Abstract
3D analytical pathology imaging examines high resolution 3D image volumes of human tissues to facilitate biomedical research and provide potential effective diagnostic assistance. Such approach – quantitative analysis of large-scale 3D pathology image volumes – generates tremendous amounts of spatially derived 3D micro-anatomic objects, such as 3D blood vessels and nuclei. Spatial exploration of such massive 3D spatial data requires effective and efficient querying methods. In this paper, we present a scalable and efficient 3D spatial query system for querying massive 3D spatial data based on MapReduce. The system provides an on-demand spatial querying engine which can be executed with as many instances as needed on MapReduce at runtime. Our system supports multiple types of spatial queries on MapReduce through 3D spatial data partitioning, customizable 3D spatial query engine, and implicit parallel spatial query execution. We utilize multi-level spatial indexing to achieve efficient query processing, including global partition indexing for data retrieval and on-demand local spatial indexing for spatial query processing. We evaluate our system with two representative queries: 3D spatial joins and 3D k-nearest neighbor query. Our experiments demonstrate that our system scales to large number of computing nodes, and efficiently handles data-intensive 3D spatial queries that are challenging in analytical pathology imaging.
Keywords: 3D Spatial Queries, 3D Digital Pathology, MapReduce, Spatial Join, k nearest neighbor search
1. INTRODUCTION
Over the last decade, three-dimensional (3D) spatial data is produced at an unprecedented rate and scale in numerous fields and scientific domains, ranging from modelling of 3D buildings for urban planning, high-resolution topography data for geographic research, to 3D digital pathology imaging for biomedical research and clinical diagnosis [2, 5]. 3D spatial objects have much more complex shapes and structures compared to 2D spatial data, resulting in spatial applications highly data- and compute-intensive.
Recently, 3D digital pathology is made possible through slicing tissues into serial sections. By registering consecutive slices, segmenting and reconstructing 3D micro-anatomic objects, it is possible to provide a 3D tissue view to explore spatial relationships and patterns among micro-anatomic objects to support biomedical research [6, 7]. For example, liver disease diagnosis and analytics rely on 3D structural features of blood vessels and their 3D spatial relationships with cells. The information-lossless 3D tissue view with microscopy imaging volumes holds significant potential to enhance the study of both normal and disease processes, and represents a new frontend for digital pathology.
A single biomedical study generates tremendous amounts of spatially derived quantifications for 3D micro-anatomic objects, and spatial explorations will then be performed on such spatial data to support biomedical research or clinical diagnosis. Common queries include feature based queries, spatial relationship based queries such as spatial joins to cross-match and compare results generated with different methods, spatial proximity based queries such as nearest neighbor search, spatial analytics such as finding spatial clusters. These queries bear significant resemblance to traditional GIS queries, but in 3D space and at extreme scale. To support efficient and scalable queries, we need to address several challenges such as the large scale data size, the 3D complex structures and representations, and the high geometric computation complexity.
Traditional spatial database management systems (SDBMS) such as PostGIS and OracleSpatial are not feasible for large scale 3D objects with complex structures and representations due to their heavy cost of I/O and limited support of geometric computations. The unique challenges of large scale 3D queries demand a highly scalable and efficient system that can run on large scale computing resources such as computing clusters or clouds. Recently, several systems have been proposed to support large scale 2D spatial queries [4, 10], but none of them provide spatial queries on 3D objects with complex structures and representations.
In this paper, we propose and develop a 3D spatial query processing framework built on MapReduce - Hadoop-GIS 3D, which takes advantage of not only traditional SDBMS based spatial indexing and querying methods for efficiency, but also distributed computing of MapReduce for scalability. We provide multi-level spatial indexing supported with 3D spatial partitioning methods: global storage indexing for minimizing data reading, cuboid indexing for reducing data for computation, object-level indexing for index based query processing, and structure-level indexing for geometric computations on 3D objects with complex structures. We provide a unique on-demand spatial querying engine which can be invoked easily with as many instances as needed at run-time, thus highly scalable. This is very different from traditional SDBMS engine which has a limited fixed set of instances. Multi-types of spatial querying pipelines are built on MapReduce, such as spatial joins and k-nearest neighbor queries. We propose skeleton based indexing and querying methods to support queries on objects with high complex 3D structures such as blood vessels. We perform extensive experiments to demonstrate the feasibility and scalability of our system.
