Abstract
Lysine methyltransferases transfer methyl groups in specific lysine sites, which regulates a variety of important biological processes in eukaryotes. In this study, we characterized a novel homolog of the yeast methyltransferase DOT1 in A. flavus, and observed the roles of dot1 in A. flavus. Deletion of dot1 showed a significant decrease in conidiation, but an increase in sclerotia formation. A change in viability to multiple stresses was also found in the Δdot1 mutant. Additionally, aflatoxin (AF) production was found severely impaired in the Δdot1 mutant. Further analysis by qRT-PCR revealed that the transcription of AF structural genes and their regulator gene aflS were prominently suppressed in the Δdot1 mutant. Furthermore, our data revealed that Dot1 is important for colonizing maize seeds in A. flavus. Our research indicates that Dot1 is involved in fungal development, aflatoxin biosynthesis and fungal virulence in A. flavus, which might provide a potential target for controlling A. flavus with new strategies.
Keywords: histone methylation, sclerotia, stress response, fungal virulence
1. Introduction
The ubiquitous fungus A. flavus is notorious for its contamination of many foodstuffs and important crops, such as maize, peanuts and rice, during pre- or post-harvest with the most carcinogenic mycotoxin, aflatoxin [1,2,3]. This fungus is also known as the second most frequent Aspergillus pathogen after A. fumigatus on immunosuppressed patients, which is also responsible for aspergillosis diseases or liver cancer for human and animals caused by the consumption of contaminated food [1,2,4]. Due to this, this fungus has caused enormous agricultural economic losses, food shortages and health problems all cross the world [1,5]. Thus, effective measures to restrict this pathogen are urgently needed, not only to protect human and animal health, but also to alleviate its deleterious effects on agricultural economic losses.
It is quite important to develop rational control strategies for A. flavus underlying the fungal virulence and mycotoxins attributes. Although numerous studies reported that pathogenesis in A. flavus was connected with fungal development, secondary metabolism and the adaptability to environmental factors [1,6,7,8], much remains to be learned about the fungal pathogenesis. In the past decade, histone posttranslational modifications (HPTMs), such as phosphorylation, ubiquitination, SUMOylation, acetylation and methylation, have been well documented as providing an important link between many biological processes, fungal secondary metabolite biosynthesis and chromatin-based regulation in eukaryotic cells [5,9,10,11]. Among these, histone methylation was one of most thoroughly studied epigenetic process in HPTMs. In particular, it has been shown that histone lysine methylation catalyzed by lysine methyltransferases in histone specific lysine sites, such as H3K4me2/3, is associated with active regions of chromatin, and H3K9me2/3 and H3K27me2/3 are related to repressive regions [12,13,14,15]. In Neurospora crassa, DIM-5, responsible for H3K9 methylation, has a close relationship with gene silencing by mediating DNA methylation on particular cytosine sites [16,17]. The H3K36 methyltransferase SET-2 in N. crassa has also been reported to regulate fungal development [16,18]. H3K4me and H3K27me are addressed as important for fungal development and many secondary metabolites biosynthesize in fungi [9,12].
Studies have demonstrated that many gene clusters of secondary metabolites, including the AF gene cluster, are located near telomeres in Aspergillus flavus [19,20]. Genes near telomeres, associated with specific chromatin structures, have position effects, resulting in gene silencing [19,21]. HPTMs such as acetylation, methylation, and ubiquitination have been linked to transcriptional silencing. The inactivation of H3K79 methylation or H3K4 methylation is quite important for heterochromatin formation and for maintaining silencing regions of chromatin in budding yeast [21,22,23]. An abnormal expression of the H3K79 methyltransferase gene dot1 in yeast and mammalian cells leads to a decrease of gene silence [23,24]. In Aspergillus, although many studies have been shown that HPTMs regulate fungal development and secondary metabolite biosynthesis [9,10,25,26,27], there has been little progress in addressing the function of histone lysine methylation in A. flavus. Here, in order to know if H3K79 is involved in AF biosynthesis, we identified a putative H3K79 methyltransferase Dot1 in A. flavus, and then observed the potential roles of the dot1 gene in A. flavus by addressing the effects of the deletion mutant on the development, conidiation, sclerotia formation, aflatoxin biosynthesis, and pathogenicity of seeds.
2. Results
2.1. Characterization of Histone H3K79 Methyltransferase in A. flavus
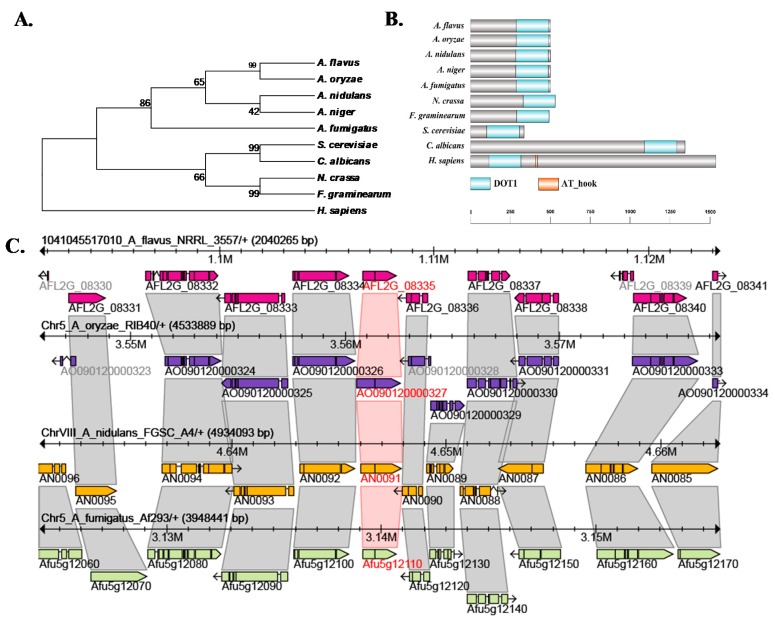
To characterize the homolog of the S. cerevisiae H3K79 methyltransferase Dot1 in A. flavus, we used BlastP analyses in the NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) with the amino acid sequence of S. cerevisiae Dot1 (EGA79271.1) as queries. AFLA_093140, encoding a 503 aa protein, was identified in A. flavus, which shared a 33% overall identity with S. cerevisiae Dot1. The Dot1 protein sequences from different fungi were downloaded, and analyzed their evolutionary relationship by MEGA5.0, which showed that Dot1 from Aspergillus Dot1 was highly conserved (Figure 1A). Dot1 from Aspergillus has two exons, and the locations and paralogs of dot1 were also quite conserved in the Aspergillus genomes (Figure 1C). The alignment of Dot1 protein sequences from different fungal species indicates that H3K79 methyltransferase only possesses the DOT1 domain (Figure 1B), however, in Homo sapiens the H3K79 methyltransferase has another domain AT_hook, which is a DNA-binding motif with a preference for A/T rich regions (Figure 1B).
