Abstract
Lysine methylation of chromosomal and nuclear proteins is a well-known mechanism of epigenetic regulation, but relatively little is known about the role of this protein modification in signal transduction. Using an RNAi-based functional screening of the SMYD family of lysine methyltransferases (KMTs), we identified SMYD2 as a KMT essential for robust bone morphogenic protein (BMP)- but not TGFβ-induced target gene expression in HaCaT keratinocyte cells. A role for SMYD2 in BMP-induced gene expression was confirmed by shRNA knockdown and CRISPR/Cas9-mediated knock-out of SMYD2. We further demonstrate that SMYD2 knockdown or knock-out impairs BMP-induced phosphorylation of the signal-transducing protein SMAD1/5 and SMAD1/5 nuclear localization and interaction with SMAD4. The SMYD2 KMT activity was required to facilitate BMP-mediated signal transduction, as treatment with the SMYD2 inhibitor AZ505 suppressed BMP2-induced SMAD1/5 phosphorylation. Furthermore, we present evidence that SMYD2 likely modulates the BMP response through its function in the cytosol. We show that, although SMYD2 interacted with multiple components in the BMP pathway, it specifically methylated the kinase domain of BMP type II receptor BMPR2. Taken together, our findings suggest that SMYD2 may promote BMP signaling by directly methylating BMPR2, which, in turn, stimulates BMPR2 kinase activity and activation of the BMP pathway.
Keywords: protein methylation, receptor, signal transduction, SMAD transcription factor, transcription regulation
Introduction
Lysine methylation has been extensively studied in the context of histone proteins as a mechanism of epigenetic regulation (1–3). With the presence of a large number of lysine methyltransferases (KMTs) 4 in mammalian genomes (4, 5), it is not surprising that an increasingly larger number of non-histone chromosomal, nuclear, and cytoplasmic proteins have been found to be lysine-methylated (6–9). Lysine methylation can be dynamically removed by the action of demethylases (10, 11), making it a feasible mechanism for signal transduction. However, until now lysine methylation has been shown to regulate signaling proteins only in limited cases. For instance, methylation of MAP3K2 by SMYD3 has been shown to increase MAPK signaling and to promote the formation of Ras-driven carcinomas (12), whereas methylation of VEGFR1 by SMYD3 activates its kinase activity (13), indicating that lysine methylation can play important roles in regulation of signal transduction.
The TGF-β/BMP superfamily of cytokines plays pleiotropic roles in embryonic development, differentiation, organ morphogenesis, and tissue homeostasis (14, 15). These cytokines bind the cell-surface type I and type II receptors that are also serine/threonine protein kinases (16, 17). Upon ligand binding, the type II receptor activates the type I receptor by phosphorylating the GS motif of the type I receptor (18). The activated type I receptor in turn phosphorylates the regulatory SMADs (R-SMADs) at the C-terminal SSXS motif. This phosphorylation disrupts inhibitory intramolecular interaction between MH2 and MH1 domains of R-SMADs and stimulates R-SMADs to form a heteromeric complex with the co-SMAD protein SMAD4, which can translocate to the nucleus and regulate target gene expression (19, 20). The specificity of the TGFβ and BMP pathways are largely mediated by distinct R-SMADs. TGFβ pathway typically activates SMAD2/3, whereas the BMP pathway typically activates SMAD1/5/8 (14, 21). Given its functional significance, the TGFβ/BMP pathway is tightly regulated at different levels through various mechanisms to ensure that signaling is controlled in a spatial and temporal manner (21, 22). However, except that the methyltransferase SET has recently been reported to potentiate TGFβ signaling by methylating SMAD7 (23), it is not known prior to our study whether other component(s) of the TGFβ/BMP-signaling pathway are also regulated by lysine methylation.
To investigate whether the TGFβ/BMP pathway is regulated by lysine methylation, we focused our attention on the SMYD family KMTs. Characterized by a split SET catalytic domain inserted with a zinc finger MIND motif, the SMYD family KMTs include five members, SMYTH-5 (24). Unlike many other KMTs that are localized mainly in the nucleus, the SMYD KMTs are abundantly cytoplasmic and thus could have a regulatory role in early stages of signal transduction (12, 24, 25). In support of this idea, SMYD3 has been shown to methylate MAP3K2, linking MAP3K2 lysine methylation to Ras-driven cancer (12). Using an RNAi-based functional screening, here we identified SMYD2 as a positive regulator for BMP2-induced but not TGFβ-induced target gene expression. We provide evidence that SMYD2 promotes BMP2-induced SMAD1/5 phosphorylation, SMAD1/5 nuclear localization, and interaction with SMAD4. We show that SMYD2 specifically methylates the kinase domain of BMP type II receptor BMPR2. Taken together, our study suggests a working model that SMYD2 may positively regulate BMP signaling by directly methylating BMPR2.
Results
RNAi-based screening reveals a role for SMYD2 in BMP- but not TGFβ-induced target gene expression
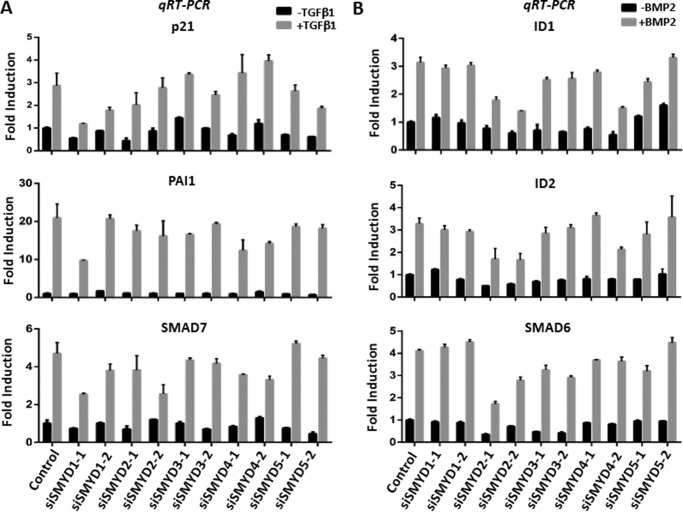
To investigate the potential function of SMYD family KMTs in TGFβ and/or BMP signaling transduction, we performed RNA interference to knock down individual SMYD in HaCaT cells, a spontaneously immortalized human keratinocyte line widely used for studies of TGFβ/BMP response, with specific siRNAs. To minimize the off-target effect of siRNA screening, two different siRNAs were employed for each SMYD gene, and the result was scored positive only when it was observed for both siRNAs. Two days after siRNA treatment, HaCaT cells were cultured with serum-free medium overnight and then treated with TGFβ1 or BMP2 for 4 h before harvesting for analysis of TGFβ or BMP-induced target gene expression by quantitative reverse transcription-PCR (qRT-PCR). Through multiple experiments, we did not observe a reproducible effect of SMYD knockdown on TGFβ1-induced expression of three representative TGFβ target genes p21, PAIL, and SMAD7 (Fig. 1A) (26). However, siRNA knockdown of SMYD2 consistently impaired basal and BMP2-induced transcriptional activation of three BMP target genes ID1, ID2, and SMAD6 (27), although knockdown of other SMYD proteins had no clear effect (Fig. 1B). These results suggest a potential role for SMYD2 in promoting BMP2-induced target gene expression.
Figure 1.
siRNA-based screening reveals a specific role for SMYD2 in BMP2-induced target gene expression. A, knockdown of SMYD family KMTs by siRNAs did not affect TGFβ1-induced target gene activation. HaCaT cells were transfected with the siRNAs against SMYD1 to SMYD5 as indicated. Two days later, cells were starved overnight with non-serum medium and then treated with 5 ng/ml TGFβ1 for 4 h. The cells were harvested for preparation of total RNAs and subsequent qRT-PCR analysis of PAI1, p21, and SMAD7 transcripts. B, knockdown of SMYD2 but not other SMYDs specifically impairs BMP2-induced target gene expression. HaCaT cells were treated as above except that TGFβ1 was replaced with 25 ng/ml BMP2, and qRT-PCR analysis was performed for BMP target genes ID1, ID2, and SMAD6.
