Abstract
The unfolded protein response (UPR) is an ancient cellular pathway that detects and alleviates protein-folding stresses. The UPR components X-box binding protein 1 (XBP1) and inositol-requiring enzyme 1α (IRE1α) promote type I interferon (IFN) responses. Here, we found that Xbp1-deficient mouse embryonic fibroblasts and macrophages had impaired antiviral resistance. Unexpectedly, this was not because of a defect in type I IFN responses, but rather an inability of Xbp1-deficient cells to undergo viral-induced apoptosis. The ability to undergo apoptosis directly limited infection in WT cells. Xbp1-deficient cells were generally resistant to the intrinsic pathway of apoptosis through an indirect mechanism involving activation of the nuclease IRE1α. We observed an IRE1α-dependent reduction in the abundance of the pro-apoptotic microRNA miR-125a, and a corresponding increase in the amounts of the members of the anti-apoptotic Bcl2 family. The activation of IRE1α by the hepatitis C virus (HCV) protein NS4B in Xbp1-proficient cells also conferred apoptosis resistance and promoted viral replication. Furthermore, we found evidence of IRE1α activation and decreased miR-125a abundance in liver biopsies from patients infected with HCV compared to those in the livers of healthy controls. Our results reveal a pro-survival role for IRE1α in virally infected cells, and suggest a possible target for IFN-independent antiviral therapy.
Introduction
Great advances have been made in our understanding of the molecular definitions of pattern recognition receptors (PRRs) and the pathogen-associated molecular patterns (PAMPs) that cells use to distinguish viruses from self (1, 2). PRR engagement results in the transcription of the genes that encode the type I interferons (IFNs) IFN-α and IFN-β, which bind to the type I IFN receptor (IFNAR) to induce the expression of hundreds of IFN-stimulated genes (ISGs). ISGs act in concert to block further viral replication and spread, as well as to support the activation of adaptive antiviral immunity (3). However, many viruses have evolved evasion mechanisms to limit PRR recognition and signal transduction, and PRR-independent mechanisms for innate sensing of viral infections remain unclear.
Endoplasmic reticulum (ER) stress occurs during infection by various viruses, presumably due to the overwhelming synthesis of viral proteins (4). The unfolded protein response (UPR) is a ubiquitous cellular pathway to detect and alleviate ER stress. The UPR is initiated by three sensors that reside within the ER: protein kinase receptor-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme 1 (IRE1) (5–7). IRE1, a highly conserved UPR sensor, oligomerizes and autophosphorylates in response to ER stress, which activates its cytosolic RNase domain and initiates a nonconventional mRNA splicing reaction of Xbp1 mRNA (8). Once processed, the spliced Xbp1 mRNA encodes a transcription factor, which controls the expression of target genes. IRE1α targets other specific mRNAs, leading to their degradation in a process termed regulated IRE1-dependent decay (RIDD) (9, 10).
ER stress synergistically enhances cytokine and IFN responses to PRR engagement through IRE1α and XBP1 (11–13). Specific activation of IRE1α also occurs during innate immune recognition of PAMPs by Toll-like receptors (TLRs) (11). In this setting, XBP1 promotes the production of inflammatory cytokines and IFN-β. Moreover, IRE1α generates ligands for RIG-I-like receptors (RLRs) during the UPR (14), which are degraded by SKIV2L RNA exosomes to prevent inappropriate activation of type I IFN responses (15). These observations prompted us to investigate the possible role of XBP1 in innate immune responses to viral infections, with the hypothesis that XBP1 could promote IFN-mediated viral resistance.
Here, we describe an unexpected role for XBP1 in antiviral resistance, not through enhancement of the IFN response, but rather by modulating susceptibility to host cell apoptosis. Xbp1-deficient cells were resistant to apoptosis during infection with vesicular stomatitis virus (VSV) and herpes simplex virus (HSV), and failure to undergo cell death resulted in increased viral replication. Xbp1 deficiency results in activation of its upstream enzyme IRE1α, which degrades specific cytosolic RNA targets (16, 17). We found that apoptosis resistance in the Xbp1-deficient cells required IRE1α. Conversely, the hepatitis C virus (HCV) nonstructural (NS) protein 4B (NS4B), which stimulates IRE1α activation (18), promoted the survival of infected cells and viral replication. Moreover, liver biopsies from patients infected with HCV showed IRE1α activation and reduced miR-125a abundance compared to healthy controls. These findings highlight the role of UPR effectors in regulating IFN-independent mechanisms of innate antiviral resistance through the induction of apoptosis to limit viral infection.
Results
Xbp1 deficiency impairs control of viral infection
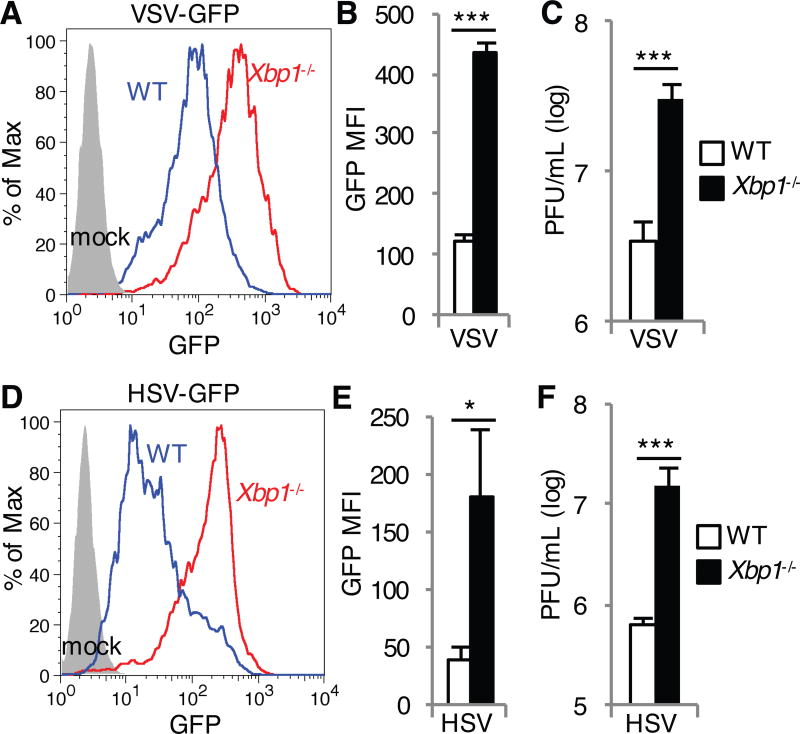
In order to determine the effect of Xbp1 deficiency on host defense against viral replication, we infected Xbp1−/− mouse embryonic fibroblasts (MEFs) with an RNA virus, vesicular stomatitis virus (VSV) and a DNA virus, herpes simplex virus (HSV). XBP1 deficiency was achieved through an insertion of the neomycin resistance gene into parts of exons 1 and 2, as well as the intervening intron (19). This insertion still allows for Xbp1 mRNA splicing, but results in a frameshift of the remaining amino acids to prevent protein production. Compared to wild-type (WT) MEFs, VSV replication was enhanced in Xbp1−/− MEFs as determined by measuring VSV-G-GFP relative abundance by flow cytometry (Fig. 1, A and B) and by plaque assays of released virus from MEF supernatant (Fig. 1C). Similarly, Xbp1−/− MEFs also supported increased replication of HSV-1-GFP as indicated by GFP abundance (Fig. 1, D and E) and viral titer in the supernatant (Fig. 1F).