Figure 1.
Characterization of H3K79 methyltransferase in A. flavus. (A) Phylogenetic relationship of Dot1 from different species was analyzed; (B) Domains from Dot1 proteins were characterized by SMART, and software DOG 2.0 were used to visualize protein domains; (C) The locations and paralogs of dot1 in Aspergillus genomes were analyzed by AspGD (http://www.aspgd.org/).
2.2. Construction of the Deleted (Δdot1) and Complemented (dot1C) Mutant Strains
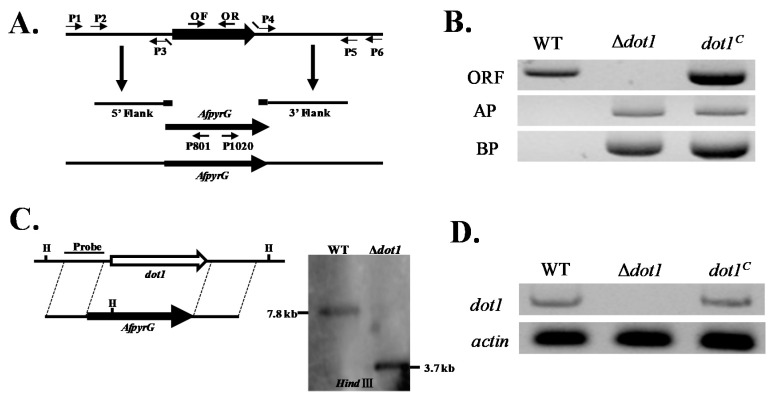
The Δdot1 mutants were generated using a homologous recombination strategy as shown in Figure 2A. The selected transformants of Δdot1 were analyzed by diagnostic PCR (Figure 2B), and the results showed that a 0.5 kb ORF fragment was amplified from WT, but none from Δdot1 (Figure 2B). With gDNA as a template, both 2.0 kb AP and 2.1 kb BP fragments were amplified in Δdot1 mutant, but not in wild type (WT) (Figure 2B). The Δdot1 mutant then was further confirmed by southern blot analyses (Figure 2C). To confirm that the phenotype changes observed in Δdot1 were due to the disruption of dot1, the Δdot1 mutant was complemented with a full-length of dot1 by a pPTRI plasmid and fused into the Δdot1 protoplast. The dot1C complemented strains were also confirmed by diagnostic PCR using the genomic DNA (Figure 2B) and further verified by RT-PCR, and the result showed that transcription level of Δdot1 was not detected in Δdot1, in contrast to the wild type (WT) and dot1C strains (Figure 2D). All these results indicated that the deletion Δdot1 and complemented dot1C mutant strains were successfully constructed.
Figure 2.
Deletion strategy and confirmation of the mutant strains. (A) Deletion strategy for Δdot1 using homologous recombination; (B) Deleted Δdot1 and complemented dot1C strains were tested by PCR analysis with genomic DNA as template; (C) Southern blot analyses of dot1 deletion mutants using PCR fragment of 5′ flanking region as probes. Genomic DNAs were extracted from WT and putative transformants. Restriction was carried out using the indicated enzymes; (D) Δdot1 and dot1C mutant strains were tested by RT-PCR with cDNA as template.
2.3. Dot1 Is Involved in Fungal Growth and Sporulation
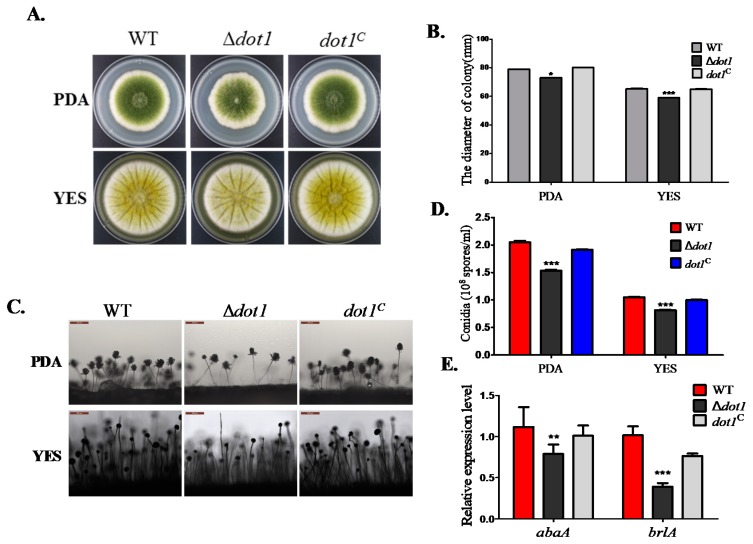
To know the potential role of the H3K79 methyltransferase gene dot1 in fungal development in A. flavus, the Δdot1, WT and dot1C strains were inoculated on YES and PDA in the dark at 37 °C, and the results showed that the Δdot1 mutant displayed less pigmentation and marginally decreased in radial growth compared to dot1C or WT strains (Figure 3A,B). The Δdot1 mutant also displayed a significant decrease in conidiation compared to the WT or dot1C strain (p < 0.01) (Figure 3D). Further examination in sporogenesis also showed that the Δdot1 mutant produced less normal conidiophore compared to the WT and dot1C strain (Figure 3C). qRT-PCR was performed to detect the transcriptional expression levels of the conidia transcriptional factors genes brlA and abaA, which showed that the transcriptional levels of these two conidia specifie genes were both down-regulated in the Δdot1 mutant (Figure 3E). These results demonstrate that Dot1 contributes to radical growth and conidiation in A. flavus.
Figure 3.
The Δdot1 mutant was altered in vegetative growth and conidiation. (A) Phenotype of the WT, Δdot1 and dot1C strains after grown on PDA and YES agar plates at 37 °C for 4 days; (B) colony diameter in on PDA and YES cultures of the WT strain and mutants were assayed; (C) Conidiophores was detected by microscope after light induction; (D) The amount of conidia from different strains were determined; (E) Transcriptional levels of conidia transcriptional factors genes in the Δdot1 mutant, WT and dot1C strains. *, ** and *** represent significantly different (p ≤ 0.05, p ≤ 0.01 or p ≤ 0.001, respectively). The experiments were conducted with technical triplicates for each strain, and were repeated three times.