Validating the role of SMYD2 in BMP-signaling pathway
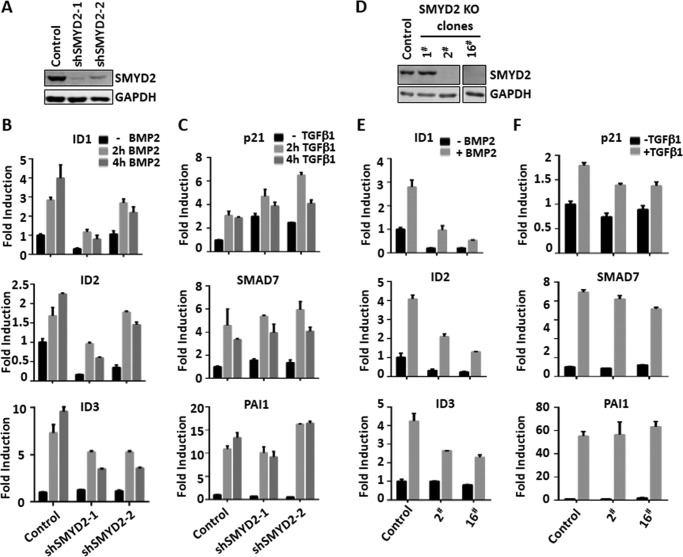
To validate the results of our RNAi-based screen, we constructed two shRNAs against SMYD2 that were targeted at different sequences of SMYD2 from the two siRNAs used in our screen in the lentiviral vector. Stable infection of HaCaT cells with lentiviral shRNAs resulted in substantial knockdown of SMYD2 as revealed by Western blot analysis (Fig. 2A). Importantly, we observed a significantly diminished induction of BMP target genes ID1, ID2, and ID3 in both shSMYD2 stably transfected HaCaT cells in comparison with the control HaCaT cells (Fig. 2B). In contrast, the expression of TGFβ downstream genes p21, PAI1, and SMAD7 was not significantly affected by SYMD2 knockdown (Fig. 2C). Thus, consistent with our siRNA screening results, SMYD2 knockdown by distinct shRNAs also selectively impaired the BMP-signaling pathway without affecting the TGFβ pathway. To further substantiate this observation, we made use of CRISPR/CAS9 technology to knock out SMYD2 in HaCaT cells. We verified deletion of various fragments in the SMYD2-coding region by DNA sequencing (data not shown) and confirmed the loss of SMYD2 proteins by Western blot analysis in isolated clones 2 and 16 (Fig. 2D). Subsequent qRT-PCR analysis demonstrated that SMYD2 knock-out indeed impaired BMP2-induced downstream gene activation but had no effect on TGFβ-induced target gene activation (Fig. 2, E and F). Together these data reveal a unique role of SYMD2 in the BMP- but not TGFβ-induced signaling pathway.
Figure 2.
Validating the role of SMYD2 in regulating BMP but not TGFβ target gene expression. A, Western blot analysis showing substantially reduced levels of SMYD2 in HaCaT cells stably infected with two different lentivirus-based shRNAs (shSMYD2-1 and -2). B, shRNA-infected HaCaT cells were serum-starved overnight and then stimulated with BMP2 for 0, 2, and 4 h as indicated. The cells were harvested, and qRT-PCR was performed to detect BMP downstream genes ID1, ID2, and ID3. C, shRNA-infected HaCaT cells were serum-starved overnight and then stimulated with TGFβ1 for 0, 2, and 4 h as indicated. The cells were harvested, and qRT-PCR was performed to detect TGFβ downstream genes p21, SMAD7, and PAI1. D, Western blot analysis showing the absence of SMYD2 proteins in SMYD2 knock-out HaCaT cell lines 2 and 16. The HaCaT knock-out cell lines were generated via CRISPR/CAS9 approach as described under “Experimental procedures.” E, SMYD2 KO 2 and 16 cell lines were serum-starved overnight and then stimulated without or with BMP2 for 4 h as indicated. The cells were harvested, and qRT-PCR was performed to detect BMP downstream genes ID1, ID2, and ID3. F, SMYD2 KO 2 and 16 cell lines were serum-starved overnight and then stimulated without or with TGFβ1 for 4 h as indicated. The cells were harvested, and qRT-PCR was performed to detect TGFβ downstream genes p21, SMAD7, and PAI1.
SMYD2 knockdown or knock-out impairs BMP2-induced SMAD1/5 phosphorylation
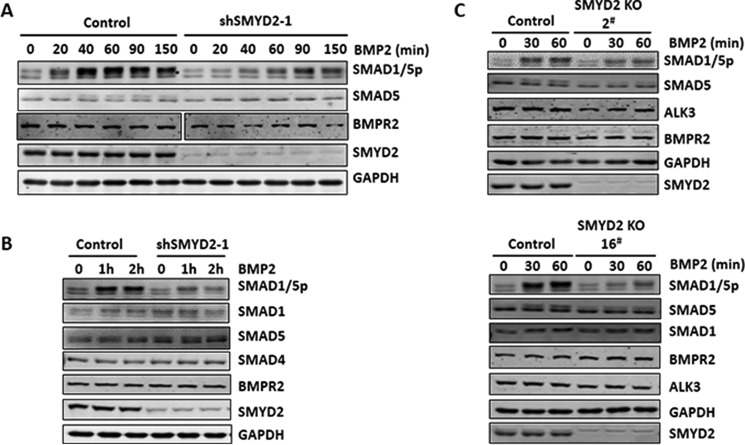
To investigate the molecular mechanism(s) by which SMYD2 regulates the BMP-signaling pathway, we first compared the status of BMP-induced SMAD1/5 phosphorylation by using a commercial antibody that recognizes both phosphorylated SMAD1 and SMAD5 in control HaCaT cells and HaCaT cells stably expressing shSMYD2-1, which down-regulated SMYD2 more effectively than shSMYD2-2 (Fig. 2A). As expected, BMP2 treatment resulted in increased levels of SMAD1/5 phosphorylation in a time course from 20 to 60 min and maintained a high level of phosphorylation up to 150 min (Fig. 3A). Knockdown of SMYD2 substantially impaired BMP-induced SMAD1/5 phosphorylation (Fig. 3A). However, no significant difference in the levels of SMAD5 and the BMP type 2 receptor BMPR2 was observed between the control and SMYD2 knockdown cells. Consistent with these observations, additional experiments in Fig. 3B show a reduced BMP2-induced SMAD1/5 phosphorylation in SMYD2 knockdown cells, yet there was no difference in the levels of SMAD1, SMAD5, and SMAD4 between the control and SMYD2 knockdown cells. We further tested the role of SMYD2 in BMP-induced SMAD1/5 phosphorylation in SMYD2 knock-out HaCaT cell lines 2 and 16. As shown in Fig. 3C, SMYD2 knock-out had no effect on the levels of SMAD1, SMAD5, type 2 receptor BMPR2, and type 1 receptor ALK3 proteins. However, SMYD2 knock-out markedly reduced the levels of BMP2-induced SMAD1/5 phosphorylation. Together, these data clearly reveal a role for SMYD2 in promoting BMP2-induced SMAD1/5 phosphorylation.
Figure 3.