Fig. 1. Xbp1 deficiency enhances the susceptibility of MEFs to HSV and VSV.
(A to F) WT and Xbp1−/− MEFs were infected with VSV-GFP at a multiplicity of infection (MOI) of 1 (A to C) or with HSV-1-GFP at an MOI of 10 (D to F). Twenty-four hours later, the extent of infection was determined by measuring the relative abundance of GFP by flow cytometry. Data are from one experiment representative of three independent experiments (A and D). The mean fluorescence intensity (MFI) of GFP in the indicated cells was then determined. Data are means ± SD from three independent experiments (B and E). Viral titers in the cell culture medium were measured by plaque assay at 48 (C) and 72 hours (F) after infection. PFU, plaque-forming units. Data are means ± SD from three independent experiments (C and F). *P < 0.05, ***P < 0.001 compared to WT, unpaired t test.
To determine whether the impaired viral control in Xbp1−/− MEFs resulted from deficient IFN responses, we measured expression of type I IFN genes and an ISG, Mx1, in MEFs infected with VSV. Unexpectedly, we observed enhanced induction of the type I IFNs, Ifna4 and Ifnb1 in Xbp1−/− MEFs infected with VSV compared to WT MEFs (fig. S1, A and B). Induction of Mx1 also increased in Xbp1−/− MEFs (fig. S1C). Moreover, Xbp1−/− MEFs were not impaired in IFN-responsiveness, as IFN-β-pretreatment prevented VSV replication in Xbp1−/− MEFs (fig. S1D). In contrast, consistent with previous reports (12, 13), the Xbp1-deficient MEF response to transfected Poly I:C (MDA5 agonist) was impaired (fig. S1E), indicating that the enhanced IFN response to VSV is specific to replicating virus. Together these findings suggest that Xbp1 contributes to protective anti-viral responses independently of type I IFNs.
Xbp1 deficiency confers resistance to virus-triggered cell death
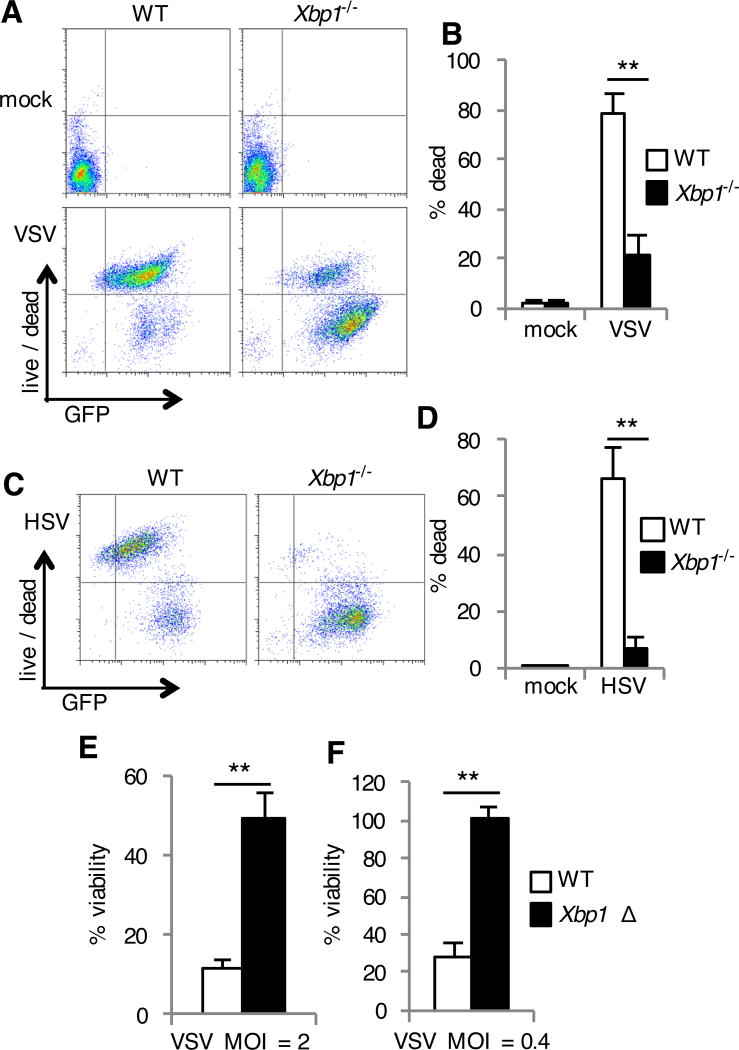
Viral infection often culminates in the death of infected host cells. To determine whether the phenotype we observed in the Xbp1-deficient MEFs is due to a difference in the death of infected cells, we evaluated cell death and abundance of virally encoded GFP following infection. During VSV infection, we found that most of the WT cells (~85%) were dead 24 hours after infection. In contrast, most Xbp1−/− MEFs (~74%) were resistant to cell death and accumulated higher amounts of viral protein as determined by measurement of GFP abundance (Fig. 2, A and B). Similarly, whereas a large proportion of HSV infected WT cells underwent cell death, infected Xbp1−/− MEFs were resistant to death (Fig. 2, C and D). To determine whether acute ablation of Xbp1 expression would have a similar effect, we treated WT MEFs with siRNA targeting Xbp1. Xbp1 knockdown strongly suppressed VSV-induced cell death and enhanced production of virally encoded GFP (fig. S2, A and B) consistent with the result of Xbp1−/− MEFs. Reconstitution of Xbp1−/− MEFs with plasmid encoded Xbp1 restored VSV-induced cell death and restricted production of virally encoded GFP (fig. S2C). To determine whether these findings extended to additional cell types, we cultured bone marrow derived macrophages (BMDMs) from mice with a tamoxifen-inducible conditional Xbp1 deletion (Xbp1 flox/flox ESR-Cre) (20). Xbp1Δ BMDMs were resistant to death during VSV infection (Fig. 2, E, F and fig. S2D), indicating that Xbp1 genetic deficiency results in protection from cell death in fibroblasts and macrophages.