2. OVERVIEW
2.1 System Architecture
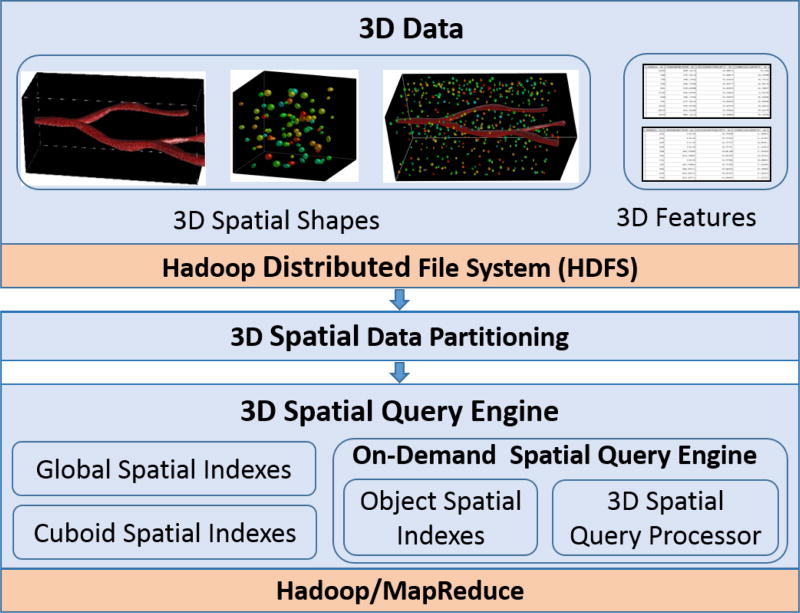
We aim to develop a highly scalable, cost-effective and efficient 3D spatial query processing system to support multiple analytical queries on large scale spatial data by taking advantage of MapReduce that runs on commodity clusters. To achieve this, we propose Hadoop-GIS 3D, a MapReduce-based system for data- and compute-intensive 3D spatial queries, and present its architecture in Fig. 1. Given 3D spatial data and features as input, we store them on Hadoop Distributed File System (HDFS) for efficient parallel access. We then spatially partition the input 3D data into cuboids, and take each cuboid as a parallel processing unit for spatial queries. We develop a stand-alone customizable query engine that can be invoked on demand to execute 3D spatial queries in parallel through MapReduce. Our 3D spatial query engine consists of multi-level spatial indexing and spatial query processor on top of Hadoop.
Figure 1.
System architecture of Hadoop-GIS 3D
2.2 3D On-demand Spatial Query Engine
To support high performance 3D spatial queries for large scale 3D dataset, we develop a 3D On-Demand Spatial Query Engine (ODSQUE), as shown in the architecture in Fig. 1. ODSQUE is a standalone 3D spatial query engine, and is generic to be extended and customized to leverage existing indexing approaches and query pipelines to support a variety of 3D spatial queries. The engine can be invoked easily with as many instances as needed at run-time, thus highly scalable. It is compiled as a shared library to be easily deployed on multiple cluster nodes to achieve better parallelization.
In the system, we take advantage of spatial access methods for query processing with multi-level indexing. At the higher level, Hadoop-GIS 3D creates global subspace based spatial indexes of partitioned cuboids for HDFS file split filtering. Thus, for certain query types such as point and window based queries, we can efficiently filter out irrelevant cuboids through global subspace indexes. The global subspace index is small to be stored as a binary file on HDFS, and shared across cluster nodes through Hadoop distributed cache mechanism. At the lower level, ODSQUE supports an object-level on demand indexing approach by building spatial index on the 3D objects within cuboids on the fly, and stores index files in main memory. As cuboid size is relatively small, object-level index creation cost on a single cuboid is very low, and the index greatly speeds up spatial query processing.
3. 3D SPATIAL DATA PARTITIONING AND MULTI-LEVEL INDEXING
3.1 Spatial Data Partitioning
Spatial data partitioning offers 3D data partitioning and generates a set of cuboids, which becomes a processing unit for querying tasks on MapReduce. A large set of such tasks can be processed in parallel without data dependance or communication requirements. Therefore, spatial partitioning significantly improves computational parallelization thus overall performance of spatial query processing.
For pathology image volumes, we perform a 3D fixed-grid partitioning algorithm which is commonly used in pathology image analysis. With the input data, we first obtain their minimal bounding boxes (MBBs) by MBB extraction. We then collect the input space information and the total number of objects to generate partition schema. The partition schema is saved as a global cache file that is shared by all the cluster nodes for further query processing. After partitioning, each object within a cuboid is assigned a corresponding cubiod id cubiod_id, and the MBBs of cuboids are maintained by a global cuboid index.