Impaired SMAD1/5 phosphorylation upon SMYD2 knockdown or knock-out. A, knockdown of SMYD2 by siRNAs impaired SMAD1/5 phosphorylation induced by BMP2. HaCaT cells were transfected with SMYD2 siRNAs (siSMYD2-1 and -2). Two days later, the cells were serum-starved overnight and stimulated with BMP2 for 0, 15, 30, and 45 min as indicated, and the levels of SMAD5, phosphorylated SMAD1/5 (SMAD1/5ph), and SMYD2 were analyzed by Western blotting. GAPDH was shown as the loading control. B, knockdown of SMYD2 by shRNAs impaired SMAD1/5 phosphorylation induced by BMP2. The stable shSMYD2-1 HaCaT cells were serum-starved overnight and stimulated with BMP2 for 0, 2, and 4 h and subsequently analyzed by Western blotting using antibodies as indicated. C, stable shSMYD2–1 HaCaT cells were serum-starved overnight and stimulated with BMP2 for 0, 2, and 4 h and subsequently analyzed by Western blotting using antibodies as indicated. D, knock-out of SMYD2 by CRISPR/CAS9 impaired SMAD1/5 phosphorylation induced by BMP2. The control and SMYD2 KO 2 and 16 cell lines were serum-starved overnight and then stimulated without or with BMP2 for 0, 30, and 60 min and subsequently analyzed by Western blotting using antibodies as indicated.
SMYD2 knockdown and knock-out impair BMP2-induced SMAD1/5 nuclear translocation, complex formation with SMAD4, and inhibition of cell proliferation
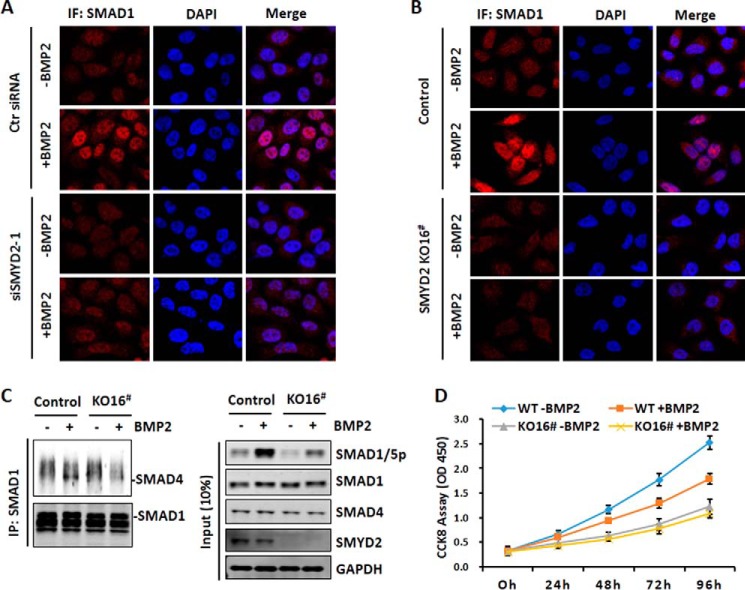
Phosphorylation of SMAD1/5 promotes translocation of SMAD1/5 from cytoplasm to nucleus and interaction with co-SMAD SMAD4 (14). Having observed that knockdown or knock-out of SMYD2 impaired BMP2-induced SMAD1/5 phosphorylation, we next tested its effect on SMAD1/5 nuclear entry and interaction with SMAD4. As shown in Fig. 4A, we observed that knockdown of SMYD2 indeed reduced BMP2-induced SMAD1 accumulation in the nucleus. Similarly, we found that BMP2-induced SMAD1 nuclear accumulation was substantially reduced in SMYD2 KO-16 cells. Furthermore, by co-immunoprecipitation assay, we observed that although BMP2 treatment stimulated the association of SMAD4 with SMAD1 in the control wild-type cells, much less SMAD4 was found to co-immunoprecipitate with SMAD1 in BMP2-treated SMYD2 KO-16 cells (Fig. 4C). Thus, consistent with an impaired BMP2-induced SMAD1/5 phosphorylation in SMYD2 knockdown or knock-out cells, BMP2-induced SMAD1 nuclear entry and the interaction with SMAD4 are also impaired upon SMYD2 knockdown or knock-out.
Figure 4.
Impaired SMAD1/5 nuclear entry and interaction with SMAD4 upon SMYD2 knockdown or knock-out. A, immunofluorescence (IF) staining analysis showing impaired BMP2-induced nuclear translocation upon knockdown of SMYD2 by siRNA. HaCaT cells were treated with or without siSMYD2-1 for 2 days. The cells were then serum-starved overnight, stimulated without or with BMP2 for 25 min, and processed for immunofluorescent staining analysis using an anti-SMAD1 antibody. B, immunofluorescence staining analysis showing impaired BMP2-induced nuclear translocation upon knock-out of SMYD2. The SMYD2 KO-16 cells were serum-starved overnight, stimulated without or with BMP2 for 25 min, and processed for immunofluorescent staining analysis. C, impairment of BMP2-induced interaction between SMAD1 and SMAD4 upon knock-out of SMYD2. Control cells and SMYD2 KO-16 cells were serum-starved overnight and stimulated with BMP2 for 40 min. The cells were harvested, and co-immunoprecipitation assay was performed using anti-SMAD1 antibody followed by Western blot analysis using anti-SMAD4 and anti-SMAD1 antibodies. D, SMYD2 knock-out diminished cell proliferation inhibition by BMP2. The wild-type and SMYD2 KO-16 cells were treated without or with 25 ng/ml BMP2 for up to 96 h, and cell proliferation was assayed by CCK8 assay at 0, 24, 48, 72, and 96 h as indicated.
We next tested whether SMYD2 knock-out affected the BMP-induced biological effect such as inhibition of cell proliferation in HaCaT cells. Compared with parent HaCaT cells, SYMD2 KO reduced cell proliferation (Fig. 4D), suggesting that SMYD2 may regulate cell proliferation through unknown mechanism. Nevertheless, although addition of BMP2 inhibited the proliferation of the wild-type HaCaT cells, addition of BMP2 had no significant inhibition on the proliferation of SMYD2 KO cells (Fig. 4D). Thus, SMYD2 not only promotes BMP2-induced gene expression but also BMP2-induced inhibition of cell proliferation.
SMYD2 regulates BMP signaling in cytoplasm and requires its KMT activity
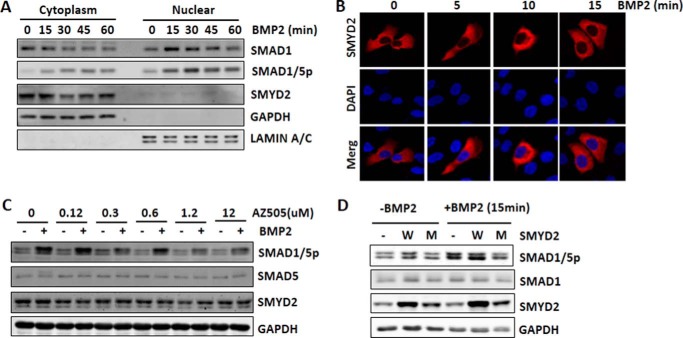
Our findings that SMYD2 knockdown or knock-out impairs BMP2-induced SMAD1/5 phosphorylation suggest that SMYD2 is likely to regulate BMP signaling at a step(s) in cytoplasm before SMAD1/5 nuclear translocation. To pinpoint its role in the BMP-signaling pathway, we first examined the subcellular localization of SMYD2 and the effect of BMP2 treatment on its subcellular localization in HaCaT cells. As expected, we observed that BMP2 treatment led to time-dependent reduction of cytosol SMAD1 and accumulation of nuclear SMAD1 (Fig. 5A). Also, as expected, BMP2 treatment led to increased levels of phosphorylated SMAD1/5 both in the cytosol and nucleus. However, SMYD2 remained in the cytoplasm through BMP2 treatment (Fig. 5A). The appropriate fractionation of the cytoplasmic and nuclear fraction was confirmed by Western blot analysis of the cytosolic marker GAPDH and the nuclear marker lamin A/C (Fig. 5A). Furthermore, by immunofluorescent staining we observed that although BMP treatment induced rapid SMAD1 nuclear accumulation (Fig. 4, A and B), SMYD2 remained in the cytoplasm throughout the treatment (Fig. 5B). Together, these data suggest that SMYD2 regulates BMP signaling in the cytoplasm and is unlikely to exert its regulatory role through a nuclear function.
Figure 5.