2.4. Dot1 Plays a Negative Role in Sclerotial Reproduction in A. flavus
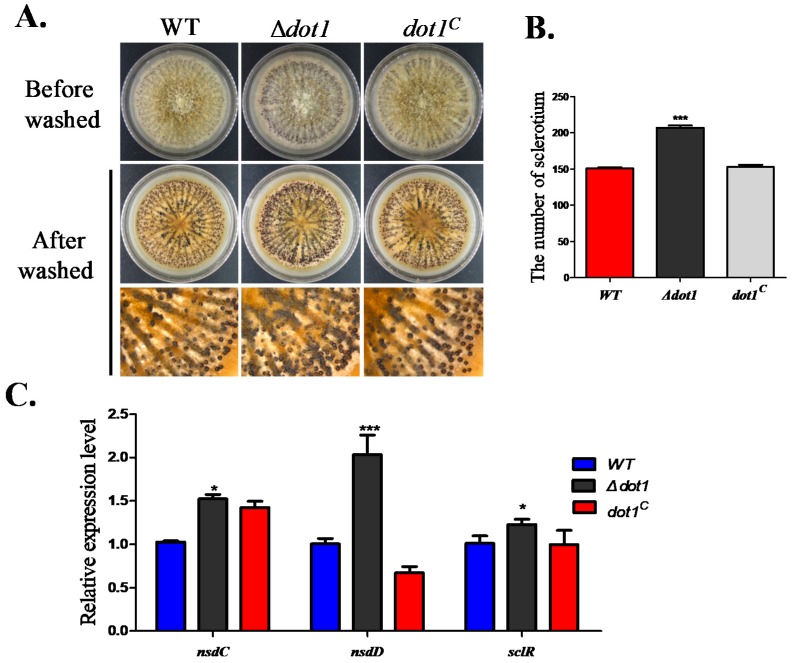
Sclerotia is considered to be a vestige of the cleistothecia, which is a survival structure for A. flavus to live through a stress environment. To determine if Dot1 participated in sclerotia formation, the Δdot1 mutant was cultured on WKM agar. The Δdot1 mutant was found to produce a larger amount of sclerotia (207 ± 1.73) than WT (150.67 ± 0.88) and the dot1C strain (152.67 ± 2.27) (p < 0.001) (Figure 4A,B). The qRT-PCR was performed to confirm the expressing levels of genes that were reported to be related with sclerotia formation, which indicated that nsdC, nsdD and sclR were all transcriptionally up-regulated in the Δdot1 mutant (Figure 4C), demonstrating that Dot1 negatively regulated sclerotia formation in A. flavus.
Figure 4.
Sclerotia reproduction analysis among the WT, Δdot1 and dot1C strains. (A) Phenotype of the WT, Δdot1 and dot1C strains were determined after being grown on sclerotia-inducing media, Wickerham medium. The plates were sprayed with 70% ethanol to allow visualization of sclerotia; (B) The amount of sclerotia was measured in (A); (C) Transcriptional levels of the sclerotial specific gene nsdC, nsdD and sclR. * and *** represent p ≤ 0.05 and p ≤ 0.001, respectively. The experiments were performed with four biological replicates for each strain, and were repeated three times.
2.5. Dot1 Response to Multiple Stresses in A. flavus
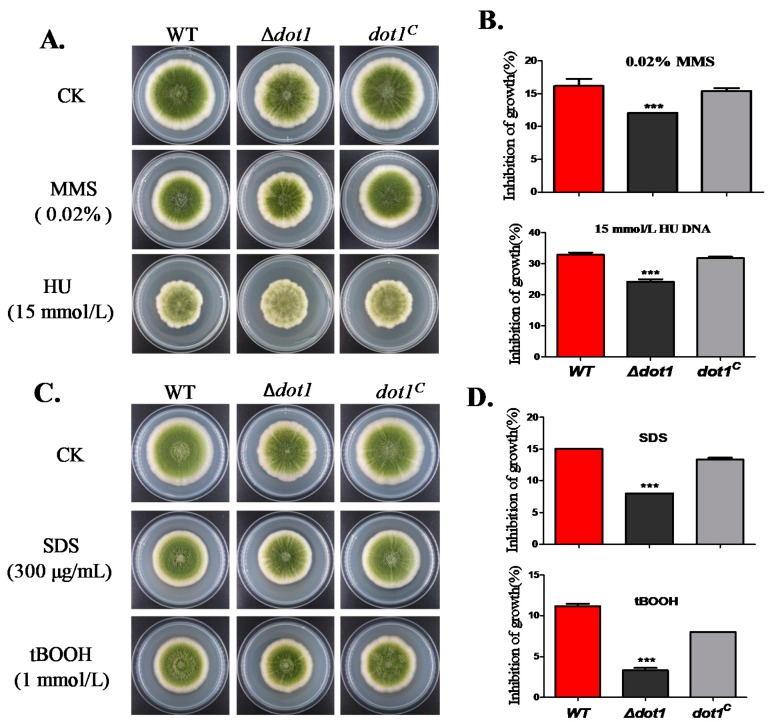
Histone post-translational modifications have been shown to contribute to DNA damage repair [28]. To determine the viability of dot1 in response to genotoxicity stress, the strains were cultured on a PDA medium within 0.02% methylmercuric sulfate (MMS) or 15 mM hydroxyurea (HU) for 3 d. As shown in Figure 5A,B, the Δdot1 mutant showed less sensitivity to genotoxicity stress than the WT and dot1C strain. Since the Δdot1 mutant showed increased resistance to genotoxicity stress, we were also interested in addressing whether Dot1 responsed to other stresses. As we can see in Figure 5C,D, the WT and dot1C strains, but not Δdot1, were much more sensitive to the cell-wall-damaging agent SDS (sodium dodecyl sulfate) and oxidative agent tBooH (tert-butyl hydroperoxide). All these results demonstrated that Dot1 has a potential role in response to multiple stresses in A. flavus.
Figure 5.
Growth of the WT, dot1 and dot1C strains under multiple stresses. (A) Phenotype of WT, Δdot1 and dot1C strains when incubated within 0.02% methylmercuric sulfate (MMS) or 15 mM hydroxyurea (HU) for 3 d. (B) The growth inhibition rate of WT, Δdot1 and dot1C strains under genome integrity stress. The inhibition rate of growth was relative to the growth rate of each untreated strain, ; (C) Morphology of WT, Δdot1 and dot1C strains when incubated within 300 µg/mL SDS or 1 mmol/L tBooH; (D) The growth inhibition was quantified in (C). *** represent significantly different p ≤ 0.001. The experiments were performed with four biological replicates for each strain, and were repeated three times.