3.2 3D Spatial Indexing
We provide a multi-level spatial indexing strategy in Hadoop-GIS 3D to accelerate spatial queries: global storage indexing, cuboid indexing, object-level indexing and structure-level indexing. A subspace is a higher level 3D box on top of cuboids and is taken as a coarse partitioning where the resulting file size is close to HDFS block size. A global storage index is created based on subspaces and used to maintain relationships between subspaces and the corresponding HDFS files for storage level filtering. Cuboid indexing is to manage cuboid CIDs generated from spatial partitioning and their corresponding MBBs. As cuboid CIDs can be taken as MapReduce keys, cuboid indexes are used to filter keys for MapReduce queries, thus serve as computational filtering to reduce the number of MapReduce tasks. Global storage indexing and cuboid indexing enable effective HDFS level data filtering and MapReduce level computation filtering, respectively. Object-level indexing is an on-demand indexing approach. The object-level index is created on-the-fly for each cuboid involved in spatial query to improve the performance of index based queries such as spatial joins. As it is small in size, object-level index can be stored in main memory for fast access during MapReduce task execution. Structure-level indexing is an intra-object level indexing built from the structural primitives of 3D objects with complicated structures such as blood vessels. For spatial queries where accurate geometrical computations are necessary, a structure level index is created on demand for each involved 3D object to speed up geometry computation.
4. 3D SPATIAL QUERY PROCESSING
4.1 3D Spatial Join
In this section, we discuss how we adapt MapReduce framework to efficiently support spatial join query for 3D analytical pathology imaging. In the Map function, each 3D object from input is converted into input key/value pair (k, v), where k is the byte offset of the line in the input file and v consists of the 3D geometry and its extracted MBB. We first load the partition schema from the cache file generated by spatial data partitioning, and build an R*-tree on the partition schema as a global index file. For each record v, we traverse the R*-tree to find out the intersecting cuboid and record its id as cuboid_id. Then an intermediate key/value pair (cuboid_id, val) is generated and emitted as output from the Map phase.
Spatial objects with same cuboid_id are grouped together during the shuffle phase, and they are sent to the same reducer for processing. Here we take a filter-and-refine strategy to reduce the computational cost of spatial predicate on 3D geometries, and the significant I/O overhead of reading and writing 3D spatial data. With the MBB of each 3D object in cubiod C, we perform 3D R*-tree bulk spatial indexing on one dataset to generate a local index file. The 3D R*-tree index file contains MBBs in its interior nodes and actual geometries (polyhedrons) in its leaf nodes, and we store it in the main memory for further query processing. To perform spatial join query, for each 3D object in the other dataset, we first query its MBB on the R*-tree as a rough filtering step to eliminate objects pairs with no MBB intersection. Next we apply spatial refinement to the polyhedron pairs by accurate 3D geometric operations for those candidates with MBB intersection. If required by users, we perform 3D geometrical computation for quantitative spatial measurements such as intersection volumes and overlap ratios. Finally, we output the execution results onto HDFS.
4.2 3D K-Nearest Neighbor Query
We propose two algorithms to support efficient k-NN query: 1-NN with 3D Voronoi diagram and k-NN with 3D R*-tree. The former takes a skeleton based approach for representing complex blood vessel objects, and the latter provides an Axis-Aligned Bounding Box (AABB) tree based method for indexing blood vessels to speed up computation.
4.2.1 3D NN with Voronoi Diagram
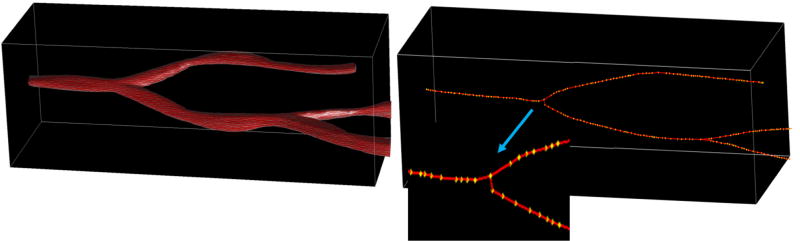
To execute the NN query of searching the nearest 3D blood vessel for each 3D cell, we need to build a 3D Voronoi diagram for the blood vessels. Due to the complex structure of 3D blood vessels, we cannot simply approximate them as 3D points. In our algorithm, we extract the skeleton of each 3D blood vessel, and represent each vessel structure by its skeleton vertices. Fig. 2 shows a 3D blood vessel structure and its extracted skeleton, and the 3D yellow dots represent skeleton vertices.