SMYD2 regulates BMP signaling transduction in cytoplasm in a KMT activity-dependent manner. A, nucleocytoplasmic fractionation showing that SMYD2 remained in cytoplasm regardless of BMP treatment. HaCaT cells were serum-starved overnight, treated with 25 ng/ml BMP2 for various times as indicated, and then subjected to nucleocytoplasmic fractionation and Western blotting with the antibodies against SMAD1, phosphorylated SMAD1/5, and SMYD2. GAPDH and lamin A/C served as control for the cytoplasmic and nuclear fraction, respectively. B, immunofluorescent staining shows that SMYD2 remained in cytoplasm upon BMP treatment. HaCaT cells were transfected with FLAG-SMYD2 for 24 h followed by serum starvation overnight. The cells were then treated with 25 ng/ml BMP2 for various times as indicated and subjected to immunofluorescent staining using anti-FLAG antibody. Nuclei were revealed by DAPI staining. C, SMYD2 selective inhibitor AZ505 diminished BMP2-induced SMAD1/5 phosphorylation. HaCaT cells were serum-starved for 4 h and then treated with increasing concentrations of SMYD2 inhibitor AZ505 for 6 h. The cells were then treated with (+) or without (−) 25 ng/ml BMP2 for 1 h before harvesting for Western blot analysis using antibodies as indicated. Note that BMP2-induced SMAD1/5 phosphorylation was inhibited by addition of 1.2 and 12 μm AZ505. D, enzymatic activity-deficient SMYD2 acted as dominant-negative inhibitor of the BMP signaling. HaCaT cells were transfected with ppY-CAGIP plasmids encoding the wild-type (W) or SMYD2 Y240A mutant (M) at conditions of high transfection efficiency. Two days later, the cells were treated without or with 25 ng/ml BMP2 for 15 min before harvesting for Western blot analysis using antibodies as indicated.
To examine whether KMT activity is required for SMYD2 to regulate the BMP-signaling pathway, we resorted to a recently identified potent and selective SMYD2 inhibitor AZ505 (28). AZ505 was shown to inhibit SMYD2 KMT activity with an IC50 of 0.12 μm, whereas the IC50 value for other KMTs is more than 83.3 μm (28). We found that treatment of HaCaT cells with 1.2 and 12 μm AZ505 was sufficient to inhibit BMP2-induced SMAD1/5 phosphorylation without affecting the protein levels of SMYD2 and SMAD5 (Fig. 5C). Considering that knockdown of SMYD2 but not other SMYD proteins impaired the BMP-signaling pathway (Fig. 1), these results provide evidence that inhibition of SMYD2 KMT activity is sufficient to impair BMP signaling transduction. To substantiate this further, we tested whether ectopic overexpression of an enzymatic inactive form of SMYD2, Y240A mutant, would act as a dominant negative to inhibit BMP2-induced SMAD1/5 phosphorylation. As shown in Fig. 5D, we observed that although ectopic expression of the wild-type SMYD2 further enhanced BMP2-induced SMAD1/5 phosphorylation, ectopic expression of SMYD2 mutant actually inhibited the BMP2-induced SMAD1/5 phosphorylation (Fig. 5D). Taking into consideration that the SMYD2 mutant was expressed only 2–3-fold more than the level of endogenous SMYD2 proteins (Fig. 5D), the observed inhibition of BMP2-induced SMAD1/5 phosphorylation provides compelling evidence for a dominant-negative effect of the SMYD2 KMT-deficient mutant.
SMYD2 interacts with multiple components in the BMP pathway
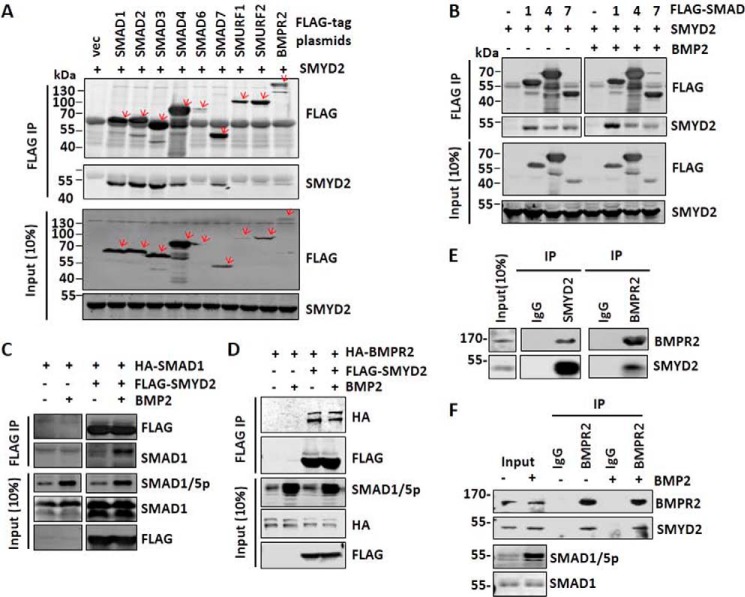
As SMYD2 requires its catalytic activity to facilitate BMP signaling transduction, we predicted that SMYD2 may regulate BMP signaling through methylating one or more components in the BMP-signaling pathway. To identify potential SMYD2 substrates, we first aimed to examine the interaction of SMYD2 with various components of the BMP/TGFβ signaling pathways, including SMADs, SMURFs, and BMPR2. To this end, 293T cells were co-transfected with SMYD2 and FLAG-tagged SMADs, SMURF1/2, or BMPR2, and co-immunoprecipitations were performed with anti-FLAG antibody (Fig. 6A). Interestingly, we found that SMYD2 co-immunoprecipitated with multiple proteins in the BMP and TGFβ pathways, including SMAD1–4, SMAD7, SMURF2, and BMPR2. The observed interaction was likely authentic because SMYD2 was not precipitated by anti-FLAG antibody when expressed alone (Fig. 6A, 1st lane). As SMYD2 specifically affected the BMP but not the TGFβ pathway, we further analyzed whether BMP2 treatment regulated the interaction of SMYD2 with SMAD1, SMAD4, and inhibitory SMAD7 by the co-immunoprecipitation assay. We confirmed that SMYD2 co-immunoprecipitated with FLAG-tagged SMAD1, SMAD4, and SMAD7 (Fig. 6B). Furthermore, we observed that BMP treatment enhanced the interaction between SMYD2 and SMAD1 but not the interaction between SMYD2 and SMAD4 and SMAD7 (Fig. 6B). Such a BMP-induced SMYD2-SMAD1 interaction was further confirmed by reciprocal co-immunoprecipitation using HA-SMAD1 and FLAG-SMYD2 (Fig. 6C). Finally, the interaction between SMYD2 and BMPR2 was also confirmed by reciprocal co-immunoprecipitation using HA-BMPR2 and FLAG-SMYD2, although in this case the interaction was not enhanced by BMP treatment (Fig. 6D).
Figure 6.
SMYD2 interacts with multiple components of the TGFβ/BMP-signaling pathways. A, co-immunoprecipitation assay showing interaction between SMYD2 and various components of the TGFβ/BMP-signaling pathways. 293T cells were co-transfected with SMYD2 and FLAG-tagged SMADs, SMURFs, and BMPR2. Two days later, the cells were harvested for immunoprecipitations (IP) using anti-FLAG antibody, followed by Western blot analysis using anti-FLAG and SMYD2 antibodies. B, co-immunoprecipitation assay showing increased interaction between SMYD2 with SMAD1 upon BMP2 treatment. 293T cells were co-transfected with SMYD2 and FLAG-SMAD1, FLAG-SMAD4, or FLAG-SMAD7 for 24 h. The cells were serum-starved overnight and treated with (+) or without (−) 25 ng/ml BMP2 for 1 h, followed by co-immunoprecipitation assay using anti-FLAG antibody. The co-immunoprecipitated SMYD2 was detected by Western blot analysis. Note that BMP treatment only enhanced the SMYD2-SMAD1 interaction. C, reciprocal co-immunoprecipitation assay confirmed a BMP-induced interaction between SMYD2 and SMAD1. 293T cells were co-transfected with FLAG-SMYD2 and HA-SMAD1 for 24 h, serum-starved overnight, and treated with or without 25 ng/ml BMP2 for 1 h, followed by co-immunoprecipitation assay using antibodies as indicated. D, BMP2 treatment did not affect the interaction between SMYD2 with BMPR2. 293T cells were co-transfected with FLAG-SMYD2 and HA-BMPR2 for 24 h, serum-starved overnight, and treated with or without 25 ng/ml BMP2 for 1 h, followed by co-immunoprecipitation assay using antibodies as indicated. E, reciprocal co-immunoprecipitation assay showing the interaction between endogenous SMYD2 and BMPR2 in HaCaT cells. F, HaCaT cells were treated without or with 25 ng/ml BMP2 for 1 h before harvesting for co-immunoprecipitation assay as above.