2.6. Dot1 Contributes to Aflatoxin Biosynthesis
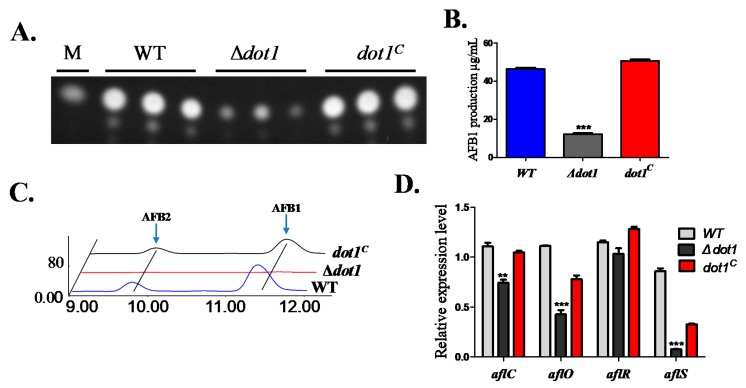
A previous study showed that histone methylation is involved in secondary metabolite synthesis in many Aspergillus species [9]. Thus, we detected aflatoxin production—which is the most important secondary metabolite in A. flavus—in PDA agar media after being cultured for 5 d at 29 °C. The TLC results indicated that AF production was severely impaired in the Δdot1 mutant (Figure 6A,B). The result was further confirmed by HPLC analysis, which also showed a significant decrease in AFB1 production, as well as AFB2 production in the Δdot1 mutant (Figure 6C). The transcript levels of the AF-regulated genes(aflR and aflS) and the biosynthesis genes (aflC and aflO) in the Δdot1 mutant were determined by qRT-PCR, which demonstrated that aflS, aflC and aflO, but not aflR, were transcriptionally inhibited in the Δdot1 mutant (Figure 6D). All these results indicated that Dot1 might be involved in regulating aflatoxin biosynthesis in A. flavus.
Figure 6.
AF production of the WT, dot1 and dot1C strains. (A) AF production was measured by Thin-Layer Chromatography (TLC) after being grown on PDA medium for 5 d at 29 °C in the dark. (B) Relative AF production in (A) was qualified. (C) HPLC analysis of aflatoxin produced by the WT, Δdot1 and dot1C strains after being grown on PDA medium for 5 d at 29 °C. (D) Transcriptional level of the AF-related genes aflR, aflS, aflC(pksA) and aflO(omtB) from WT, Δdot1 and dot1C strains. * and *** represent p ≤ 0.05 and p ≤ 0.001, respectively. The experiments were performed with three biological replicates for each strain, and were repeated three times.
2.7. Dot1 Contributes to t Crop Seeds Colonization
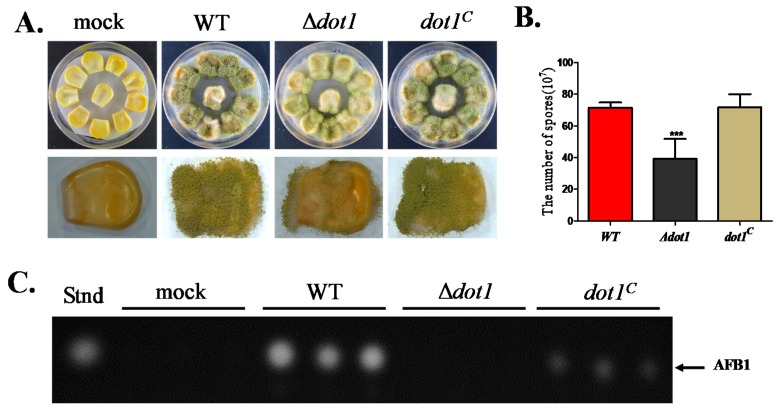
A. flavus is notorious for its contamination of many important crops and plant seeds with aflatoxins. To assay the role of Dot1 in pathogenicity, maize corn seeds were inoculated with spore suspension from WT, Δdot1 and dot1C strains. The Δdot1 mutant showed a reduced ability to colonize maize seeds (Figure 7A), which also exhibited a significant drop in conidia production (Figure 7B). Mycotoxin production from the infected maize seeds was also detected, which showed that AF production was blocked in the Δdot1 mutant (Figure 7C). These data suggested that Dot1 was important for colonizing maize seeds.
Figure 7.
Seed infection of the WT, Δdot1 and dot1C strains. (A) Phenotype of fungal strains grown on living maize corn seeds; (B) The amount of conidia was determined from the infected seeds; (C) Mycotoxin production from infected maize corn seed was detected by TLC. *** represents p ≤ 0.001. The experiments were performed with five biological replicates for each strain, and were repeated three times.
2.8. Subcellular Localization of Dot1 in A. flavus
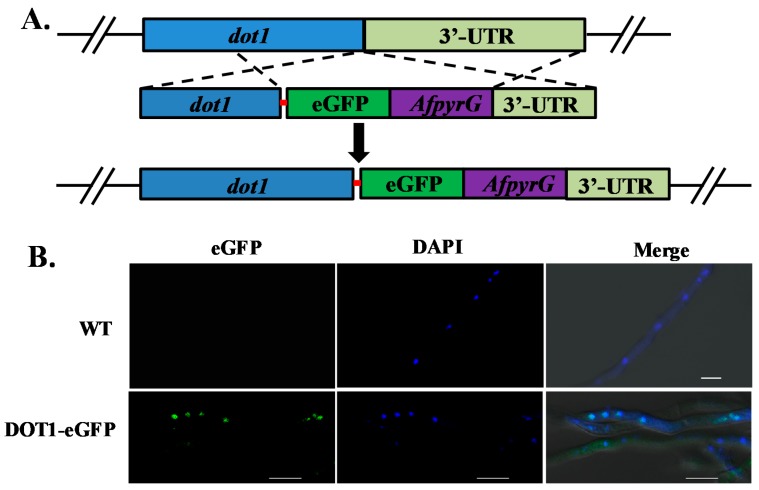
In order to determine the localization pattern of Dot1, we constructed an eGFP tag with a 5 Gly–Ala linker, and inserted the tag at the C-terminus of the Dot1 protein (Figure 8A). AfpyrG was used as the selective marker to screen the Dot1::eGFP transformants, which displayed no phenotype difference with the WT strain (data not shown), suggesting that Dot1-eGFP was fully functional. In the hyphae growth period, Dot1-eGFP was found to be mainly accumulated in the nucleus (Figure 8B), which was verified by 40, 6-diamidino-2-phenylindole (DAPI) stain, while no green fluorescence signal was found in the WT strain. All these results showed that Dot1 mainly functions in the nucleus.
Figure 8.
Location of Dot1-eGFP in A. flavus. (A) Construction strategy of dot1(p)-dot1-egfp strain. A 5 Gly–Ala linker was shown as a red line. (B) Fluorescent image of Dot1-eGFP during the hyphae growth period, the nucleus was stained by 40, 6-diamidino-2-phenylindole (DAPI).