Fig. 2. Xbp1-deficient cells are resistant to cell death during infection with VSV and HSV.
(A to D) WT and Xbp1−/− MEFs were left uninfected (mock) or were infected with VSV-GFP (A and B) or HSV-1-GFP (C and D) for 24 hours. Cell death was then assessed with a membrane-impermeant, amine-reactive fluorescent dye, which was measured by flow cytometry. Data are from one experiment representative of three experiments (A and C). The percentages of dead cells were then determined. Data are means ± SD from three independent experiments (B and D). (E and F) BMDMs were cultured from Xbp1flox/flox ESR Cre+ (Xbp1 Δ), or Cre- littermate (WT) mice in the presence of tamoxifen. Cells were infected with VSV-GFP at the indicated multiplicity of infection (MOI) for 24 hours. Viability was then assessed by measuring MTS reduction. Data are means ± SD of three replicates and are representative of three experiments (E and F).**P < 0.01 compared to WT, unpaired t test.
These results suggested that there should not be an Xbp1-dependent antiviral phenotype against viruses that do not trigger death of infected cells. We found that infection with a VSV-G pseudotyped lentivirus encoding GFP did not result in host cell death (fig. S3A). In this case, we did not observe enhanced production of virally encoded GFP in Xbp1−/− MEFs (fig. S3, A and B). These findings suggest that impaired control of VSV and HSV infection by Xbp1−/− MEFs was directly related to their resistance to cell death.
Apoptosis limits the replication of VSV and HSV
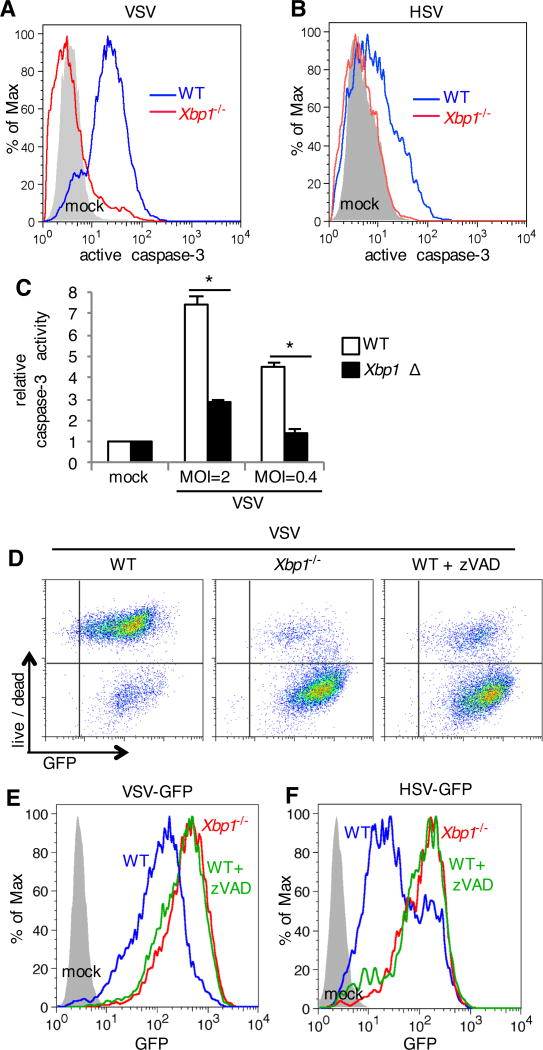
The endpoint of cell death can result from numerous upstream signaling pathways. VSV infection induces apoptotic cell death in Jurkat cells (21) and MCF-7 breast adenocarcinoma cells (22). WT MEFs infected with VSV demonstrated active caspase-3 (Fig. 3A), indicative of apoptotic cell death in these cells. In contrast, Xbp1−/− MEFs failed to activate caspase-3 during VSV infection (Fig. 3A). Similarly, HSV-infected WT MEFs contained active caspase-3 and Xbp1−/− MEFs were resistant to activation of this apoptotic effector (Fig. 3B). These findings were not limited to MEFs, because caspase-3/7 activation also occurred in VSV infected BMDMs and Xbp1Δ BMDMs were resistant to VSV induced caspase-3 activation (Fig. 3C).
Fig. 3. Apoptosis induced by VSV and HSV limits viral infection.
(A to B) WT and Xbp1−/− MEFs were left uninfected (mock) or were infected with VSV-GFP (A) or HSV-1-GFP (B) for 24 hours. Cells were stained with an antibody to active (cleaved) caspase-3, which was measured by flow cytometry. Data are from one experiment representative of three experiments. (C) BMDMs were cultured from Xbp1flox/flox ESR Cre+ (Xbp1 Δ), or Cre- littermate (WT) mice in the presence of tamoxifen. Cells were infected with VSV-GFP at the indicated multiplicity of infection (MOI) for 7 hours. Caspase-3 activity was then assessed by measuring fluorometric substrate cleavage, and is shown relative to uninfected cells. Data are means ± SD of three replicates and are representative of three experiments. (D to F) MEFs were infected in the presence of zVAD to inhibit caspase activity. Twenty-four hours after infection, cell death was assessed with a membrane impermeant, amine-reactive fluorescent dye, which was measured by flow cytometry. The extent of infection was determined by measuring the relative abundance of GFP by flow cytometry. Data are from one experiment representative of three independent experiments. *P < 0.01 compared to WT, unpaired t test.
