by circles with different patterns. Different patterns indicates the degree of methylation of each CpG within the cell culture. The methylation under 5-Aza treatment is shown for comparison.
During the differentiation, the methylation rate of the CpG site within the Runx2 (at −307 bp) site was rather low (<5%). The co-treatment of dexamethasone plus vitamin C and application of Wnt3a did not alter the methylation (Fig. 8B). The most methylated site was within the binding site for SP1 at −1,698 bp (Fig. 8B). In addition, the promoter constructs of Runx2 and SP1, i.e. pRunx2-Luc and pSP1-Luc, were transfected into cultured osteoblasts. The transcriptional activity of pSP1-Luc, but not pRunx2-Luc, was greatly affected by application of 5-Aza or TSA (Fig. 9A). In parallel, the effects of 5-Aza and TSA were maintained in affecting the activity of pAChEΔRunx2-Luc-transfected osteoblasts during differentiation (Fig. 9B). Thus, the Runx2-mediated transcription rate of the ACHE gene should be rather independent to the epigenetic regulation; however, this regulation was significant in the SP1-mediated transcription.
Figure 9.
Epigenetic regulation of AChE during osteoblastic differentiation may be mediated by the SP1 site within the ACHE promoter. A, cultured osteoblasts were transiently transfected with pRunx2-Luc (left panel) and pSP1-Luc (right panel) for 24 h before the application of dexamethasone plus vitamin C (Dex. 20 nm, and Vit. C 250 μm) for 3 days. B, cultured osteoblasts were transiently transfected with pAChEΔRunx2-Luc for 24 h before the application of dexamethasone plus vitamin C as in A (left panel) or Wnt3a (200 ng/ml) (right panel) for 3 days. During the whole process, 5-Aza (5 μm) or TSA (5 nm) were included. Cell lysates were collected on different days as indicated and subjected to luciferase assay. Values are expressed as fold-increase to control reading, and are mean ± S.E., n = 4, each with triplicate samples, *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Discussion
The non-cholinergic function of AChE in bone development has been proposed. Supporting this notion, AChE was detected in avian cartilages (27) and in rat chondrocytes (28), and a specific form of AChE (PRiMA-linked) was revealed in osteosarcoma MG-63 cells (29) and primary osteoblasts (22). Moreover, the amount of AChE secreted by cultured osteoblasts was shown to increase after osteoblastic differentiation (21). In an in vitro study, the coating of the AChE protein onto the culture plate could facilitate the adhesion of primary osteoblasts as well as osteoblastic cell lines (16). Furthermore, the chondrogenic expression of AChE in rats was in accord with the development of lower limbs (30). These studies strongly supported the existence and a possible function of AChE in bone tissue and osteoblastic cell lines. Here, we provided different lines of evidence to support AChE playing role in the development of osteoblasts. More important, the expression of AChE in osteoblasts was under a specific regulation profile.
The elevation of AChE expression, induced by Wnt3a, indicated that AChE might be regulated by the Wnt/β-catenin-signaling pathway. Osteoblastic differentiation is predominantly regulated by Wnts, which are mammalian homologs of Drosophila-secreted morphogen wingless and bone morphogenetic proteins, which are members of the TGF-β superfamily. To confirm the response of AChE to Wnt signaling activation, LiCl was applied onto the culture. As a bipolar disorder drug, LiCl has been used for decades in patients. After LiCl treatment, a reduced risk of fracture was observed in patients (31). The osteogenic function of LiCl was driven by suppressing osteoblast apoptosis and adipogenesis while enhancing osteogenesis, i.e. inhibition of GSK-3β. As expected, the application of LiCl further supported that AChE could be enhanced by activation of Wnt/β-catenin-signaling pathway.
Runx2 is the first transcription factor required for the determination of osteoblast lineage, and a downstream transcription factor of the Wnt/β-catenin-signaling pathway. The early steps of the differentiation of mesenchymal stem cells to osteoblasts require the function of Runx2 to induce the differentiation of osteoblasts (32). Runx2 has been identified as a controller of the osteoblastic lineage differentiation. In the absence of Runx2, osteoblasts did not form (2, 3). As identified here, the binding site of Runx2 was found on the ACHE promoter, which suggested that activated Runx2 could control the primary expression of AChE in osteoblasts. By overexpression of Runx2 in osteoblasts, the expression of AChE and its transcriptional rate were increased. In addition, the Runx2-induced AChE expression was downstream of the Wnt-signaling pathway. Several lines of studies supported that the activation of Runx2 could regulate downstream target genes to promote osteogenesis. Despite an extensive search for osteoblast-specific factors, only expression and function of osteocalcin and Runx2 are limited to the osteoblast lineage (32, 33). The runt domain of Runx2 that was found to selectively bind to the osteoblast-specific elements of the osteocalcin promoter has subsequently been identified in genes of many proteins known to play significant roles in osteoblastic differentiation, including type I collagen, osteonectin, and osteopontin (34, 35). These proteins are commonly used as biomarkers of osteoblastic differentiation at different stages. As Runx2-binding sites have been identified in the upstream promoter region of the ACHE gene and the coincidence of AChE expression with the development of active osteoblast, it further supported that AChE could be used as a biomarker for osteoblastic differentiation.