We further characterized whether the endogenous SMYD2 and BMPR2 interacted with each other by co-immunoprecipitation assay. The representative results in Fig. 6E show reciprocal co-immunoprecipitation of SMYD2 and BMPR2. Furthermore, this interaction appears to be constitutive and not affected by BMP2 treatment (Fig. 6F).
SMYD2 methylates BMPR2 at its kinase domain
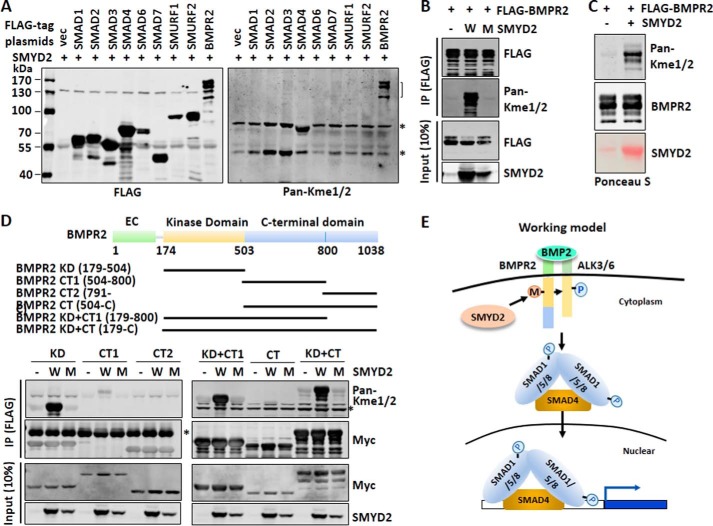
Next, we investigated which component(s) of BMP/TGFβ pathway could be methylated by SMYD2. 293T cells were co-transfected with SMYD2 and FLAG-tagged SMADs, SMURFs, or BMPR2 for 48 h. Subsequently FLAG-SMADs, SMURFs, or BMPR2 were immunoprecipitated and examined for methylation by Western blot analysis using a pan-specific mono/dimethylated lysine antibody (Fig. 7A). Despite broad interaction as observed above, none of the proteins except for BMPR2 were found to be methylated by SMYD2 under our experimental conditions (Fig. 7A). We confirmed that SMYD2 methylated BMPR2, and this methylation was dependent on the enzymatic activity of SMYD2, as was not observed when the enzyme-inactive SMYD2 Y240A mutant was co-expressed with BMPR2 (Fig. 7B). In addition, under the same conditions SMYD2 did not appear to methylate type I receptors ALK1, ALK3, ALK5, ALK6, and ALK7 (data not shown). These results indicate that SMYD2 specifically methylates BMPR2. To lend further support that SMYD2 could methylate BMPR2, we expressed and purified recombinant His6-SMYD2 from bacteria and performed in vitro methylation assay with FLAG-BMPR2 immunoprecipitated from transfected 293T cells as substrate. The in vitro methylation reaction showed that purified SMYD2 methylated BMPR2 (Fig. 7C).
Figure 7.
SMYD2 methylates BMPR2 at its kinase domain. A, Western blot analysis using a pan-mono/di-methylated lysine antibody revealed specific methylation of BMPR2 by SMYD2. 293T cells were co-transfected with SMYD2 and FLAG-SMADs, SMURFs, or BMPR2 for 48 h. Various FLAG-tagged proteins were immunoprecipitated from the whole-cell extracts with anti-FLAG antibody, and methylation was detected by Western blot analysis using a pan-mono/di-methylated lysine antibody (Pan-Kme1/2). The methylated BMPR2 is indicated by a bracket. Asterisks indicate two nonspecific bands, which were likely Hsp70, an abundant chaperone protein often heavily methylated endogenously, and IgG heavy chain. B, methylation of BMPR2 by SMYD2 required SMYD2 catalytic activity. 293T cells were co-transfected with FLAG-BMPR2 and either wild-type SMYD2 (W) or SMYD2 Y240A mutant (M) for 48 h. The subsequent immunoprecipitation (IP)-Western blot analysis was as above. C, SMYD2 methylated BMPR2 in vitro. FLAG-BMPR2 was expressed in 293T cells via transient transfection and purified by anti-FLAG antibody. The immunoaffinity-purified BMPR2 was subjected to in vitro methylation reactions using bacterially expressed and purified His6-SMYD2 (SMYD2). The methylation of FLAG-BMPR2 was detected by Western blot analysis using Pan-Kme1/2. D, SMYD2 methylates the BMPR2 kinase domain in vivo. The upper panel shows various truncated BMPR2 constructs. Various Myc-tagged SMYD2 constructs were co-transfected with the wild-type (W) or SMYD2-Y240A mutant (M) for 48 h, followed by immunoprecipitation with anti-Myc antibody and Western blot analysis with Pan-Kme1/2 and other indicated antibodies. E, working model illustrating how SMYD2 may potentiate BMP signaling transduction by methylating BMPR2. SMYD2 interacts with and methylates BMPR2 within its kinase domain (KD). This methylation may promote the BMPR2 kinase activity and therefore phosphorylation of the type 1 receptor. The activated type 1 receptor then phosphorylates SMAD1/5 and promotes SMAD1/5 nuclear entry and interaction with SMAD4, and consequently it promotes BMP-induced target gene expression.
We also mapped which domain of BMPR2 could be methylated by SMYD2. As SMYD2 is cytoplasmic and thus most likely methylates the intracellular portion of BMPR2, we divided the intracellular part of BMPR2 into three fragments, namely kinase domain (residues 179–504), C-terminal I (CT1, 504–800), and C-terminal II (CT2, 791-C). We also generated additional BMPR2 constructs without or with the kinase domain as illustrated in Fig. 7D. When these constructs were co-expressed with SMYD2 or SMYD2 Y240A mutant in 293T cells, strong methylation was detected for the constructs containing the BMPR2 kinase domain, whereas a weak methylation signal was also detected for CT domain (Fig. 7D). Together, these results demonstrate that SMYD2 preferentially methylates BMPR2 within its kinase domain.
Discussion
In recent years, although there have been more and more reports on regulatory functions of lysine methylation on non-histone proteins (6–8), very few have linked lysine methylation to membrane receptors and their downstream signaling. In this study, we chose to explore whether primarily the cytoplasmically localized SMYD family KMTs regulate TGFβ/BMP signal transduction. By using functional siRNA screening, we identified SMYD2 as a regulator specific for the BMP but not TGFβ pathway. Through various functional assays, we demonstrate that SMYD2 has a role in BMP-induced SMAD1/5 phosphorylation. Consequently, loss of SMYD2 also impairs SMAD1 nuclear localization and SMAD1-SMAD4 interaction. Furthermore, we show that SMYD2 methylates the BMP type 2 receptor BMPR2 at its kinase domain. Together, these results indicate that SMYD2 likely promotes BMP signaling through methylating BMPR2, which in turn facilitates SMAD1/5 phosphorylation, nuclear entry, and interaction with SMAD4 and consequently BMP target gene expression.