3. Discussion
Histone methylation, as an important epigenetic modification, plays a vital role in the recruitment of other chromatin remodeling complexes and transcriptional machinery [16,29]. As an important HPTM, histone lysine methylation catalyzed by Dot1 is reported to be found from yeast to humans, which has been shown to be required for the DNA damage checkpoint [28,30,31]. However, studies of H3K79 methyltransferase in filamentous fungi are still rare, and no previous publications have yet reported the existence of these in Aspergillus. Here, we functionally characterized Dot1 in A. flavus, which shares a 33% overall identity with yeast Dot1. The locations and paralogs of dot1 in Aspergillus genomes and the phylogenetic analysis of the Dot1 constructed in this study revealed that Dot1 was quite conserved in biological functions among Aspergillus.
Histone lysine methylation has been shown to provide an epigenetic layer for transcriptional regulation, and the methylation could lead to chromatin regions being active or repressive [12,13,14,15]. In this study, we found that disruption of the H3K79 methyltransferase gene dot1 reduced A. flavus conidiation through down-regulating the transcriptional levels of the key transcription factors BrlA and AbaA [32]. Conidiation and sclerotial formation have been shown to stay balanced in Aspergillus [1]. Here, we also found that inactivation of dot1 up-regulated the expression levels of the sclerotia-related genes nsdC, nsdD and sclR, as a result, increasing sclerotia reproduction. Sclerotia is considered to be a survival structure for adapting to stress environments, here we also found that the Δdot1 mutant showed less sensitivity to genotoxicity stress, cell wall-damaging agents and oxidative stress. All this suggested that methyltransferase Dot1 is involved in fungal development and stress response.
In A. flavus, an AF gene cluster has been shown to be located near the telomeres [19,20], which are associated with specific chromatin structures [19,21]. Our former studies had shown that DNA methyltransferase and histone acetylation contribute to AF biosynthesis in A. flavus [10,33], but not for arginine methyltransferase [25], which indicates that HPTMs are involved in the regulation of AF biosynthesis. HPTMs such as acetylation, methylation, and ubiquitination have been linked to transcriptional silencing in the telomere region. The inactivation of H3K79 methylation or H3K4 methylation is quite important for heterochromatin formation and the maintenance of the silencing regions of chromatin in budding yeast [21,22,23]. An abnormal expression of the H3K79 methyltransferase gene dot1 in yeast and mammalian cells leads to a decrease in gene silence [23,24]. Here, we found that the most important secondary metaboliteaflatoxin in A. flavus was severely impaired in the dot1 deficient mutant compared to the WT strain. In A. nidulans, studies have shown that H3K9 methylation and heterochromatin protein-binding are involved in the activation of the sterigmatocystin gene cluster [34]. Simultaneously, we found that the transcription of genes relevant to aflatoxin biosynthesis, including the structural genes aflC and aflO and the regulator gene aflS, were prominently suppressed in the Δdot1 mutant. However, the expression levels of the AF global regulator gene aflR showed no difference between the Δdot1 mutant and WT strain. Dot1 might affect AF biosynthesis by regulation of the activation of AflS, which could affect the activity of AflR in A. flavus. Therefore, it could be suggested that Dot1 is involved in activating the aflatoxin biosynthesis-relevant gene cluster.
In many plant pathogens, including Magnaporthe oryzae and Fusarium graminearum, the histone lysine methylation is known to function in fungal virulence [35,36]. Many seed crops, such as maize corn and peanuts, can be potentially colonized by A. flavus, which could sporulate on injured seeds and subsequently contaminate the hosts with aflatoxin. Here the dot1C complemented strain, although normal in its phenotype, had a defect in the activation of aflS and AF production during colonization, indicating that Dot1 in the complemented strain might not fully function. However, we found that the Δdot1 mutant shows a significant reduction in pathogenicity in A. flavus. Although the physiological significance of these H3K79 methylation events remains unknown, the reduction of conidiation and aflatoxin biosynthesis as a result of the inactivation of Dot1 might be related to fungal virulence in A. flavus.
4. Conclusions
We identified a novel and functional methyltransferase Dot1 in A. flavus, and we found that Dot1 contributes to conidiation, AF metabolism and fungal virulence in A. flavus. Our preliminary results suggest valuable information that could advance our understanding of H3K79 methylation modification in the regulation of AF biosynthesis and fungal pathogenicity in A. flavus, and would provide a potential target for new control strategies of this fungal pathogen.
5. Materials and Methods
5.1. Strain and Culture Conditions
The A. flavus strains used in this study are listed in Table 1. Potato dextrose agar (PDA, BD Difco, Franklin, NJ, USA) was used for the growth and conidiation assays, supplemented with the appropriate amounts of uridine (5 mM), uracil (5 mM), or pyrithiamine (100 ng/mL) when necessary. The modified Wickerham medium (WKM) was used for the sclerotial production analysis [37]. After being grown for 7 days, the cultures in the WKM plates were washed with 70% ethanol to visualize sclerotia. PDA agar supplemented with 100 μg/mL sodium dodecyl sulfate (SDS), 2.0 mM/L tert-butyl hydroperoxide (tBooH), 15 mM hydroxyurea (HU), or 0.02% methyl methanecsulfonate (MMS) were used to determine sensitivities to multiple stresses. All the experiments were performed with technical triplicates, and were repeated three times.
Table 1.
A. flavus strains used in this study.
| Strain | Genotype Description | Reference |
|---|---|---|
| A. flavus CA14 PTs | Δku70, ΔpyrG | [38] |
| wild-type | Δku70, ΔpyrG::AfpyrG | This study |
| Δdot1 | Δku70, Δdot1::AfpyrG | This study |
| Δdot1C | Δku70, Δdot1:: AfpyrG, dot1(p)::dot1::ptrA | This study |
| Gdot1 | Δku70, dot1(p)::dot1-egfp::AfpyrG | This study |
5.2. Generation of Gene Deletion and Complementation Strains
The dot1 deficient mutant (Δdot1) and the Δdot1 complemented strain (dot1C) were generated by using a previously described method [33]. The primers used for gene knockout are listed in Table 2. A 1189-bp 5′ flanking fragment of dot1 was amplified with primers dot1/P1 and dot1/P3, and a 1033-bp 3′ flanking fragment of dot1 was amplified using primers dot1/P4 and dot1/P6. A pyrG selective marker was amplified from A. fumigatus genomic DNA with primers pyrG/F and pyrG /R. The nested primers dot1/P2 and dot1/P5 were used to generate the dot1 deletion construct containing the 5′ and 3′ flanking region and the pyrG selection marker. The fusion PCR products were purified and transformed into the protoplasts of A. flavus CA14 PTs strain using an established procedure (Yang et al., 2016). Transformants were screened by PCR with primer dot1-ORF/F and dot1-ORF/R (Table 2), and further confirmed by Southern blot analysis.