Some viruses induce apoptosis as a means of viral transmission and avoidance of the immune system (23). In other cases, apoptosis is beneficial for the host and limits viral replication. We observed decreased abundance of virally encoded GFP in the population of dead cells during HSV infection of WT MEFs (Fig. 2C), suggesting that apoptosis may limit viral replication. To test this hypothesis, we added a caspase inhibitor, zVAD, to infected cells. zVAD prevented death of VSV infected MEFs (Fig. 3D) and lead to increased abundance of virally encoded GFP (Fig. 3, D and E), phenocopying the result obtained with Xbp1−/− MEFs. Similarly, inhibition of apoptosis with zVAD increased abundance of virally encoded GFP in HSV infected MEFs, similar to the values attained in Xbp1−/− MEFs (Fig. 3F). In addition, overproduction of the anti-apoptotic protein BCL2 limited death of VSV infected cells and lead to increased abundance of virally encoded GFP (fig. S3C). Together these findings suggest that Xbp1-deficient cells are resistant to virus-induced apoptosis and apoptosis directly limits viral replication.
Resistance to virally induced apoptosis in Xpb1− − cells is independent of Beclin 1 and CHOP
ER stress has been associated with autophagy, which regulates cell survival (24). In particular, XBP1 promotes transcription of the gene encoding the autophagy component, Beclin-1 (25). Consistent with these data, we found decreased Beclin-1 in Xbp1-deficient cells (fig. S4A). However, Beclin-1 siRNA knockdown did not affect VSV infection or induction of cell death in either WT or Xbp1-deficient MEFs (fig. S4B).
Our finding that Xbp1-deficient cells are resistant to VSV and HSV-induced apoptosis suggested the possibility that apoptosis during viral infection may directly result from XBP1-mediated transcriptional activity. During ER stress, XBP1 partially contributes to the induction of the apoptosis mediator CHOP (26), suggesting that XBP1-mediated CHOP induction may play a role in virus-induced apoptosis. As a functional control, transfection with Chop siRNA efficiently prevented death of MEFs treated with the ER stress inducing agents, tunicamycin and thapsigargin (fig. S5, A and B). In contrast, Chop knockdown did not prevent death of VSV or HSV infected MEFs (fig. S5B). Further arguing against a direct role for XBP1-mediated transcriptional activity in virally induced cell death, we did not observe Xbp1 splicing (fig. S6A) or induction of the UPR responsive genes Hspa5 (encoding BiP) and Chop during VSV infection (fig. S6, B and C), consistent with published observations for VSV (27) and HSV (28, 29).
Xbp1-deficient cells are resistant to the intrinsic pathway of apoptosis
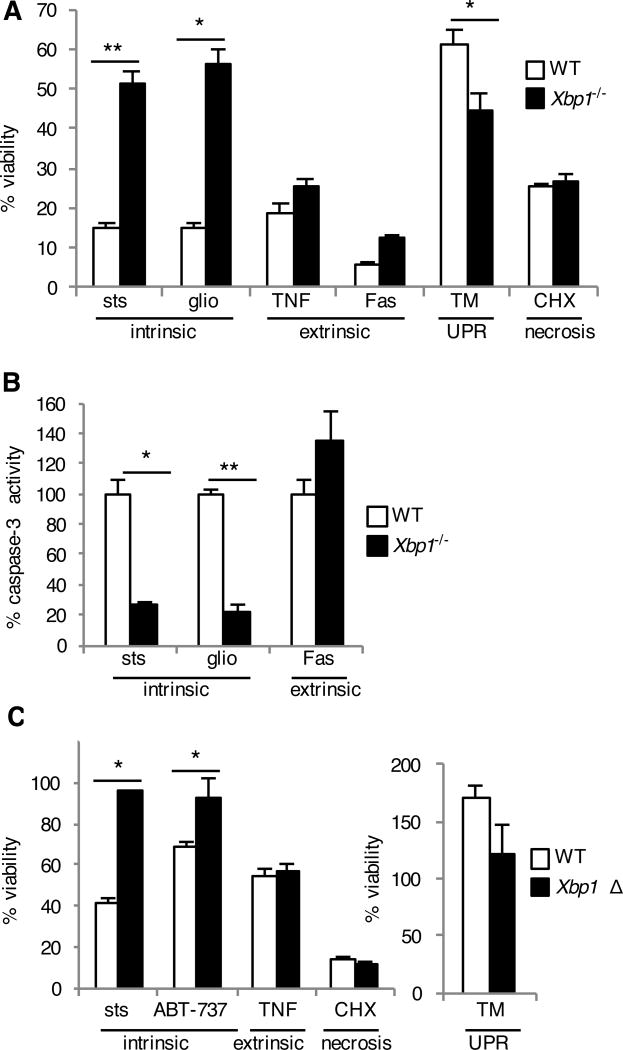
As we did not find evidence for XBP1-mediated transcriptional activity in promoting apoptosis during infection, we hypothesized that Xbp1-deficient cells may be inherently resistant to apoptosis in general. We therefore treated Xbp1−/− MEFs with a panel of apoptosis-inducing stimuli. Staurosporine and gliotoxin trigger the intrinsic or mitochondrial pathway of apoptosis (30–33), whereas ligation of the tumor necrosis factor (TNF) and Fas receptors initiates the extrinsic apoptotic pathway (34). Xbp1−/− MEFs were specifically resistant to stimuli that induced the intrinsic pathway of apoptosis, both as demonstrated by increased viability (Fig. 4A) and impaired caspase-3 activation (Fig. 4B). Consistent with previous studies demonstrating a protective role of Xbp1 during ER stress (26, 35–37), Xbp1−/− MEFs were slightly more susceptible to death induced by tunicamycin (Fig. 4A). In contrast, there was no difference between WT and Xbp1−/− MEFs in necrotic death induced by high dose cycloheximide (Fig. 4A). These findings were further verified using Xbp1Δ BMDMs, which demonstrated resistance to staurosporine and the selective Bcl-2 inhibitor ABT-737 (38), but not TNF-induced apoptosis (Fig. 4C). Thus, Xbp1 genetic deficiency results in specific protection from the intrinsic pathway of apoptosis.
Fig. 4. Xbp1-deficient cells are resistant to the intrinsic pathway of apoptosis.
(A and B) WT and Xbp1−/− MEFs were treated with staurosporine (sts), gliotoxin (glio), tumor necrosis factor + low dose cycloheximide (TNF), Fas antibody + low dose cycloheximide (Fas), tunicamycin (TM) to induce the unfolded protein response (UPR) or high dose cycoheximide (CHX). Twenty-four hours later, viability was assessed by measuring MTS reduction (A). Seven hours after treatment, caspase-3 activity was assessed by measuring fluorometric substrate cleavage, and is shown relative to WT cells (B). Data are means ± SD of three replicates and are representative of three experiments. (C) BMDMs were cultured from Xbp1flox/flox ESR Cre+ (Xbp1 Δ), or Cre- littermate (WT) mice in the presence of tamoxifen and treated with cell death inducers as in (A). Twenty-four hours later, viability was assessed by measuring MTS reduction. Data are means ± SD of three replicates and are representative of three experiments. *P < 0.05, **P < 0.001 compared to WT, unpaired t test.