In gene regulation, DNA methylation is an important mechanism underlying the cascade of events turning mesenchymal stem cell into osteoblasts (24). Studies showed that tissue-specific expression of Runx2 and osteocalcin correlated with their promoter methylation status, indicating the crucial role of DNA methylation in osteoblast-specific gene regulation (36, 37). Apart from Runx2, several transcription factor-binding elements were determined to be important for AChE transcription in neuron or muscle, e.g. intronic E- and N-box motifs (38), SP1, EGR-1, AP2, cyclic AMP-responsive element and Elk-binding site (39, 40). In addition, the epigenetic regulation of the AChE transcript was shown to be involved during muscle and neuron differentiation, and this specific regulation was proposed to be mediated by DNA methylation at a SP1 site located on the ACHE promoter (25). Here, we provided several lines of evidence supporting the hypothesis of ACHE gene transcription being controlled by DNA methylation during osteoblastic differentiation. The expression of AChE was increased during the differentiation under treatment of 5-Aza and TSA, which indicated that transcriptional activation of the ACHE promoter could be blocked by DNA methylation. During the differentiation, the methylation rate on the ACHE promoter was rather similar between the induction by dexamethasone plus vitamin C or Wnt3a, suggesting the epigenetic regulation probably triggered by the process of osteoblastic differentiation. Moreover, this regulation could be mainly mediated by the regions of SP1 sites in the ACHE promoter. This DNA methylation of SP1 site of ACHE gene transcription was also revealed in muscle differentiation (25). Nevertheless, these lines of evidence indicated that DNA methylation is one of the potential mechanisms to regulate AChE expression and histone modification might be a factor in epigenetic regulation of AChE during osteoblastic differentiation.
Our preliminary result of the decrease in differentiation makers in osteoblasts from ACHE−/− mice provided strong evidence for the participation of AChE in osteoblastic differentiation. Particularly, a defect in Wnt/β-catenin signaling cascade transduction was observed in osteoblasts from ACHE−/− mice, which may indicate the possible involvement of AChE in the Wnt/β-catenin-signaling pathway to regulate bone differentiation. 3 In addition, a preliminary result showed that osteoblastic differentiation markers could be down-regulated by antisense inhibition of AChE. This indicated that AChE might be involved in bone differentiation through a non-enzymatic mechanism, but via the structural or special role of the protein to perform a regulation in osteoblast activity. According to the studies of Spieker et al. (19), inhibition of AChE by BW284c51, or by the monoclonal antibody, delayed cartilage formation in developing embryonic chicken limbs, which could possibly be due to an enzymatic side activity of AChE. It is possible that AChE may act differently at distinct points during bone formation.
Experimental procedures
Chemicals
Vitamin C, dexamethasone, LiCl, and p-nitrophenyl phosphate were purchased from Sigma. Recombinant human Wnt3a and DKK-1 were purchased from R & D Systems (Minneapolis, MN). All culture medium and reagents were from Life Technologies.
Extraction of protein from bone
Calvarias and femurs from rats were carefully cleaned from adhering connective tissue. The prepared bones were placed in an earthen bowl to grind into powder under liquid nitrogen. The bone powders were re-suspended by gentle shaking at 4 °C in a lysis buffer (0.1 g of bone tissue/ml of buffer) containing: 50 mm Tris, pH 8.0, 150 mm NaCl2, 2 mm EDTA, 1% Triton X-100, 1% SDS, 0.5% sodium deoxycholate with addition of protease inhibitors (Sigma) (10 μg/ml of leupeptin, 10 μg/ml of aprotinin, and 2.5 mm benzamidine HCl). The supernatants were collected after centrifugation at 13,200 rpm for 20 min at 4 °C.