One of the interesting observations in our study is that although SMYD2 interacts with multiple components of the BMP and TGFβ signaling pathways (Fig. 6), it only methylates BMPR2 (Fig. 7). So far SMYD2 has been identified as a KMT catalyzing specific lysine monomethylation in diverse proteins (28, 29), including the famous tumor suppressor proteins p53 (30), RB (31, 32), and PTEN (33). A recent comprehensive, large-scale proteomic study of lysine mono-methylation has identified several hundreds of potential SMYD2 methylation sites, and a subset of 35 sites was confirmed by SMYD2 knockdown (34). Although several studies have provided insight into the diversity of SMYD2 substrates, these studies also underscore a striking substrate selectivity for SMYD2, manifested as the binding of substrate peptides to the deep pocket of SMYD2 and extensive interaction of substrate peptides with SMYD2 (28, 29). We suggest that the multiple interactions with BMP pathway components may help the recruitment of SMYD2 into the signaling transduction process and facilitate its methylation on BMPR2. The physiological role for the interaction between SMYD2 and multiple TGFβ/BMP pathway components, if any, remains to be investigated in future study. In this regard, although in HaCaT cells SMYD2 does not appear to regulate the TGFβ pathway, it was reported that in mouse macrophages Smyd2 promotes TGFβ production through regulation of transcription (35). Thus, SMYD2 is likely to exert cell type-specific function in a context-dependent manner.
It is well-established that the BMP signaling cascades begin with binding of ligands and activation of type 2 receptor kinase activity (14). Given our compelling evidence that SMYD2 methylates BMPR2 and remains in the cytoplasm upon BMP induction and that loss of SMYD2 impairs BMP2-induced SMAD1/5 phosphorylation, it is tempting to propose a working model in Fig. 7E as to how SMYD2 selectively regulates the BMP pathway. In this model, SMYD2 methylates BMPR2 at its kinase domain to promote its phosphorylation on a type 1 receptor. The activated type 1 receptor then phosphorylates SMAD1/5 and promotes SMAD1/5 nuclear entry and interaction with SMAD4, and consequently it promotes BMP-induced target gene expression. SMYD2-catalyzed BMPR2 methylation is likely functionally important for the BMP-signaling pathway, because inhibition of SMYD2 using a SMYD2-specific inhibitor AZ505 or ectopic expression of a KMT-deficient SMYD2 mutant both impaired BMP2-induced SMAD1/5 phosphorylation (Fig. 5, C and D). At this stage, it is not known whether SMYD2 constitutively methylates BMPR2 or methylates BMPR2 in a BMP-induced manner. So far, we have not been able to convincingly detect endogenous methylated BMPR2 in the presence or absence of BMP treatment. Although SMYD2 interacts with BMPR2 constitutively (Fig. 6, E and F), we could not rule out the possibility that SMYD2 methylates BMPR2 only when it is activated by ligands. In this regard, it is noteworthy that arginine methylation by PRMT1 has been shown to regulate BMP signaling (21). PRMT1 was shown to bind BMPR2, and BMP treatment activates complex formation between type I and II receptors and brings PRMT1 and inhibitory SMAD6 into proximity. PRMT1 then methylates SMAD6 on Arg-74 and Arg-81 and releases SMAD6 from the receptors, allowing SMAD1/5 to bind type I receptor and be activated (21).
The identification of SMYD2 as a positive regulator of the BMP pathway and BMPR2 as a substrate for SMYD2 methylation provides new insight into the signaling pathway of BMP. However, multiple questions remain to be addressed. For example, it is necessary in the future to identify the methylation site(s) in the BMPR2 kinase domain and to investigate how methylation regulates BMPR2 kinase activity. In addition, the finding that BMPR2 can be methylated by SMYD2 also raises the question whether BMPR2 is regulated by dynamic methylation/demethylation and, if it does, the demethylase(s) involved.
A previous study has reported lysine methylation of membrane receptor VEGFR1 by SMYD3 (13). Methylation at Lys-831 of VEGFR1 by SMYD3 enhances VEGFR1 tyrosine autophosphorylation and kinase activity (13). Here, we show BMPR2 methylation by SMYD2 and a novel role for SMYD2 in promoting the BMP-signaling pathway. Thus, consistent with their primary cytoplasmic localization, the SMYD family KMTs may play unique roles in methylation of membrane receptors and regulation of signaling transduction.
Experimental procedures
Cell lines and reagents
Human HaCaT and 293T cell lines are from the ATCC. The culture medium for HaCaT cells was RPMI 1640 (Gibco) and 10% fetal bovine serum, and the culture medium for 293T cells was high glucose-DMEM (Gibco) plus 10% fetal bovine serum. Reagents include the following: total RNA extraction (Biorezyme); reverse transcription, qRT-PCR, and DNA purification kits (TransGen); DNA transfection kit and protease and phosphatase inhibitor mixtures (BioTools); Lipofectamine 2000 (Invitrogen); BMP2 (GenScript); TGFβ1 (BD Biosciences); restriction enzymes and T4 DNA ligase and PNK (New England Biolabs); G10-agarose (Biowest); FLAG M2 and HA affinity purification beads (Abmart); and Ni+ affinity purification column (Qiagen). The sequence information for siRNAs and shRNAs is as follows: siSMYD1-1, 5′-GGA GGA UGG UGG ACG GCU AUA-3′; siSMYD1-2, 5′-AGA ACG AAU UCA UGU ACU ACA-3′; siSMYD2-1, 5′-GGA AAG AAG GAU UGU CCA AAU-3′; siSMYD2-2, 5′-GGC AGA AGU CAG AGC UGU ACA-3′; siSMYD3-1, 5′-ACU GUU CGA UUG UGU UCA AUG-3′; siSMYD3-2, 5′-GGA GUC AAA UAU UAA CAA ACU-3′; siSMYD4-1, 5′-CCA AGA UUA UGU UAC GUA AAG-3′; siSMYD4-2, 5′-GGC GAU GAC CAC CAU ACA ACA-3′; siSMYD5-1, 5′-GCA ACU GGA GAG UUU CUU AAC-3′; siSMYD5-2, 5′-GGA GGA AAU UGU CCA UAA ACU-3′; shSMYD2–1, 5′-CGG CAA AGA TCA TCC ATA TAT-3′; and shSMYD2-2, 5′-GCT GTG AAG GAG TTT GAA TCA-3′. Guide RNA sequence for CRISPR/CAS9 SMYD2 knock-out is 5′-GTT AGT CTT ACA GTC TCC GA-3′. Primers for RT-PCRs are as follows: GAPDH-qPCR-F, 5′-AGC CTC AAG ATC ATC AGC AAT G-3′, and GAPDH-qPCR-R, 5′-ATG GAC TGT GGT CAT GAG TCC TT-3′; ID1-qPCR-F, 5′-CTG CTC TAC GAC ATG AAC GG-3′, and ID1-qPCR-R, 5′-GAA GGT CCC TGA TGT AGT CGA T-3′; ID2-qPCR-F, 5′-AGC ACT GTG TGG CTG AAT AAG-3′, and ID2-qPCR-R, 5′-AGT AAG AGA ACA CCC TGG GA-3′; ID3-qPCR-F, 5′-ATC CTA CAG CGC GTC ATC G-3′, and ID3-qPCR-R, 5′-CTT CCG GCA GGA GAG GTT C-3′; SMAD6-qPCR-F, 5′-CCT ACT CTC GGC TGT CTC CT-3′, and SMAD6-qPCR-R, 5′-GAA TTC ACC CGG AGC AGT GA-3′; PAI-1-qPCR-F, 5′-ACC GCA ACG TGG TTT TCT CA-3′, and PAI-1-qPCR-R, 5′-TTG AAT CCC ATA GCT GCT TGA AT-3′; p21-qPCR-F, 5′-TGA GCC GCG ACT GTG ATG-3′, and p21-qPCR-R, 5′-GTC TCG GTG ACA AAG TCG AAG TT-3′; SMAD7-qPCR-F, 5′-TGC TCC CAT CCT GTG TGT TAA G-3′, and SMAD7-qPCR-R, 5′-TCA GCC TAG GAT GGT ACC TTG G-3′.