The resistance of Xbp1-deficient cells to apoptosis results from the activation of IRE1α
Xbp1 deficiency results in activation of its upstream enzyme IRE1α, which degrades specific cytosolic RNA targets (16, 17). Although they cannot make XBP1s protein, Xbp1-deficient cells transcribe mRNA that contains the IRE1α cleavage sites. Consistent with previous reports, we observed IRE1α activation in Xbp1-deficient cells, indicated by Xbp1 mRNA splicing (fig. S7A). The magnitude of IRE1α activation in Xbp1-deficient cells was not as robust as the response to canonical UPR stimulation with thapsigargin, nor were classical RIDD substrates diminished (fig. S7B), consistent with other studies (39).
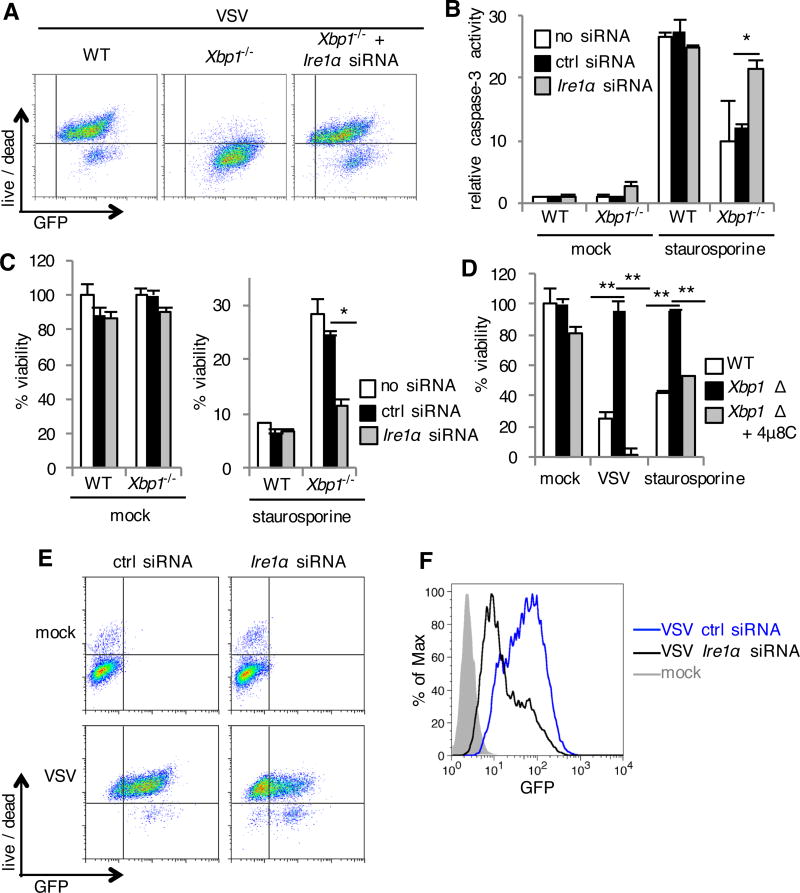
To determine whether IRE1α is involved in resistance to apoptosis, we silenced Ire1α using siRNA (fig. S7C). As a functional control, Ire1α siRNA efficiently prevented Xbp1 mRNA splicing (fig. S7D). IRE1α knockdown in Xbp1−/− MEFs reversed resistance to VSV-induced cell death (Fig. 5A and fig. S7E). Expression of human IRE1α, resistant to mouse Ire1α siRNA, prevented this reversal (fig. S7F). IRE1α knockdown alone was minimally cytotoxic, but restored caspase-3 activation in Xbp1−/− MEFs in response to staurosporine (Fig. 5B) and sensitized these cells to staurosporine-induced cell death (Fig. 5C). To determine whether the RNase activity of IRE1α mediates resistance to apoptosis, we treated Xbp1-deficient cells with the selective IRE1α nuclease inhibitor 4µ8C (40). 4µ8C reversed resistance of Xbp1-deficient cells to apoptosis both during infection with VSV and treatment with staurosporine (Fig. 5D). As a negative control, the structurally similar compound AMC had no effect at equimolar concentration (fig. S2D). These findings indicate that IRE1α’s RNAse activity contributes to resistance to the intrinsic pathway of apoptosis observed in the setting of Xbp1 deficiency.
Fig. 5. The resistance of Xbp1-deficient cells to apoptosis results from the activation of IRE1α.
(A, E and F) WT and Xbp1−/− MEFs were transfected with siRNA targeting Ire1α or control siRNA (ctrl siRNA). Cells were then left uninfected (mock) or infected with VSV-GFP for 24 hours. Cell death was then assessed with a membrane impermeant, amine-reactive fluorescent dye, and the relative abundance of GFP was measured by flow cytometry. Data are from one experiment representative of three (A) or two (E and F) independent experiments. (B and C) siRNA transfected MEFs were left untreated (mock) or treated with staurosporine. Seven hours later, caspase-3 activity was assessed by measuring fluorometric substrate cleavage, and is shown relative to untreated WT cells (B). Twenty-four hours after treatment, viability was assessed by measuring MTS reduction (C). (D) BMDMs were cultured from Xbp1flox/flox ESR Cre+ (Xbp1 Δ), or Cre- littermate (WT) mice in the presence of tamoxifen and the IRE1α inhibitor 4µ8C. Cells were then infected with VSV-GFP at a multiplicity of infection of two or treated with staurosporine. Viability was assessed 24 hours later by measuring MTS reduction. Data are means ± SD of three replicates and are representative of three experiments (B, C and D). *P < 0.01, **P < 0.001, unpaired t test.
We consistently observed faint Xbp1 mRNA splicing in WT cells (fig. S7, A and C), suggesting that IRE1α has some basal activity (41) which could regulate apoptotic responses in WT cells. In line with this, IRE1α knockdown in WT MEFs was not toxic alone (Fig. 5, B, C and E), but sensitized cells to VSV-induced cell death and limited viral replication (Fig. 5, E and F).