Cell culture
Rat primary cultured osteoblasts were prepared by the method previously described with minor modifications (41). In brief, postnatal day 1 rats were decapitated to collect calvarias. Tissues were sequentially digested by 1% of trypsin for 10 min, 0.2% of collagenase Type I (Life Technologies) for 20 min, and 0.2% of collagenase for another 40 min. The sequential digestions were placed at 37 °C under constant agitation. After the digestion, the supernatant was collected and centrifuged for 3 min at 1,500 rpm. Osteoblastic cells were re-suspended in modified Eagle's medium α (MEMα) supplemented with 10% fetal bovine serum (FBS; Life Technologies), 2 mm l-glutamine, 100 units/ml of penicillin, and 100 μg/ml of streptomycin, and maintained in a water-saturated atmosphere at 37 °C in 5% CO2, 95% air. Osteoblastic differentiation was induced by the co-treatment of vitamin C (250 μm) and dexamethasone (20 nm).
Qualitative PCR and real-time quantitative PCR
Total RNA from cultured osteoblasts was isolated by RNAzol® RT reagent (Molecular Research Center, Cincinnati, OH), and 5 μg of RNA was reverse-transcribed by Moloney murine leukemia virus reverse transcriptase (Invitrogen), according to the manufacturer's instructions. For qualitative analysis, standard PCR was performed for detecting AChET, PRiMA I, PRiMA II, ColQ-1, and ColQ-1a. Real-time PCRs of AChE, PRiMA, Runx2, and 18S rRNA transcripts were performed on equal amounts of reverse-transcribed products using Roche SYBR Green according to the manufacturer's instructions (Roche Applied Science). Primers employed were as follows: AChE catalytic subunit (5′-CTG GGG TGC GGA TCG GTG TAC CCC-3′ and 5′-TCA CAG GTC TGA GCA GCG TTC CTG-3′; NM_015831); PRiMA I (5′-TCT GAC TGT CCT GGT CAT CAT TTG CTA C-3′ and 5′-TCA CAC CAC CGC AGC GTT CAC-3′; NM_133364); PRiMA II (5′-TCT GAC TGT CCT GGT CAT CAT TTG CTA C-3′ and 5′-TCC GAT CCT CTG TGG GCC AAT C-3′; NM_178023); ColQ-1 (5′-GGT GGT CCT GAA TCC AAT G-3′ and 5′-GAA GGT TCT TCA TGT CTG G-3′; NM_009937); ColQ-1a (5′-CTT CTC CTC ATC ATT TCG G-3′ and 5′-GAA GGT TCT TCA TGT CTG G-3′; NM_009937); Runx2 (5′-AAC TTC CTG TGC TCC GTG CT-3′ and 5′-GAC TGT TAT GGT CAA GGT GAA-3′, NM_001146038); and 18S rRNA (5′-GAC TGT TAT GGT CAA GGT GAA-3′ and 5′-GAT AGT CAA GTT CGA CCG TC-3′; NR_003286). The SYBR Green signals were detected by 7500 Fast Real-time PCR system (Applied Biosystems, Foster City, CA). The relative levels of transcript expression were quantified using the 2−ΔΔCt method, where the values were normalized by the 18S as a control. The PCR products were analyzed by gel electrophoresis, and the specificity of amplification was confirmed by a melting curve.
DNA constructions and transfection
The DNAs (∼2.2 kb) encompassing the human ACHE promoters (40) with and without the Runx2-binding site were subcloned into pGL3 vector (BD Biosciences Clontech, Palo Alto, CA) upstream of a firefly luciferase gene, designated as pAChE-Luc and pAChEΔRunx2-Luc, respectively (40). A plasmid containing the Runx2 cDNA was a kind gift from Dr. Pääbo and Dr. Kuhlwilm (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany) (42). The promoter constructs of pRunx2-Luc and pSP1-Luc were described (25, 41). Transient transfection of osteoblasts with the cDNA constructs was performed with a jetPRIME reagent (Polyplus Transfection, NY), according to the manufacturer's instructions. The transfection efficiency was consistently 20–30% in the osteoblasts culture, as determined by another control plasmid having a β-galactosidase gene under a cytomegalovirus (CMV) enhancer promoter.
SDS-PAGE and Western blot analysis
For reducing SDS-PAGE, the protein lysate was denatured in the presence of 2% SDS and 100 mm β-mercaptoethanol. Anti-AChE antibody E-19 (1:10,000, Santa Cruz Biotechnology, Santa Cruz, CA), anti-PRiMA antibody (1:2000, self-generated) (13), anti-Runx2 antibody (1:100, Abcam Ltd., Cambridge, UK), anti-β-catenin antibody (1:1,000, Santa Cruz Biotechnology), anti-GSK-3β antibody (1:1,000, Santa Cruz Biotechnology), anti-phosphorylated GSK-3β antibody (1:1,000, Santa Cruz Biotechnology), and anti-GAPDH antibody (1:50,000, Invitrogen) were used for Western blot analyses. The immune complexes were visualized using the enhanced chemiluminescence (ECL) method in strictly standardized conditions (13). The intensities of bands were quantified by ImageJ2x analysis software. The labeling intensities of the protein bands were in the non-saturating range of calibration curves.