Figure 2.
A 3D blood vessel and its 3D skeleton with one close-up view of a subpart. The yellow dots are skeleton vertices.
In the Voronoi-based 3D NN query, the input 3D cells are partitioned into cuboids and distributed among cluster nodes for NN query. For 3D blood vessels, we first extract their skeletons using the Mean Curvature Skeleton (MCS) algorithm [9] by a Mapper-only job. In the Mapper, each record of 3D blood vessel is converted into a key/value pair (k, v), where k is the byte offset of the line in the input file and is taken as the id of the blood vessel, and v is the 3D geometry record. Next we apply the MCS algorithm to each record and emit the key/value pair (k, val) as output where val is the skeleton vertices. We take the 3D skeleton vertices of all blood vessels as input sites for Voronoi construction and store the 3D Voronoi diagram as a global cache file for efficient access by computing nodes. With the 3D Voronoi diagram, we perform NN search for every cell within the cuboids and output the final query results to HDFS.
4.2.2 3D K-NN with R*-tree
To support efficient k-NN query, R-tree based algorithms introduce two metrics mindist and minmaxdist for space search speedup [8]. In our system, we build 3D R*-tree for k-NN query by extending SpatialIndex library [3].
Similarly to the NN query process with 3D Voronoi diagram, we partition the 3D cells into cuboids and distribute them into cluster nodes for parallel processing. We extract the MBB of each 3D blood vessel and store both the MBB and the geometry of all records as a global cache file. As the number of blood vessels is much smaller (hundreds or thousands) than that of cells (millions) in a typical 3D pathology image volume, the storage is almost negligible. We construct the 3D R*-tree on the cache file and perform k-NN query for each given 3D cell to obtain its k nearest neighbors. We compute the accurate distances between the queried 3D cell and its k nearest 3D blood vessels by building an AABB tree on each of the k nearest 3D blood vessels to speed up distance calculation. After we obtain the results for each cuboid, we perform result aggregation and output results on HDFS.
5. EXPERIMENTAL RESULTS
We study the performance of our 3D spatial query system on a cluster environment. The cluster has five nodes with 124 cores (Intel(R) Xeon(R) CPU E5-2650 v3 at 2.30GHz). Each node comes with 5TB hard drive at 7200rmp and 128GB memory. Cluster nodes are connected via a 1Gb network and the OS for each node is CentOS 6.7 (64 bit). We use Apache Hadoop 2.7.1 as our MapReduce platform, and adopt Boost 1.57.0 and CGAL 4.8 [1] libraries for 3D geometric computation and spatial measurement in the spatial query engine. We also extend the SpatialIndex library 1.8.1 [3] for index building. Datasets are uploaded to HDFS and the replication factor is set as 3 for each datanode.
We evaluate the performance of 3D spatial join with five datasets of various data sizes (90GB to 460GB), and test k-NN query with 3D datasets containing both nuclei and blood vessels. The number of nuclei in 3D datasets is from 0.6 million to 3 million and the average number of 3D vertices per nucleus is roughly 2,000. Besides 3D nuclei, the datasets for k-NN query testing also contain several thousand 3D blood vessels.
5.1 3D Spatial Join Performance
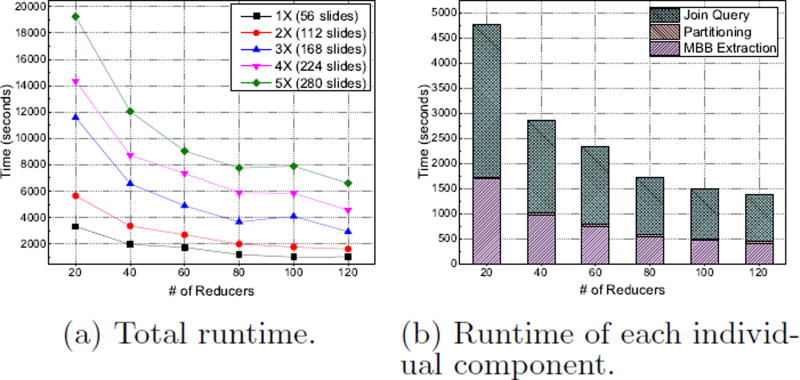
We run 3D spatial join query on our datasets and evaluate its performance with varying number of reducers. As shown in Fig. 3, it takes 958 seconds to process 1× dataset (90GB) with 120 reducers, demonstrating the efficiency of our system. The system also presents good scalability as we can see a continuous decline of query time when the number of reducers increases in Fig. 3(a). It achieves a nearly linear speedup, e.g., on average the total query runtime is reduced to half as the number of reducers increases from 20 to 40. Fig. 3(a) also presents the processing time for join query on 5× dataset (280 slides) is roughly 5 times of that on 1× dataset (56 slides), showing the linear increase of query processing time with the increase of dataset size.