IRE1α targets the pro-apoptotic miRNA miR-125a
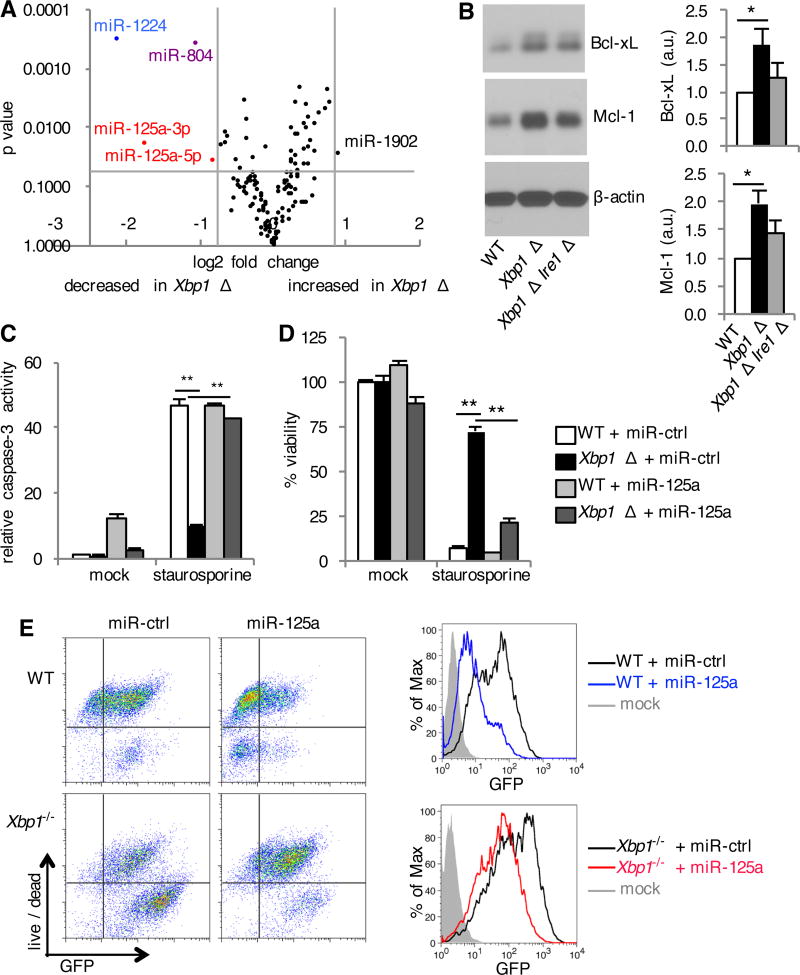
Studies of coding genes targeted by IRE1α for RIDD have not revealed obvious candidates to explain our observed IRE1α-mediated resistance to apoptosis (9, 42). Thus, we focused on microRNAs, which have also been shown to be targeted by RIDD (43). To this end, we performed microRNA profiling of Xbp1-deficient cells and found four microRNAs that were decreased in Xbp1-deficient cells with active IRE1α (Fig. 6A). miR-125a was represented twice among these differentially expressed microRNAs. Quantitative PCR confirmed the decrease in miR-125a in Xbp1-deficient cells, which was restored by reconstitution with plasmid encoded Xbp1 (fig. S8A). miR-125a has been well described to sensitize cells to apoptosis and has been suggested to negatively regulate anti-apoptotic Bcl-2 family members including Bcl-xL and Mcl-1 (44–46). In accordance with decreased miR-125a, we found an increase in anti-apoptotic Bcl-xL and Mcl-1 in Xbp1-deficient cells (Fig. 6B). In order to test whether these effects were dependent on IRE1α, we crossed Xbp1flox/flox×ER-Cre+ mice to Ern1flox/flox mice. BMDMs obtained from Xbp1flox/flox × Ern1flox/flox × ER-Cre+ mice were treated with tamoxifen to generate XBP1 Δ IRE1α Δ cells. We found that the protein amounts of both Bcl-xL and Mcl-1 in XBP1 Δ IRE1α Δ BMDMs were reduced compared to XBP1 Δ cells, albeit not to the amount observed in the WT cells (Fig. 6B). These results indicated that anti-apoptotic Bcl-xL and Mcl-1 are increased in Xbp1-deficient cells in a manner largely dependent on IRE1α. Consistent with our observation of sensitization to apoptosis by IRE1α knockdown in WT cells (Fig. 5E), IRE1α Δ cells had increased miR-125a (fig. S8B). Inhibition of the RNase activity of IRE1α with the selective IRE1α nuclease inhibitor 4 µ 8C was sufficient to increase miR-125a (fig. S8B).
Fig. 6. IRE1α mediates reduction in pro-apoptotic miR-125a.
(A) BMDMs were cultured from Xbp1flox/flox ESR Cre+ (Xbp1 Δ), or Cre- littermate (WT) mice in the presence of tamoxifen. Volcano plot demonstrating distribution of microRNAs between WT and Xbp1 Δ BMDMs measured using the NanoString nCounter assay. Data are from one experiment with quadruplicates. (B) BMDMs were cultured from Xbp1flox/flox ESR Cre+ (Xbp1 Δ), Xbp1flox/flox Ern1flox/flox ESR Cre+ (Xbp1 Δ Ire1α Δ) or Cre- littermate (WT) mice in the presence of tamoxifen. The relative abundance of Bcl-xL, Mcl-1 and β-actin in the cell lysates was determined by Western blotting and densitometry. The ratio of Bcl-xL or Mcl-1 to β-actin is shown, normalized to WT. Data are means ± SD from three independent experiments. a.u., arbitrary units.(C and D) WT and Xbp1−/− MEFs were transfected with negative control microRNA mimetic (miR-ctrl) or miR-125a mimetic. Cells were left untreated (mock) or treated with staurosporine. Seven hours later, caspase-3 activity was assessed by measuring fluorometric substrate cleavage, and is shown relative to untreated WT cells (C). Twenty-four hours after treatment, viability was assessed by measuring MTS reduction (D). Data are means ± SD of three replicates and are representative of two experiments. (E) MicroRNA transfected MEFs were infected with VSV-GFP for 24 hours. Cell death was then assessed with a membrane impermeant, amine-reactive fluorescent dye, which was measured by flow cytometry. The extent of infection was determined by measuring the relative abundance of GFP by flow cytometry. Data are from one experiment representative of two independent experiments. *P < 0.01, **P < 0.001, unpaired t test.