Sucrose density gradient analysis and Ellman assay
Separation of molecular forms of AChE was performed by sucrose density gradient analysis, as described previously (13). In brief, continuous 5–20% sucrose gradients containing detergent-containing buffer (10 mm HEPES, pH 7.5, 1 mm EDTA, 1 mm EGTA, 0.2% Triton X-100, and 150 mm NaCl) were prepared in 12-ml polyallomer ultracentrifugation tubes. Samples of cell extracts (0.2 ml) containing equal amounts of protein were mixed with sedimentation markers, alkaline phosphatase (6.1 S; Roche Applied Science) and β-galactosidase (16 S; Roche Applied Science), and loaded onto the gradients. Centrifugation was carried out in a SW41 Ti Rotor (Beckman, Palo Alto, CA) for 18 h at 38,000 rpm at 4 °C. Approximately 45 fractions of 0.3 ml each were collected and subjected to enzymatic activity assays. AChE activity was determined using the methods of Ellman (43), a colorimetric assay of cholinesterase activity, with minor modifications to allow microplate analysis for high-throughput screening. The cell lysates were incubated with 0.1 mm tetra-isopropylpyrophosphoramide (Sigma) for 10 min to inhibit butyrylcholinesterase activity. The amounts of AChE forms were determined by summation of the enzymatic activities corresponding to the peaks of their respective sedimentation profiles. The sedimentation values of the enzymes were calculated from the positions of the sedimentation markers.
Lectin binding analysis
Three hundred μl of cell lysates (at ∼1 μg/μl) were added onto 100 μl (hydrated volume) of agarose (control) (Vector Laboratories, Burlingame, CA) or Con A (Sigma) or SNA (Vector Laboratories), immobilized in agarose. The enzyme/lectin mixture was incubated overnight at 4 °C with gentle mixing. The unbound and precipitated AChE were separated by centrifugation at 1,000 × g for 15 min at 4 °C in a Beckman model J2–21M/E centrifuge using a JA-20 rotor, and which were subjected to further assays.
Nuclear localization assay of β-catenin
Nuclear extract was collected according to Andrews et al. (44). In brief, primary cultured osteoblast in a 100-mm tissue culture plate was washed by 1× PBS, and collected in a Eppendrof followed by centrifugation with 14,000 rpm for 10 min at 4 °C. The cell pellet was re-suspended by 800 ml of ice-cold buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm dithiothreitol (DTT), and 0.5 mm PMSF), and allowed to swell on ice for 15 min. Fifty μl of 10% Nonidet P-40 was added, and the mixture was vigorously vortex for 10 s, followed by centrifugation with 14,000 rpm for 30 s at 4 °C. The pellet was re-suspended in 100 μl of ice-cold buffer C (20 mm HEPES, pH 7.9, 0.4 m NaCl, 1 mm EGTA, 1 mm EDTA, 1 mm DTT, and 1 mm PMSF), and vigorously vortex for 15 min at 4 °C. The mixture was centrifuged with 14,000 rpm for 15 min at 4 °C. Supernatant was kept in −80 °C. The recognition of histone-1 by antibody (1:1,000, Cell Signaling Technology, Inc.) served as a control for the enrichment of nuclear fraction.
Chromatin immunoprecipitation (ChIP) assay
Primary cultured osteoblasts were cultured with or without Wnt3a treatment. After 48 h, cells were gently fixed with formaldehyde and lysed by sonication. The specific Runx2-DNA complex was immune precipitated using anti-Runx2 antibody. Primers flanking the Runx2-binding site of the ACHE promoter (with reference to AF134349.1 Rattus norvegicus chromosome 12) were used (sense: GAG CTG TCA GTG TGT CCT TCC GTC; antisense: GGG GCA GAC ACC CAC GTG ACA). The quantitative ChIP assay was performed using the Qiagen EpiTect ChIP OneDay Kit (Qiagen, UK), and the results were normalized with each pulldown GAPDH Ct value.