Figure 3.
System Scalability of Spatial Join Query
We present in Fig. 3(b) the detailed run time of each component in spatial join query pipeline for 3× dataset (168 slides) with 80 reducers. For the MBB extraction and spatial join query modules, the runtime is linearly decreasing as more reducers are used. While for spatial partitioning step, as we use the 3D fixed-grid partitioning algorithm, its runtime is negligible and almost consistent with different number of reducers. Overall, each component in the 3D spatial join query process shows good scalability, making the system well-suited for large scale 3D spatial datasets.
5.2 3D Nearest Neighbor Query Performance
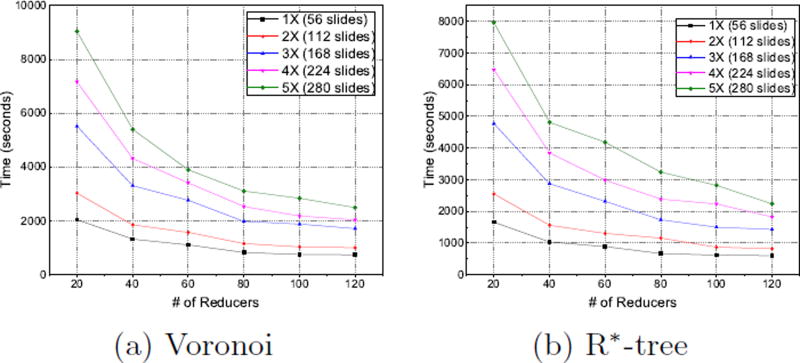
For 3D nearest neighbor query performance testing, we evaluate the example query“return the distance to the nearest blood vessel for each nuclei” on the datasets containing both 3D nuclei and blood vessels, and report the results in Fig. 4. As shown in both figures, the execution time is linearly decreased when the number of reducers increases, exhibiting good scalability of the system for nearest neighbor query by Voronoi and R*-tree.
Figure 4.
System Scalability of NN Query
6. CONCLUSION
3D analytical pathology imaging provides high potential to facilitate biomedical research and computer aided diagnosis. In this paper, we present a MapReduce based system to support high performance 3D spatial queries on large scale datasets, and demonstrate its scalability and efficiency on supporting two representative spatial query types. The system is generic and applicable to similar 3D spatial queries from other domains such as scientific simulation and remote sensing.
Acknowledgments
This research is supported in part by grants from National Science Foundation ACI 1443054 and IIS 1350885, National Institute of Health K25CA181503, and the Emoy University Research Committee.
References
- 1.The computational geometry algorithms library (cgal) http://www.cgal.org/
- 2.Openstreetmap-3d. http://www.osm-3d.org/
- 3.Spatial index library. http://libspatialindex.github.com.
- 4.Aji A, Wang F, Vo H, Lee R, Liu Q, Zhang X, Saltz J. Hadoop gis: a high performance spatial data warehousing system over mapreduce. Proceedings of the VLDB Endowment. 2013;6(11):1009–1020. [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Janabi S, Huisman A, Van Diest PJ. Digital pathology: current status and future perspectives. Histopathology. 2012;61(1):1–9. doi: 10.1111/j.1365-2559.2011.03814.x. [DOI] [PubMed] [Google Scholar]
- 6.Liang Y, Wang F, Treanor D, Magee D, Teodoro G, Zhu Y, Kong J. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015. Springer; 2015. A 3d primary vessel reconstruction framework with serial microscopy images; pp. 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang Y, Wang F, Treanor D, Magee D, Teodoro G, Zhu Y, Kong J. 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI) IEEE; 2015. Liver whole slide image analysis for 3d vessel reconstruction; pp. 182–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roussopoulos N, Kelley S, Vincent F. ACM sigmod record. ACM; 1995. Nearest neighbor queries; pp. 71–79. [Google Scholar]
- 9.Tagliasacchi A, Alhashim I, Olson M, Zhang H. Computer Graphics Forum. Wiley Online Library; 2012. Mean curvature skeletons; pp. 1735–1744. [Google Scholar]
- 10.Xie D, Li F, Yao B, Li G, Zhou L, Guo M. Simba: Efficient in-memory spatial analytics. 2016 [Google Scholar]