Finally, we wished to examine the extent to which miR-125a degradation by IRE1α is responsible for the pro-survival phenotype of the Xbp1-deficient cells. Reconstitution of miR-125a using the micro RNA mimetic was sufficient to restore caspase-3 activation in Xbp1−/− MEFs in response to staurosporine (Fig. 6C) and sensitize Xbp1-deficient cells to the intrinsic pathway of apoptosis (Fig. 6D and fig. S8C). miR-125a reconstitution also reversed resistance of Xbp1−/− MEFs to VSV-induced cell death (Fig. 6E). Finally, neutralizing miR-125a in WT cells with an miRNA hairpin inhibitor resulted in an anti-apoptotic state resembling that observed in Xbp1-deficient cells (fig. S8D). Together, these findings suggest that the IRE1α-dependent decrease in miR-125a contributes to resistance to apoptosis.
IRE1α Activation During HCV Infection Mediates Resistance to Apoptosis
Some viruses encode genes that promote IRE1α activation. Hepatitis C virus (HCV) non-structural protein NS4B activates IRE1α (18) and IRE1α activation is also seen in HCV infected cells (47). Curiously, in cells expressing NS4B, IRE1α splices Xbp1 mRNA, but XBP1 targets are not transcribed (18), suggesting that the virus uses IRE1α for another reason. Furthermore, HCV has been suggested to cause resistance to the intrinsic pathway of apoptosis (48, 49), although the mechanism of this effect remains unknown.
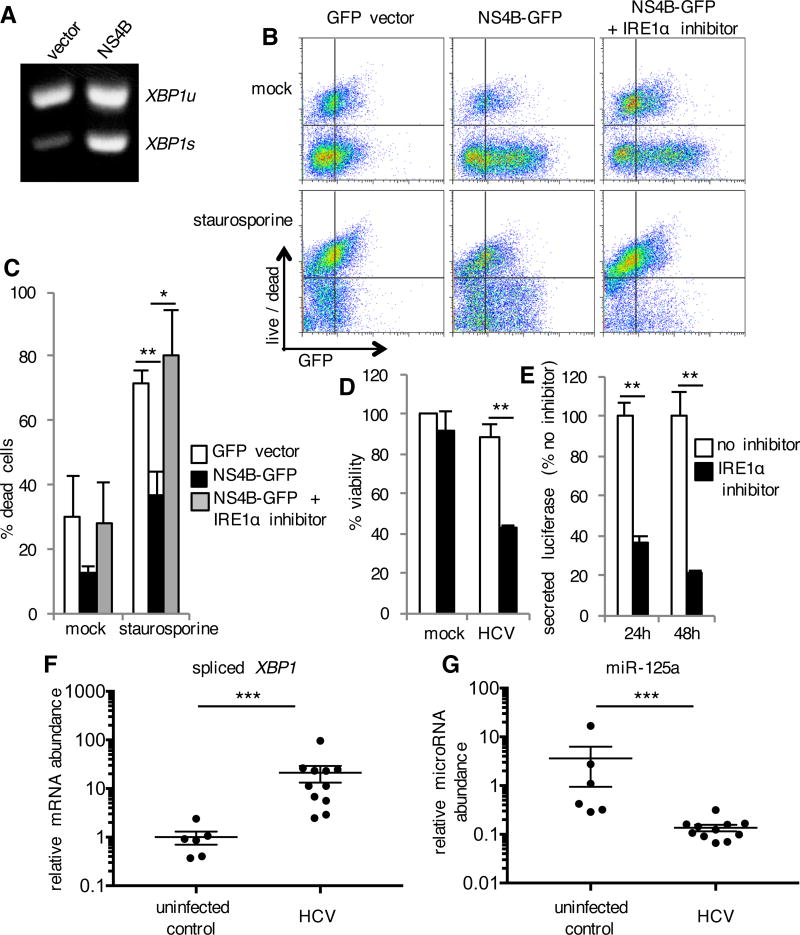
Consistent with previous reports (18), we detected IRE1α activation indicated by spliced XBP1 mRNA in cells transiently transfected with an NS4B expression plasmid (Fig. 7A). miR-125a was decreased (fig. S8E) and NS4B expression induced IRE1α-dependent resistance to staurosporine (Fig. 7, B and C). We infected Huh-7.5 human hepatoma cells with trans-packaged HCV replicons encoding Gaussia luciferase, which allowed us to quantitate HCV replication over time (50). IRE1α inhibition alone was not cytotoxic, but sensitized HCV-infected cells to death (Fig. 7D). Furthermore, inhibition of IRE1α decreased secretion of virally encoded luciferase, a marker of viral replication (Fig. 7E). These findings suggest that IRE1α activation during HCV infection may promote viral replication by inhibiting death of infected cells. To determine if these findings could extend to human HCV infection, we quantified spliced XBP1 mRNA in human liver tissue of patients infected with HCV. We detected HCV-associated IRE1α activation, as indicated by an increase in spliced XBP1 mRNA in liver tissue from HCV infected patients compared to HCV negative controls (Fig. 7F). In addition to IRE1α activation, HCV infected patients also exhibited significantly reduced miR-125a (Fig. 7G), suggesting that IRE1α activation during human HCV infection may confer resistance to apoptosis.
Fig. 7. IRE1α-Mediated Apoptosis Resistance Induced by HCV NS4B.
(A) HeLa cells were transfected with constructs expressing GFP alone (vector) or together with HCV NS4B. RNA was isolated 72 hours later. XBP1 mRNA maturation from unspliced (u) to spliced (s) was analyzed by RT-PCR. Data are from one experiment representative of two independent experiments. (B and C) Transfected cells were treated with IRE1α inhibitor 2 for 24 hours and then staurosporine was added. Twenty hours later, cell death was assessed with a membrane impermeant, amine-reactive fluorescent dye, which was measured by flow cytometry. Data are from one experiment representative of three independent experiments (B). The percentage of dead cells among transfected GFP positive cells is shown in (C). Data are means ± SD from three independent experiments. (D and E) Huh-7.5 cells were infected with trans-packaged HCV encoding luciferase. Forty-eight hours later, viability was determined by measuring cellular ATP (D). Secretion of virally encoded luciferase was measured 24 and 48 hours after infection (E). Data are means ± SD of three replicates and are representative of two experiments. (F and G) RNA was isolated from liver tissue of HCV infected patients (n=11) and HCV negative controls (n=6). Expression of spliced XBP1 and miR-125a (relative to an internal control) were determined by quantitative RT-PCR. Data are mean ± SEM. *P < 0.05, **P < 0.01, unpaired t test. ***P < 0.01, Mann-Whitney test.