Bioinformatics analysis
The presence of CpG islands on the 5′-flanking region of rat ACHE gene was analyzed with the CpG plot tool of European Bioinformatic Institute (http://www.ebi.ac.uk/Tools/deqstata/emboss cpgplot) (45). 4
Bisulfite conversion and pyrosequencing
Genomic DNAs isolated from cultured rat osteoblast during osteoblastic differentiation were subjected to bisulfite conversion by the EpiTect® Bisulfite Kit (Qiagen, Germany) according to the manufacturer's instructions. Bisulfite-converted DNAs were amplified by specific biotin-labeled primer (HPLC purified) sets flanking the CpGs of interest. Primers were designed by Qiagen Gene Global Assay Design software (Qiagen, Germany). To determine the methylation rate of the CpG sites in osteoblast, the amplicons were sequenced with sequencing primer sets by Biotage-Qiagen PSQ 96MA through sub-contraction with Center of Genomic Studies at The University of Hong Kong. The primers for bisulfite-converted DNA and pyrosequencing were as follows: ACHE CpG 1–4 (5′-TTA GTT GAG GGG GTT TTT AGT TAG-3′ and 5′-AAA ATC AAA AAC CTA ATA CTT AAA CAT CTA-3′) and sequencing primer (5′-ATA CTT AAA CAT CTA TAA CCA C-3′); ACHE CpG 5–8 (5′-GGT TTG TTT GAA TTT ATT ATT GGA GTG TG-3′ and 5′-AAC TAC CAC CTC CCC TTC TC-3′) and sequencing primer (5′-GGA GTG TGT TTG GGT-3′); ACHE CpG 9–11 (5′-TTT TTA TTT TTT TTA AAG TTT GGG GAT ATT-3′ and 5′-CAA CCC TCA AAA TAA AAT AAT ACA TTC-3′) and sequencing primer (5′-GTT TGG GGA TAT TGG AA-3′); ACHE CpG 12–16 (5′-AGG GGT TTG AGT TTT GGT GA-3′ and 5′-CAC CTT ACC CCA CCC TAC T-3′) and sequencing primer (5′-GGG TGA GGT TGG TGT AAA A-3′); ACHE CpG 17–18 (5′-GGT TGG AGA AGT AGG AAT TAT AGT AGT-3′ and 5′-ACC CCT ATT ACA TCC CCA TAT T-3′) and sequencing primer (5′-TTT TTA GAT ATT TTT ATA TTA AGG-3′); ACHE CpG 19 (5′-TTT GTG TTG TAT ATT AGG GGT TTT AG-3′ and 5′-CTC CCT AAC CCA ACA AAT TTT AA-3′) and sequencing primer 5′-TGT ATA TTA GGG GTT TTA GT-3′).
Other assays
For the ALP assay, the cells were collected in lysis buffer (10 mm HEPES, pH 7.5, 1 mm EDTA, 1 mm EGTA, 150 mm NaCl, and 0.5% Triton X-100) with addition of the following protease inhibitors (Sigma): 10 μg/ml of leupeptin, 10 μg/ml of aprotinin, and 2.5 mm benzamidine HCl. The cell lysate was obtained by vortexing for 15 min and centrifuged for 10 min at 16,000 × g at 4 °C. ALP activity was measured by mixing the cell lysate with 5 mm p-nitrophenyl phosphate (Sigma) in a buffer containing 0.1 m glycine (pH 10.4), 1 mm MgCl2, and 1 mm ZnCl2 at 37 °C, and absorbance was measured at 405 nm. Luciferase assay was performed using a commercial kit (Thermo Fisher Scientific, Waltham, MA). In brief, cell cultures were washed with PBS and re-suspended in 100 mm potassium phosphate buffer (pH 7.8) containing 0.2% Triton X-100 and 1 mm DTT. Forty μl of lysate per sample was used in the luciferase assay. The luminescent reaction was quantified in a GloMax® 96 Microplate Luminometer, and the activity was expressed as absorbance (up to 560 nm) per mg of protein. Protein concentrations were measured routinely by the Bradford method with a kit from Bio-Rad. Statistical tests were performed using one-way analysis of variance; differences from basal or control values were classified as *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Author contributions
M. X. designed and performed most of the experiments, analyzed the results, and wrote most of the paper. C. B. helped to do primary culture and conducted sucrose gradient of bone tissue. E. L. helped to conduct experiments on binding of Runx2 on ACHE promoter by ChIP assay. T. D. conducted experiments searching for Runx2 function. K. T. conceived the idea for the project, and wrote the paper with M. X.
Supplementary Material
Acknowledgments
We thank Dr. Pääbo and Dr. Kuhlwilm (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany) for providing the plasmid containing the Runx2 cDNA.
This work was supported by Hong Kong Research Council Theme-based Research Scheme Grant T13–607/12R, General Research Fund Grants 663012, 662713, M-HKUST604/13, the TUYF Charitable Trust grant TUYF15SC01, and Science and Technology Plan of Shenzhen Grants JCYJ20160229205726699 (to K. T.) and JCYJ20160229205812004 (to T. D.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Fig. 1.