Plasmids and antibodies
The ppY-CAGIP-SMYD2 and ppY-CAGIP-SMYD2-Y240A mutant, the FLAG-SMAD1–7, FLAG-SMURF1/2, FLAG-BMPR2, HA-BMPR2, and Myc-BMPR2 truncation constructs, and the HA-SMAD1 and FLAG-SMYD2 were either constructed by standard recombinant DNA techniques or provided by Dr. Xin-Hua Feng. The antibodies include mouse HA, FLAG, Myc, SMYD2, and GAPDH (Abmart), rabbit SMYD2 (ABclonal), SMAD1 and BMPR2 (Cell Signaling), SMAD4, SMAD5, and SMAD1/5ph (Abcam), ALK3 (Protein Tech), and pan-lysine mono/dimethylation (Pan-Kme1/2) (PTM Biolabs).
siRNA transfection and TGFβ1/BMP2 treatment
The siRNA transfection was performed using Lipofectamine 2000 essentially according to the manufacturer's instructions. The final concentration for each siRNA was 100 nm. For siRNA-based screen, HaCaT cells were transfected with the siRNAs against SMYD1 to SMYD5 as indicated. Two days later, cells were starved overnight with non-serum medium and then treated with 5 ng/ml TGFβ1 or 25 ng/ml of BMP2 for 2 or 4 h as indicated.
Total RNA isolation and qRT-PCR
Total RNA isolation and qRT-PCR were all performed according to manufacturers' procedures.
Co-immunoprecipitation assay
For co-immunoprecipitation assay, cells were lysed in buffer containing 50 mm Tris·HCl (pH 7.5), 150 mm KCl, 1% Nonidet P-40, 8% glycerol, 1 mm EDTA, 1 mm DTT, and protease and phosphatase inhibitor mixtures. The cell lysates were centrifuged at 12,000 rpm for 20 min at 4 °C. The supernatants were diluted with up to 3 volumes of 50 mm Tris·HCl (pH 7.5), 150 mm KCl, 8% glycerol, 1 mm EDTA, 1 mm DTT and protease inhibitor mixture, mixed with antibody-loaded protein A beads, and rotated at 4 °C for 3 h or overnight. The beads were then retrieved by centrifugation at 2000 rpm for 1 min at 4 °C, washed three or four times with buffer containing 50 mm Tris·HCl (pH 7.5), 150 mm KCl, 0.1% Triton X-100, 1 mm EDTA, 1 mm DTT, and protease inhibitor mixture by rotation at 4 °C for 5 min. The beads were boiled in 20 ml of SDS-loading buffer for 5 min, and samples were subjected to standard SDS-PAGE and Western blot analysis.
Immunofluorescence staining
Immunofluorescent staining was performed essentially as described (36). In brief, HaCaT cells were washed three times with PBS and fixed in 4% paraformaldehyde in PBS for 15–20 min. The cells were then washed three times with PBS and permeabilized with 1% Triton X-100 in PBS for 10 min, followed by washing three times with PBS and blocking with 5% BSA, 0.2% Triton X-100 in PBS for 1 h. Then the cells were stained with the indicated primary and corresponding secondary antibodies, and nuclei were revealed by DAPI staining.
Nucleocytoplasmic fractionation
HaCaT cells were washed twice with cold PBS and lysed on ice for 15–20 min in lysis buffer (10 mm HEPES-NaOH (pH 7.9), 10 mm KCl, 1.5 mm MgCl2, 0.5 mm β-mercaptoethanol, and protease and phosphatase inhibitors). Then, Nonidet P-40 was added to a final concentration of 0.5%. 2 min later, cells were centrifuged at 16,000 × g for 10–15 min. The corresponding supernatant was the cytoplasmic fraction. The pellet was washed twice with cold PBS, lysed in nuclear lysis buffer (10 mm Tris·HCl (pH 7.6), 420 mm NaCl, 0.5% Nonidet P-40, and 1 mm DTT, 1 mm PMSF, 2 mm MgCl2, and protease and phosphatase inhibitors) on ice for 20 min, and centrifuged at 16,000 × g for 10–15 min to obtain the supernatant, which was the nuclear fraction. Then, lower salt buffer (10 mm Tris·HCl (pH 7.6), 1 mm DTT, 1 mm PMSF, 2 mm MgCl2, and protease and phosphatase inhibitors) was added to adjust the final concentration of NaCl to 150 mm.
In vitro methylation assay
In vitro methylation assay was performed essentially as described (37). In brief, 0.5 μg of bacterially purified His-SMYD2 and immunoprecipitated FLAG-BMPR2 were incubated in 25 mm Tris·HCl (pH 8.0), 1 mm DTT, 1 mm PMSF, 67 μm S-adenosylmethionine at 37 °C for 2 h. FLAG-BMPR2 methylation was then detected by SDS-PAGE and Western blotting with Pan-Kme1/2 antibody.
Stable SMYD2 knockdown and knock-out cell lines
To knock down SMYD2 by shRNAs, SMYD2 shRNAs with the following targeting sequences 5′-CGG CAA AGA TCA TCC ATA TAT-3′ (shSMYD2-1) and 5′-GCT GTG AAG GAG TTT GAA TCA-3′ (shSMYD2-2) were cloned into lentiviral vector pLKO.1-puro. HaCaT cells were infected with lentivirus expressing shRNAs of SMYD2 and control vector. Two days after infection, the stable shRNA-infected cells were selected with addition of puromycin and cultured for 2 more days and used for subsequent experiments.
To generate SMYD2 knock-out cell lines by CRISPR/CAS9, a small guide RNA (5′-G TTA GTC TTA CAG TCT CCGA-3′) targeting exon3 of SMYD2 was cloned into pLKO.1-puro-based sgRNA expression vector. Guide RNA of SMYD2 was selected through an on-line search at http://crispr.mit.edu. 5 Stable cell lines expressing sgRNA was established by infecting HaCaT cells with sgRNA lentivirus and cell cultivation with puromycin. A second infection of adenovirus expressing Cas9 resulted in SMYD2 knock-out clones. The individual SMYD2 knock-out clones were isolated through limiting dilution and verified by DNA sequencing and Western blot analysis.
Cell proliferation assay
One thousand cells were seeded per well into a 96-well plate, and cells were cultured for 24, 48, 72, 96, or 120 h. The cells were labeled with Cell Counting Kit-8 (CCK8) (Bimake, B34302) for 2 h, and proliferation rates were determined at an absorbance of 450 nm.
Author contributions
S. G. and Z. W. performed most of the experiments. X. H. and W. W. helped in siRNA screening and RT-PCR analysis. P. C. helped in in vitro methylation assay. J. L., X. F., J. D., and J. W. supervised the experiments and conceived the ideas. J. D. and J. W. wrote the manuscript with S. G.
Acknowledgment
We thank members of the Wong laboratory for valuable discussions.
This work was supported by Ministry of Science and Technology of China Grant 2015CB910402 (to J. W.), National Natural Science Foundation of China Grant 91419303, and Science and Technology Commission of Shanghai Municipality Grants 14XD1401700 and 11DZ2260300. The authors declare that they have no conflicts of interest with the contents of this article.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- KMT
- lysine methyltransferase
- BMP
- bone morphogenic protein
- R-SMAD
- regulatory SMAD
- qRT-PCR
- quantitative real time PCR
- CT
- C-terminal
- F
- forward
- R
- reverse.