Table 2.
Primers used for gene deletion and complementation.
| Primers | Sequence (5′-3′) | Application |
|---|---|---|
| dot1/P1 | CATTGAGATGGACGAGGACG | dot1 deletion and probe |
| dot1/P3 | GGGTGAAGAGCATTGTTTGAGGCGTTGACCGAGCGGAAATGT | |
| dot1/P4 | GCATCAGTGCCTCCTCTCAGACCACTACGCGGTTACCGAGAC | |
| dot1/P6 | CGACAGAAAGCCATAATGAAAT | |
| dot1/P2 | CGAGGACGACAATGACTACAC | |
| dot1/P5 | GAGTTGAAGGGAAAGGCTAAA | |
| PyrG/F | GCCTCAAACAATGCTCTTCACCC | pyrG selective marker |
| PyrG/R | GTCTGAGAGGAGGCACTGATGC | |
| P801/R | CAGGAGTTCTCGGGTTGTCG | |
| dot1-ORF/F | TTCTTACAGTATCCGAGTGC | dot1 mutant screen |
| dot1-ORF/R | TTCATGTCCAGGAAGTGGTT | |
| CM-dot1/F | CTATGACCATGATTACGCCAACTATGACCATGATTACGCCAAGCTTAGCCTAAGCAGCAGGTGAAGC | dot1 complementation construct |
| CM-dot1/R | CCAGTGAATTCGAGCTCGGTACCGATGGCAAGGATGGGCAAAG | |
| eGFP-pyrG/F | GGAGCTGGTGCAGGCGCTGGAGCCGGTGCCATGGTGAGCAAGGGCGAGGA | egfp |
| eGFP-pyrG/R | GGGTGAAGAGCATTGTTTGAGGCTTACTTGTACAGCTCGTCCATG | |
| dot1-eGFP/P1 | ATTCTTACAGTATCCGAGTGC | dot1-gfp tag construct |
| dot1-eGFP/P3 | GGCTCCAGCGCCTGCACCAGCTCCACCCATGCTCTCTGCAAAAG | |
| dot1-eGFP/P2 | AAAATTACATACCGGAGGACG |
To complete Δdot1, a 2.9-kb PCR product (1.6-kb dot1 coding sequence, 0.8-kb upstream sequence and 0.5-kb dot1 terminator region) was amplified from A. flavus wild-type genomic DNA using primers dot1CM/F and dot1CM/R. The purified PCR product and the linearization pPTRI (Takara, Tokyo, Japan) vector were then recombined with T4 DNA ligase. The recombinant pPTR-dot1 was transformed into Δdot1 protoplasts. The pyrithiamine-resistant transformants were screened by PCR, and confirmed by reverse transcription PCR (RT-PCR).
5.3. Aflatoxin Analysis
The WT, Δdot1 and dot1C strains were incubated into 20 mL potato dextrose broth (PDB) medium in the dark at 29 °C for 5 days. Then the cultures were used for AF extraction. AF extraction was performed as previously described in [33]. Thin layer chromatography (TLC) and high performance liquid chromatography (HPLC) were both used to measure AF production with the method described in [33].
5.4. Maize Corn Infection Assay
The maize seed colonization assay was performed as described previously in [6,39]. The maize seeds were co-cultured with conidia of the WT, Δdot1 and dot1C strains 29 °C for 5 d. After five days incubation, the infected seeds were collected in 50 mL Falcon tubes, then mixed with 15 mL of sterile 0.05% Tween 80, followed by 2 min vortex to release the spores. A 100 μL aliquot of spores was removed, diluted, and counted haemocytometrically. 15 mL of chloroform were added to the Falcon tubes, and the tubes were shaken at 29 °C for 30 min. 10 mL of the organic layer was removed, dried down, and resuspended in 1 mL chloroform. Finally, AF production was detected by TLC.
5.5. Gene Expression Detection
The gene expression level was detected by qRT-PCR. 48 h old mycelia of A. flavus were collected, washed, and lyophilized from PDA medium. Total RNA was isolated according to the protocol previously described by Yang et al. [6]. SYBR Green Supermix (Takara) was used for the qRT-PCR reaction with the PikoReal 96 Real-time PCR system. The 2−ΔΔCT method was used to calcite the relative quantification of each transcript [40]. The qRT-PCR primers are listed in Table 3.
Table 3.
Primers used for RT-PCR.
| Primers | Sequence (5′-3′) | Application |
|---|---|---|
| brlA/QF | GCCTCCAGCGTCAACCTTC | brlA qRT-PCR |
| brlA/QR | TCTCTTCAAATGCTCTTGCCTC | |
| abaA/QF | CACGGAAATCGCCAAAGAC | abaA qRT-PCR |
| abaA/QR | TGCCGGAATTGCCAAAG | |
| nsdC/QF | GCCAGACTTGCCAATCAC | nsdC qRT-PCR |
| nsdC/QR | CATCCACCTTGCCCTTTA | |
| nsdD/QF | GGACTTGCGGGTCGTGCTA | nsdD qRT-PCR |
| nsdD/QR | AGAACGCTGGGTCTGGTGC | |
| sclR/QF | CAATGAGCCTATGGGAGTGG | sclR qRT-PCR |
| sclR/QR | ATCTTCGCCCGAGTGGTT | |
| aflC/QF | GTGGTGGTTGCCAATGCG | aflC qRT-PCR |
| aflC/QR | CTGAAACAGTAGGACGGGAGC | |
| aflR/QF | AAAGCACCCTGTCTTCCCTAAC | aflR qRT-PCR |
| aflR/QR | GAAGAGGTGGGTCAGTGTTTGTAG | |
| aflS/QF | CGAGTCGCTCAGGCGCTCAA | aflS qRT-PCR |
| aflS/QR | GCTCAGACTGACCGCCGCTC | |
| aflO/QF | GATTGGGATGTGGTCATGCGATT | aflO qRT-PCR |
| aflO/QR | GCCTGGGTCCGAAGAATGC | |
| Actin/QF | ACGGTGTCGTCACAAACTGG | actin qRT-PCR |
| Actin/QR | CGGTTGGACTTAGGGTTGATAG |
5.6. Generation of Dot1-eGFP Strain
The former approach for protein location was performed to generate the Dot1-GFP strain [6,41]. To generate the dot1-eGFP construct, a 1068 bp dot1 ORF without the termination codon (TAG) and a 1033-bp 3′ flanking fragment of dot1 was amplified from A. flavus WT strain gDNA using the primer pairs dot1-eGFP, P1-dot1-eGFP/P3 and dot1/P4, dot1/P6, respectively. The primers dot1-eGFP/P2 and dot1-eGFP/P5 were used to generate the Dot1-eGFP fusion PCR cassettes containing the upstream and downstream region, egfp and the pyrG selection marker. The purified fusion PCR products were inserted into the protoplasts of the A. flavus CA14 PTs strain, and the selected transformants were then confirmed by PCR.