M L. Xu, C. W. Bi, E Y. Liu, T. T. Dong, and K. W. Tsim, unpublished results.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- Runx2
- Runt-related transcription factor 2
- AChE
- acetylcholinesterase
- ACh
- acetylcholine
- PRiMA
- proline-rich membrane anchor
- DKK-1
- Dickkopf protein-1
- 5-Aza
- 5-azacytidine
- TSA
- trichostatin
- ColQ
- collagen Q
- ALP
- alkaline phosphatase
- Con A
- Canavalia ensiformis lectin
- SNA
- Sambucus nigra lectin
- LiCl
- lithium chloride
- SP1
- specificity protein 1
- EGR-1
- early growth response protein 1
- AP2
- activating protein-2
- obs/exp
- observed to expected
- GSK
- glycogen synthase kinase.
References
- 1. Frost H. M. (1990) Skeletal structural adaptations to mechanical usage (SATMU): 4. mechanical influences on intact fibrous tissues. Anat. Rec. 226, 433–439 [DOI] [PubMed] [Google Scholar]
- 2. Stains J. P., and Civitelli R. (2003) Genomic approaches to identifying transcriptional regulators of osteoblast differentiation. Genome Biol. 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krane S. M. (2005) Identifying genes that regulate bone remodeling as potential therapeutic targets. J. Exp. Med. 201, 841–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yi P., Melnyk S., Pogribna M., Pogribny I. P., Hine R. J., and James S. J. (2000) Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J. Biol. Chem. 275, 29318–29323 [DOI] [PubMed] [Google Scholar]
- 5. Massoulié J., Bon S., Perrier N., and Falasca C. (2005) The C-terminal peptides of acetylcholinesterase: cellular trafficking, oligomerization and functional anchoring. Chem. Biol. Interact. 157–158, 3–14 [DOI] [PubMed] [Google Scholar]
- 6. Lapidot-Lifson Y., Prody C. A., Ginzberg D., Meytes D., Zakut H., and Soreq H. (1989) Coamplification of human acetylcholinesterase and butyrylcholinesterase genes in blood cells: correlation with various leukemias and abnormal megakaryocytopoiesis. Proc. Natl. Acad. Sci. U.S.A. 86, 4715–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaufer D., Friedman A., Seidman S., and Soreq H. (1998) Acute stress facilitates long-lasting changes in cholinergic gene expression. Nature 393, 373–377 [DOI] [PubMed] [Google Scholar]
- 8. Meshorer E., Erb C., Gazit R., Pavlovsky L., Kaufer D., Friedman A., Glick D., Ben-Arie N., and Soreq H. (2002) Alternative splicing and neuritic mRNA translocation under long-term neuronal hypersensitivity. Science 295, 508–512 [DOI] [PubMed] [Google Scholar]
- 9. Luk W. K., Chen V. P., Choi R. C., and Tsim K. W. (2012) N-linked glycosylation of dimeric acetylcholinesterase in erythrocytes is essential for enzyme maturation and membrane targeting. FEBS J. 279, 3229–3239 [DOI] [PubMed] [Google Scholar]
- 10. Bon S., and Massoulié J. (1997) Quaternary associations of acetylcholinesterase. I. Oligomeric associations of T subunits with and without the amino-terminal domain of the collagen tail. J. Biol. Chem. 272, 3007–3015 [DOI] [PubMed] [Google Scholar]
- 11. Leung K. W., Xie H. Q., Chen V. P., Mok M. K., Chu G. K., Choi R. C., and Tsim K. W. (2009) Restricted localization of proline-rich membrane anchor (PRiMA) of globular form acetylcholinesterase at the neuromuscular junctions: contribution and expression from motor neurons. FEBS J. 276, 3031–3042 [DOI] [PubMed] [Google Scholar]
- 12. Perry C., Sklan E. H., Birikh K., Shapira M., Trejo L., Eldor A., and Soreq H. (2002) Complex regulation of acetylcholinesterase gene expression in human brain tumors. Oncogene 21, 8428–8441 [DOI] [PubMed] [Google Scholar]
- 13. Xie H. Q., Liang D., Leung K. W., Chen V. P., Zhu K. Y., Chan W. K., Choi R. C., Massoulié J., and Tsim K. W. (2010) Targeting acetylcholinesterase to membrane rafts: a function mediated by the proline-rich membrane anchor (PRiMA) in neurons. J. Biol. Chem. 285, 11537–11546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grisaru D., Sternfeld M., Eldor A., Glick D., and Soreq H. (1999) Structural roles of acetylcholinesterase variants in biology and pathology. Eur. J. Biochem. 264, 672–686 [DOI] [PubMed] [Google Scholar]