References
- 1. Greer E. L., and Shi Y. (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nimura K., Ura K., and Kaneda Y. (2010) Histone methyltransferases: regulation of transcription and contribution to human disease. J. Mol. Med. 88, 1213–1220 [DOI] [PubMed] [Google Scholar]
- 3. Patel D. J., and Wang Z. (2013) Readout of epigenetic modifications. Annu. Rev. Biochem. 82, 81–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schneider R., Bannister A. J., and Kouzarides T. (2002) Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem. Sci. 27, 396–402 [DOI] [PubMed] [Google Scholar]
- 5. Falnes P. Ø., Jakobsson M. E., Davydova E., Ho A., and Mal̸ecki J. (2016) Protein lysine methylation by seven-β-strand methyltransferases. Biochem. J. 473, 1995–2009 [DOI] [PubMed] [Google Scholar]
- 6. Lanouette S., Mongeon V., Figeys D., and Couture J. F. (2014) The functional diversity of protein lysine methylation. Mol. Syst. Biol. 10, 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hamamoto R., Saloura V., and Nakamura Y. (2015) Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat. Rev. Cancer 15, 110–124 [DOI] [PubMed] [Google Scholar]
- 8. Zhang X., Huang Y., and Shi X. (2015) Emerging roles of lysine methylation on non-histone proteins. Cell. Mol. Life Sci. 72, 4257–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pradhan S., Chin H. G., Estève P. O., and Jacobsen S. E. (2009) SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics 4, 383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bannister A. J., and Kouzarides T. (2005) Reversing histone methylation. Nature 436, 1103–1106 [DOI] [PubMed] [Google Scholar]
- 11. Mosammaparast N., and Shi Y. (2010) Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79, 155–179 [DOI] [PubMed] [Google Scholar]
- 12. Mazur P. K., Reynoird N., Khatri P., Jansen P. W., Wilkinson A. W., Liu S., Barbash O., Van Aller G. S., Huddleston M., Dhanak D., Tummino P. J., Kruger R. G., Garcia B. A., Butte A. J., Vermeulen M., et al. (2014) SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature 510, 283–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kunizaki M., Hamamoto R., Silva F. P., Yamaguchi K., Nagayasu T., Shibuya M., Nakamura Y., and Furukawa Y. (2007) The lysine 831 of vascular endothelial growth factor receptor 1 is a novel target of methylation by SMYD3. Cancer Res. 67, 10759–10765 [DOI] [PubMed] [Google Scholar]
- 14. Feng X. H., and Derynck R. (2005) Specificity and versatility in tgf-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 21, 659–693 [DOI] [PubMed] [Google Scholar]
- 15. Miyazono K., Kamiya Y., and Morikawa M. (2010) Bone morphogenetic protein receptors and signal transduction. J. Biochem. 147, 35–51 [DOI] [PubMed] [Google Scholar]
- 16. Liu F., Ventura F., Doody J., and Massagué J. (1995) Human type II receptor for bone morphogenic proteins (BMPs): extension of the two-kinase receptor model to the BMPs. Mol. Cell. Biol. 15, 3479–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rosenzweig B. L., Imamura T., Okadome T., Cox G. N., Yamashita H., ten Dijke P., Heldin C. H., and Miyazono K. (1995) Cloning and characterization of a human type II receptor for bone morphogenetic proteins. Proc. Natl. Acad. Sci. U.S.A. 92, 7632–7636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Macias M. J., Martin-Malpartida P., and Massagué J. (2015) Structural determinants of Smad function in TGF-β signaling. Trends Biochem. Sci. 40, 296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi Y., and Massagué J. (2003) Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 20. ten Dijke P., and Hill C. S. (2004) New insights into TGF-β-Smad signalling. Trends Biochem. Sci. 29, 265–273 [DOI] [PubMed] [Google Scholar]
- 21. Xu J., Wang A. H., Oses-Prieto J., Makhijani K., Katsuno Y., Pei M., Yan L., Zheng Y. G., Burlingame A., Brückner K., and Derynck R. (2013) Arginine methylation initiates BMP-induced Smad signaling. Mol. Cell 51, 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Massagué J. (2012) TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elkouris M., Kontaki H., Stavropoulos A., Antonoglou A., Nikolaou K. C., Samiotaki M., Szantai E., Saviolaki D., Brown P. J., Sideras P., Panayotou G., and Talianidis I. (2016) SET9-mediated regulation of TGF-β signaling links protein methylation to pulmonary fibrosis. Cell Rep. 15, 2733–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Spellmon N., Holcomb J., Trescott L., Sirinupong N., and Yang Z. (2015) Structure and function of SET and MYND domain-containing proteins. Int. J. Mol. Sci. 16, 1406–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Donlin L. T., Andresen C., Just S., Rudensky E., Pappas C. T., Kruger M., Jacobs E. Y., Unger A., Zieseniss A., Dobenecker M. W., Voelkel T., Chait B. T., Gregorio C. C., Rottbauer W., Tarakhovsky A., and Linke W. A. (2012) Smyd2 controls cytoplasmic lysine methylation of Hsp90 and myofilament organization. Genes Dev. 26, 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Elbendary A., Berchuck A., Davis P., Havrilesky L., Bast R. C. Jr., Iglehart J. D., and Marks J. R. (1994) Transforming growth factor β1 can induce CIP1/WAF1 expression independent of the p53 pathway in ovarian cancer cells. Cell Growth Diiffer. 5, 1301–1307 [PubMed] [Google Scholar]
- 27. Ogata T., Wozney J. M., Benezra R., and Noda M. (1993) Bone morphogenetic protein 2 transiently enhances expression of a gene, Id (inhibitor of differentiation), encoding a helix-loop-helix molecule in osteoblast-like cells. Proc. Natl. Acad. Sci. U.S.A. 90, 9219–9222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ferguson A. D., Larsen N. A., Howard T., Pollard H., Green I., Grande C., Cheung T., Garcia-Arenas R., Cowen S., Wu J., Godin R., Chen H., and Keen N. (2011) Structural basis of substrate methylation and inhibition of SMYD2. Structure 19, 1262–1273 [DOI] [PubMed] [Google Scholar]
- 29. Xu S., Zhong C., Zhang T., and Ding J. (2011) Structure of human lysine methyltransferase Smyd2 reveals insights into the substrate divergence in Smyd proteins. J. Mol. Cell Biol. 3, 293–300 [DOI] [PubMed] [Google Scholar]
- 30. Huang J., Perez-Burgos L., Placek B. J., Sengupta R., Richter M., Dorsey J. A., Kubicek S., Opravil S., Jenuwein T., and Berger S. L. (2006) Repression of p53 activity by Smyd2-mediated methylation. Nature 444, 629–632 [DOI] [PubMed] [Google Scholar]
- 31. Cho H. S., Hayami S., Toyokawa G., Maejima K., Yamane Y., Suzuki T., Dohmae N., Kogure M., Kang D., Neal D. E., Ponder B. A., Yamaue H., Nakamura Y., and Hamamoto R. (2012) RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia 14, 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saddic L. A., West L. E., Aslanian A., Yates J. R. 3rd, Rubin S. M., Gozani O., and Sage J. (2010) Methylation of the retinoblastoma tumor suppressor by SMYD2. J. Biol. Chem. 285, 37733–37740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakakido M., Deng Z., Suzuki T., Dohmae N., Nakamura Y., and Hamamoto R. (2015) Dysregulation of AKT pathway by SMYD2-mediated lysine methylation on PTEN. Neoplasia 17, 367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ahmed H., Duan S., Arrowsmith C. H., Barsyte-Lovejoy D., and Schapira M. (2016) An integrative proteomic approach identifies novel cellular SMYD2 substrates. J. Proteome Res. 15, 2052–2059 [DOI] [PubMed] [Google Scholar]
- 35. Xu G., Liu G., Xiong S., Liu H., Chen X., and Zheng B. (2015) The histone methyltransferase Smyd2 is a negative regulator of macrophage activation by suppressing interleukin 6 (IL-6) and tumor necrosis factor α (TNF-α) production. J. Biol. Chem. 290, 5414–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu X., Gao Q., Li P., Zhao Q., Zhang J., Li J., Koseki H., and Wong J. (2013) UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat. Commun. 4, 1563. [DOI] [PubMed] [Google Scholar]
- 37. Fang L., Zhang L., Wei W., Jin X., Wang P., Tong Y., Li J., Du J. X., and Wong J. (2014) A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol. Cell 55, 537–551 [DOI] [PubMed] [Google Scholar]