5.7. Microscopic Determination of Dot1-eGFP Location
To assay the subcellular location pattern of Dot1-eGFP, 12 h growth mycelial was collected and analyzed using a Leica SP8 confocal laser scanning microscope. Dual-channel imaging was used to sequentially image cells labeled with DAPI (excitation: 405 nm, emission bandwidth: 420–460 nm), and eGFP (excitation: 488 nm, emission bandwidth: 525/565 nm).
5.8. Statistical Analysis
Statistical analysis was performed using ANOVA and least significant difference (LSD) tests to determine significant differences among group means. A p-values less than 0.05 was regarded as statistically significant.
Acknowledgments
Funding was provided for this research from (No. 2013CB127802), the grants of the National Natural Science Foundation of China (No. 31172297). We especially thanked Dr Perng Kuang Chang (Southern Regional Research Center, United States Department of Agriculture, New Orleans, USA), Prof. Yang Liu (Institute of Food Science and Technology CAAS), Prof. Kong Qing for their kindness to provide the strains. We thanked Dr. Yanyun Li (FAFU, Life Science College) for her help in taking the confocal images. We thanked Center for Molecular Cell and Systems Biology, Life Science College of FAFU, for providing facilities and support.
Author Contributions
Kunlong Yang and Shihua Wang designed the experiments and wrote the manuscript. Linlin Liang, Yinghang Liu and Kunlong Yang performed all the experiments. Guinan Lin, Zhangling Xu, Huahui Lan and Xiuna Wang performed a few experiments and data analysis. All authors read and approved the final manuscript.
Conflicts of Interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Amaike S., Keller N.P. Aspergillus flavus. Ann. Rev. Phytopathol. 2011;49:107–133. doi: 10.1146/annurev-phyto-072910-095221. [DOI] [PubMed] [Google Scholar]
- 2.Yang K., Zhuang Z., Zhang F., Song F., Zhong H., Ran F., Yu S., Xu G., Lan F., Wang S. Inhibition of aflatoxin metabolism and growth of Aspergillus flavus in liquid culture by a DNA methylation inhibitor. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2015;32:554–563. doi: 10.1080/19440049.2014.972992. [DOI] [PubMed] [Google Scholar]
- 3.Abbas H.K., Shier W.T., Plasencia J., Weaver M.A., Bellaloui N., Kotowicz J.K., Butler A.M., Accinelli C., de la Torre-Hernandez M.E., Zablotowicz R.M. Mycotoxin contamination in corn smut (ustilago maydis) galls in the field and in the commercial food products. Food Control. 2017;71:57–63. doi: 10.1016/j.foodcont.2016.06.006. [DOI] [Google Scholar]
- 4.Hedayati M.T., Pasqualotto A.C., Warn P.A., Bowyer P., Denning D.W. Aspergillus flavus: Human pathogen, allergen and mycotoxin producer. Microbiology. 2007;153:1677–1692. doi: 10.1099/mic.0.2007/007641-0. [DOI] [PubMed] [Google Scholar]
- 5.Nie X., Yu S., Qiu M., Wang X., Wang Y., Bai Y., Zhang F., Wang S. Aspergillus flavus sumo contributes to fungal virulence and toxin attributes. J. Agric. Food Chem. 2016;64:6772–6782. doi: 10.1021/acs.jafc.6b02199. [DOI] [PubMed] [Google Scholar]
- 6.Yang K., Liu Y., Liang L., Li Z., Qin Q., Nie X., Wang S. The high-affinity phosphodiesterase pdeh regulates development and aflatoxin biosynthesis in Aspergillus flavus. Fungal Genet. Biol. FG B. 2017;101:7–19. doi: 10.1016/j.fgb.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Chalivendra S.C., DeRobertis C., Chang P.K., Damann K.E. Cyclopiazonic acid is a pathogenicity factor for Aspergillus flavus and a promising target for screening germplasm for ear rot resistance. Mol. Plant Microbe Interact. MPMI. 2017;30:361–373. doi: 10.1094/MPMI-02-17-0026-R. [DOI] [PubMed] [Google Scholar]
- 8.Bosso L., Lacatena F., Varlese R., Nocerino S., Cristinzio G., Russo D. Plant pathogens but not antagonists change in soil fungal communities across a land abandonment gradient in a mediterranean landscape. Acta Oecol. 2017;78:1–6. doi: 10.1016/j.actao.2016.11.002. [DOI] [Google Scholar]
- 9.Gacek-Matthews A., Berger H. Kdmb, a jumonji histone h3 demethylase, regulates genome-wide h3k4 trimethylation and is required for normal induction of secondary metabolism in aspergillus nidulans. PLoS Genet. 2016;12:e1006222. doi: 10.1371/journal.pgen.1006222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan H., Sun R., Fan K., Yang K., Zhang F., Nie X.Y., Wang X., Zhuang Z., Wang S. The Aspergillus flavus histone acetyltransferase aflgcne regulates morphogenesis, aflatoxin biosynthesis, and pathogenicity. Front. Microbiol. 2016;7:1324. doi: 10.3389/fmicb.2016.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macheleidt J., Mattern D.J., Fischer J., Netzker T., Weber J., Schroeckh V., Valiante V., Brakhage A.A. Regulation and role of fungal secondary metabolites. Annu. Rev. Genet. 2016;50:371–392. doi: 10.1146/annurev-genet-120215-035203. [DOI] [PubMed] [Google Scholar]
- 12.Connolly L.R., Smith K.M., Freitag M. The fusarium graminearum histone h3 k27 methyltransferase kmt6 regulates development and expression of secondary metabolite gene clusters. PLoS Genet. 2013;9:e1003916. doi: 10.1371/journal.pgen.1003916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brakhage A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 15.Gacek A., Strauss J. The chromatin code of fungal secondary metabolite gene clusters. Appl. Microbiol. Biotechnol. 2012;95:1389–1404. doi: 10.1007/s00253-012-4208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strauss J., Reyes-Dominguez Y. Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet. Biol. FG B. 2011;48:62–69. doi: 10.1016/j.fgb.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis Z.A., Honda S., Khlafallah T.K., Jeffress J.K., Freitag M., Mohn F., Schubeler D., Selker E.U. Relics of repeat-induced point mutation direct heterochromatin formation in neurospora crassa. Genome Res. 2009;19:427–437. doi: 10.1101/gr.086231.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adhvaryu K.K., Morris S.A., Strahl B.D., Selker E.U. Methylation of histone h3 lysine 36 is required for normal development in neurospora crassa. Eukaryot. Cell. 2005;4:1455–1464. doi: 10.1128/EC.4.8.1455-1464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer J.M., Mallaredy S., Perry D.W., Sanchez J.F., Theisen J.M., Szewczyk E., Oakley B.R., Wang C.C., Keller N.P., Mirabito P.M. Telomere position effect is regulated by heterochromatin-associated proteins and nkua in aspergillus nidulans. Microbiology. 