- 15. Ofek K., and Soreq H. (2013) Cholinergic involvement and manipulation approaches in multiple system disorders. Chem. Biol. Interact. 203, 113–119 [DOI] [PubMed] [Google Scholar]
- 16. Genever P. G., Birch M. A., Brown E., and Skerry T. M. (1999) Osteoblast-derived acetylcholinesterase: a novel mediator of cell-matrix interactions in bone? Bone 24, 297–303 [DOI] [PubMed] [Google Scholar]
- 17. Grisaru D., Lev-Lehman E., Shapira M., Chaikin E., Lessing J. B., Eldor A., Eckstein F., and Soreq H. (1999) Human osteogenesis involves differentiation-dependent increases in the morphogenically active 3′ alternative splicing variant of acetylcholinesterase. Mol. Cell. Biol. 19, 788–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu M. L., Bi C. W., Kong A. Y., Dong T. T., Wong Y. H., and Tsim K. W. (2016) Flavonoids induce the expression of acetylcholinesterase in cultured osteoblasts. Chem. Biol. Interact. 259, 295–300 [DOI] [PubMed] [Google Scholar]
- 19. Spieker J., Ackermann A., Salfelder A., Vogel-Höpker A., and Layer P. G. (2016) Acetylcholinesterase regulates skeletal in ovo development of chicken limbs by ACh-dependent and -independent mechanisms. PLoS ONE 11, e0161675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cowles E. A., DeRome M. E., Pastizzo G., Brailey L. L., and Gronowicz G. A. (1998) Mineralization and the expression of aatrix proteins during in vivo bone development. Calcif. Tissue Int. 62, 74–82 [DOI] [PubMed] [Google Scholar]
- 21. Inkson C. A., Brabbs A. C., Grewal T. S., Skerry T. M., and Genever P. G. (2004) Characterization of acetylcholinesterase expression and secretion during osteoblast differentiation. Bone 35, 819–827 [DOI] [PubMed] [Google Scholar]
- 22. Xu M. L., Luk W. K., Lau K. M., Bi C. W., Cheng A. W., Gong A. G., Lin H., and Tsim K. W. (2015) Three N-glycosylation sites of human acetylcholinesterase share similar glycan composition. J. Mol. Neurosci. 57, 486–491 [DOI] [PubMed] [Google Scholar]
- 23. Chen V. P., Choi R. C., Chan W. K., Leung K. W., Guo A. J., Chan G. K., Luk W. K., and Tsim K. W. (2011a) The assembly of proline-rich membrane anchor (PRiMA)-linked acetylcholinesterase enzyme: glycosylation is required for enzymatic activity but not for oligomerization. J. Biol. Chem. 286, 32948–32961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hupkes M., Jonsson M. K., Scheenen W. J., van Rotterdam W., Sotoca A. M., van Someren E. P., van der Heyden M. A., van Veen T. A., van Ravestein-van Os R. I., Bauerschmidt S., Piek E., Ypey D. L., van Zoelen E. J., and Dechering K. J. (2011) Epigenetics: DNA demethylation promotes skeletal myotube maturation. FASEB J. 25, 3861–3872 [DOI] [PubMed] [Google Scholar]
- 25. Lau K. M., Gong A. G., Xu M. L., Lam C. T., Zhang L. M., Bi C. W., Cui D., Cheng A. W., Dong T. T., and Tsim K. W., Lin H. (2016) Transcriptional activity of acetylcholinesterase gene is regulated by DNA methylation during C2C12 myogenesis. Brain Res. 1642, 114–123 [DOI] [PubMed] [Google Scholar]
- 26. Dokmanovic M., Perez G., Xu W., Ngo L., Clarke C., Parmigiani R. B., and Marks P. A. (2007) Histone deacetylase inhibitors selectively suppress expression of HDAC7. Mol. Cancer Ther. 6, 2525–2534 [DOI] [PubMed] [Google Scholar]
- 27. Layer P. G. (1990) Cholinesterases reveal early patterns of neurogenesis in the chick. Acta Histochem. Suppl. 38, 145–150 [PubMed] [Google Scholar]
- 28. Umezu Y., Nagata N., Doi Y., Furukawa H., Sagara T., Hayashida T., Ogata H., and Fujimoto S. (1993) Cytochemical and immunocytochemical demonstration of acetylcholinesterase of the prenatal rat lower limb. Arch. Histol. Cytol. 56, 217–224 [DOI] [PubMed] [Google Scholar]