2010;156:3522–3531. doi: 10.1099/mic.0.039255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller N.P., Turner G., Bennett J.W. Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi Y.H., Schulze J.M., Jackson J., Hentrich T., Seidel C., Jaspersen S.L., Kobor M.S., Shilatifard A. Dot1 and histone h3k79 methylation in natural telomeric and hm silencing. Mol. Cell. 2011;42:118–126. doi: 10.1016/j.molcel.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osborne E.A., Dudoit S., Rine J. The establishment of gene silencing at single-cell resolution. Nat. Genet. 2009;41:800–806. doi: 10.1038/ng.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacoste N., Utley R.T., Hunter J.M., Poirier G.G., Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone h3 methyltransferase. J. Biol. Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 24.Ng H.H., Feng Q., Wang H., Erdjument-Bromage H., Tempst P., Zhang Y., Struhl K. Lysine methylation within the globular domain of histone h3 by dot1 is important for telomeric silencing and sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., He Y., Li X., Fasoyin O.E., Hu Y., Liu Y., Yuan J., Zhuang Z., Wang S. Histone methyltransferase aflrmta gene is involved in the morphogenesis, mycotoxin biosynthesis, and pathogenicity of Aspergillus flavus. Toxicon. 2017;127:112–121. doi: 10.1016/j.toxicon.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Gacek-Matthews A., Noble L.M., Gruber C., Berger H., Sulyok M., Marcos A.T., Strauss J., Andrianopoulos A. Kdma, a histone h3 demethylase with bipartite function, differentially regulates primary and secondary metabolism in aspergillus nidulans. Mol. Microbiol. 2015;96:839–860. doi: 10.1111/mmi.12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Govindaraghavan M., Anglin S.L., Osmani A.H., Osmani S.A. The set1/compass histone h3 methyltransferase helps regulate mitosis with the cdk1 and nima mitotic kinases in aspergillus nidulans. Genetics. 2014;197:1225–1236. doi: 10.1534/genetics.114.165647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossodivita A.A., Boudoures A.L., Mecoli J.P., Steenkiste E.M., Karl A.L., Vines E.M., Cole A.M., Ansbro M.R., Thompson J.S. Histone h3 k79 methylation states play distinct roles in uv-induced sister chromatid exchange and cell cycle checkpoint arrest in saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:6286–6299. doi: 10.1093/nar/gku242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croken M.M., Nardelli S.C., Kim K. Chromatin modifications, epigenetics, and how protozoan parasites regulate their lives. Trends Parasitol. 2012;28:202–213. doi: 10.1016/j.pt.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ontoso D., Acosta I., van Leeuwen F., Freire R., San-Segundo P.A. Dot1-dependent histone h3k79 methylation promotes activation of the mek1 meiotic checkpoint effector kinase by regulating the hop1 adaptor. PLoS Genet. 2013;9:e1003262. doi: 10.1371/journal.pgen.1003262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bani Ismail M., Shinohara M., Shinohara A. Dot1-dependent histone h3k79 methylation promotes the formation of meiotic double-strand breaks in the absence of histone h3k4 methylation in budding yeast. PLoS ONE. 2014;9:e96648. doi: 10.1371/journal.pone.0096648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han S., Adams T.H. Complex control of the developmental regulatory locus brla in aspergillus nidulans. Mol. Genet. Genom. MGG. 2001;266:260–270. doi: 10.1007/s004380100552. [DOI] [PubMed] [Google Scholar]
- 33.Yang K., Liang L., Ran F., Liu Y., Li Z., Lan H., Gao P., Zhuang Z., Zhang F., Nie X., et al. The dmta methyltransferase contributes to Aspergillus flavus conidiation, sclerotial production, aflatoxin biosynthesis and virulence. Sci. Rep. 2016;6:23259. doi: 10.1038/srep23259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reyes-Dominguez Y., Bok J.W., Berger H., Shwab E.K., Basheer A., Gallmetzer A., Scazzocchio C., Keller N., Strauss J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in aspergillus nidulans. Mol. Microbiol. 2010;76:1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pham K.T., Inoue Y., Vu B.V., Nguyen H.H., Nakayashiki T., Ikeda K., Nakayashiki H. Moset1 (histone h3k4 methyltransferase in magnaporthe oryzae) regulates global gene expression during infection-related morphogenesis. PLoS Genet. 2015;11:e1005385. doi: 10.1371/journal.pgen.1005385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y., Liu N., Yin Y., Chen Y., Jiang J., Ma Z. Histone h3k4 methylation regulates hyphal growth, secondary metabolism and multiple stress responses in fusarium graminearum. Environ. Microbiol. 2015;17:4615–4630. doi: 10.1111/1462-2920.12993. [DOI] [PubMed] [Google Scholar]
- 37.Chang P.K., Scharfenstein L.L., Mack B., Ehrlich K.C. Deletion of the Aspergillus flavus orthologue of a. Nidulans flug reduces conidiation and promotes production of sclerotia but does not abolish aflatoxin biosynthesis. Appl. Environ. Microbiol. 2012;78:7557–7563. doi: 10.1128/AEM.01241-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang P.K., Scharfenstein L.L., Wei Q., Bhatnagar D. Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J. Microbiol. Methods. 2010;81:240–246. doi: 10.1016/j.mimet.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Tsitsigiannis D.I., Keller N.P. Oxylipins act as determinants of natural product biosynthesis and seed colonization in aspergillus nidulans. Mol. Microbiol. 2006;59:882–892. doi: 10.1111/j.1365-2958.2005.05000.x. [DOI] [PubMed] [Google Scholar]
- 40.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Yang K., Qin Q., Liu Y., Zhang L., Liang L., Lan H., Chen C., You Y., Zhang F., Wang S. Adenylate cyclase acya regulates development, aflatoxin biosynthesis and fungal virulence in Aspergillus flavus. Front. Cell. Infect. Microbiol. 2016;6:190. doi: 10.3389/fcimb.2016.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]