- 29. Choi R. C., Zhu T., Xie H. Q., Chu K. Y., Lau F., Chen V. P., Lin H. Q., Wan D. C,. and Tsim K. W. (2010) Osteogenic effect of PRiMA-linked acetylcholinesterase in cultured MG63 osteosarcoma cells. J. Mol. Neurosci. 40, 1–235 [Google Scholar]
- 30. Bruder S. P., Fink D. J., and Caplan A. I. (1994) Mesenchymal stem cells in bone development, bone repair, and skeletal regeneration therapy. J. Cell. Biochem. 56, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vestergaard P., Rejnmark L., and Mosekilde L. (2005) Reduced relative risk of fractures among users of lithium. Calcif. Tissue Int. 77, 1–8 [DOI] [PubMed] [Google Scholar]
- 32. Karsenty G. (2008) Transcriptional control of skeletogenesis. Annu. Rev. Genomics Hum. Genet. 9, 183–196 [DOI] [PubMed] [Google Scholar]
- 33. Ducy P., Zhang R., Geoffroy V., Ridall A. L., and Karsenty G. (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89, 747–754 [DOI] [PubMed] [Google Scholar]
- 34. Sato M., Yasui N., Nakase T., Kawahata H., Sugimoto M., Hirota S., Kitamura Y., Nomura S., and Ochi T. (1998) Expression of bone matrix proteins mRNA during distraction osteogenesis. J. Bone Miner. Res. 13, 1221–1231 [DOI] [PubMed] [Google Scholar]
- 35. Tyson K. L., Reynolds J. L., McNair R., Zhang Q., Weissberg P. L., and Shanahan C. M. (2003) Osteo/chondrocytic transcription factors and their target genes exhibit distinct patterns of expression in human arterial calcification. Arterioscler. Thromb. Vasc. Biol. 23, 489–494 [DOI] [PubMed] [Google Scholar]
- 36. Villagra A., Gutiérrez J., Paredes R., Sierra J., Puchi M., Imschenetzky M., Wijnen Av, Lian J., Stein G., Stein J., and Montecino M. (2002) Reduced CpG methylation is associated with transcriptional activation of the bone-specific rat osteocalcin gene in osteoblasts. J. Cell. Biochem. 85, 112–122 [PubMed] [Google Scholar]
- 37. Kang M. I., Kim H. S., Jung Y. C., Kim Y. H., Hong S. J., Kim M. K., Baek K. H., Kim C. C., and Rhyu M. G. (2007) Transitional CpG methylation between promoters and retroelements of tissue-specific genes during human mesenchymal cell differentiation. J. Cell. Biochem. 102, 224–239 [DOI] [PubMed] [Google Scholar]
- 38. Angus L. M., Chan R. Y., and Jasmin B. J. (2001) Role of intronic E- and N-box motifs in the transcriptional induction of the acetylcholinesterase gene during myogenic differentiation. J. Biol. Chem. 276, 17603–17609 [DOI] [PubMed] [Google Scholar]
- 39. Siow N. L., Choi R. C., Cheng A. W., Jiang J. X., Wan D. C., Zhu S. Q., and Tsim K. W. (2002) A cyclic AMP-dependent pathway regulates the expression of acetylcholinesterase during myogenic differentiation of C2C12 cells. J. Biol. Chem. 277, 36129–36136 [DOI] [PubMed] [Google Scholar]
- 40. Choi R. C., Siow N. L., Cheng A. W., Ling K. K., Tung E. K., Simon J., Barnard E. A., and Tsim K. W. (2003) ATP acts via P2Y1 receptors to stimulate acetylcholinesterase and acetylcholine receptor expression: transduction and transcription control. J. Neurosci. 23, 4445–4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guo A. J., Choi R. C., Cheung A. W., Chen V. P., Xu S. L., Dong T. T., Chen J. J., and Tsim K. W. (2011) Baicalin, a flavone, induces the differentiation of cultured osteoblasts: an action via the Wnt/β-catenin signaling pathway. J. Biol. Chem. 286, 27882–27893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kuhlwilm M., Davierwala A., and Pääbo S. (2013) Identification of putative target genes of the transcription factor RUNX2. PLoS ONE 8, e83218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ellman G. L., Courtney K. D., Andres V. Jr., Feather-Stone R. M. (1961) A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 7, 88–95 [DOI] [PubMed] [Google Scholar]
- 44. Andrews N. C., and Faller D. V. (1991) A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19, 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y. M., Buso N., and Lopez R. (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W580–W584 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.