Discussion
In this study, we examined the IRE1α / XBP1 branch of the UPR in innate antiviral defense. We uncovered an unexpected role for Xbp1 and IRE1α in modulating susceptibility to the intrinsic pathway of apoptosis, which is induced during VSV and HSV infection and plays a critical role in limiting viral replication. Xbp1−/− MEFs were protected from virally induced cell death and, as a consequence, sustained more viral replication despite an increased IFN response compared to that of WT cells. In Xbp1−/− cells with active IRE1α, the abundance of the pro-apoptotic miRNA miR-125a decreased, conferring resistance to the intrinsic apoptotic pathway. IRE1α activation by HCV NS4B in WT cells also conferred resistance to apoptosis and promoted viral replication. Therefore, Xbp1-deficient cells with active IRE1α gain apoptosis resistance, and IRE1α silencing or inhibition is a model for loss of apoptosis resistance. Finally, we observed IRE1α activation and substantially less miR-125a in the liver biopsies from HCV infected patients, suggesting the in vivo relevance of the survival strategy used by HCV. These results highlight a previously unappreciated role of the XBP1-IRE1α axis in regulation of apoptosis, and its consequences in viral susceptibility.
Previous studies showed that, after the engagement of TLRs and RLRs, XBP1 plays an important role in enhancing cytokine and IFN production in macrophages and dendritic cells (11–13). Given the observation that transfection with poly(I:C) induced less IFN production in Xbp1−/− MEFs compared to that in WT MEFs, our results are consistent with these previous findings that Xbp1 promotes RLR signaling for IFN induction in MEFs. However, despite this impairment in IFN production downstream of RLRs, infected Xbp1-deficient MEFs produced high amounts of IFN. We speculate that the enhanced IFN response observed in Xbp1−/− MEFs may result from both prolonged cellular survival as well as an accumulation of viral PAMPs. Apoptotic caspases can cleave and inactivate signaling proteins important for the IFN response, suggesting that the apoptotic process directly antagonizes the IFN response (51–54). In addition, IRE1α can cleave host RNA for RLR stimulation (15). Therefore, our results highlight a unique consequence of IRE1α activation, whereby Xbp1 deficiency results in robust pro-survival response, leading to prolonged RLR stimulation that mitigates impairment in RLR signaling to produce enhanced IFN responses. The pro-survival signals induced through IRE1α activation are so dominant that they overcome ISG-mediated antiviral functions and enable virus replication.
In addition to protection from virus-induced death, we found that Xbp1 deficiency conferred resistance to the intrinsic pathway of apoptosis triggered by a variety of chemical inducers. IRE1α activation in Xbp1-deficient cells contributed to this apoptosis resistance, as siRNA knockdown of Ire1α expression or inhibition of IRE1α nuclease function rendered Xbp1-deficient cells susceptible to apoptosis. RIDD has been shown to target both coding and non-coding RNAs including microRNAs (42). IRE1α was found to reduce the amount of miR-125a, leading to enhancement of its target gene expression. The targets include pro-survival members of the Bcl2 family (44–46). miRNA controls target genes at the transcriptional and translational levels (55). We found increased pro-survival proteins Bcl-XL and Mcl-1. The biological targets and functions of the other miRNAs identified in this study, namely miR-1224 and miR-804, have not yet been well described. The relevance of these other miRNA in the IRE1α -dependent phenotypes will be investigated in future studies.
Our study revealed an unexpected role of IRE1α in controlling apoptosis. Many studies of the UPR have been performed using high doses of pharmacological inducers of ER stress, such as tunicamycin and thapsigargin, which inevitably lead to cell death. In these experimental settings of irremediable ER stress, various mechanisms have been proposed to induce apoptosis (56). The net effect of IRE1α activity in promoting cell death compared to cell survival has been controversial, with some studies suggesting that IRE1α promotes cell survival (57–59) and others suggesting that IRE1α promotes cell death (60–62). Specifically, IRE1α was proposed to induce apoptosis by degrading microRNAs that lead to increased caspase-2 (60), although there are conflicting data challenging this observation (63). IRE1α has been linked to apoptosis in cells irreversibly damaged by the activation of the JNK/ASK1 pathway (64, 65). IRE1α appears to have opposing roles in regulating apoptosis, pro-survival in the absence of irreversible UPR, and pro-apoptotic under irreversible UPR. This double-edged nature of IRE1α may be important to consider for the use of IRE1α inhibitors in the treatment of various human diseases (66).
We speculate that IRE1α may represent an ancient form of cell protection that has been subverted by some viruses for their own replicative advantage. HCV and other members of the Flaviviridae family induce ER stress and activate IRE1α. Our results suggest that HCV NS4B induces IRE1α-dependent protection from apoptosis, which may favor the development of chronic infection and hepatocellular carcinoma. In the setting of avian coronavirus infection, IRE1α also promotes cell survival in association with JNK and AKT regulation (67). Influenza virus has been shown to induce only the IRE1α arm of the UPR (not ATF6 or PERK) and inhibitors of IRE1α reduce virus replication (68). Therefore, our findings predict that the IRE1α-RIDD pathway could be exploited as a novel target of intervention against viral infections.
Materials and Methods
Cells and viruses
Xbp1+/+ and Xbp1−/− MEFs were gifts of Dr. Laurie H. Glimcher (Weill Cornell Medical College, New York, NY). H1-HeLa cells stably overexpressing the antiapoptotic protein BCL2 have been previously described (69). MEFs and H1-HeLa cells were propagated in high-glucose DMEM (Gibco) and supplemented with 10% heat-inactivated FBS, 1% Hepes, and 100 U/mL penicillin, and 100 µg/mL streptomycin (Gibco). Huh-7.5 cells were propagated in high-glucose DMEM with 10% heat-inactivated FBS and 1 mM nonessential amino acids (Invitrogen). Xbp1flox/flox (20) (a gift of Dr. Laurie H. Glimcher, Weill Cornell Medical College, New York, NY), Ern1flox/flox (70) (RIKEN BRC, Japan) and CAGGCre-ER™ (The Jackson Laboratory) mice were bred in the Yale animal facility. All procedures performed in this study complied with federal guidelines and institutional policies set by Yale Animal Care and Use Committee. Bone marrow derived macrophages were prepared from male and female 6 to 12 week old mice according to a previously described method (71) and cultured in 0.2µM 4-hydroxytamoxifen (Sigma) during days 2–4 of differentiation to induce Cre-mediated recombination. The following genotype combinations of BMM that were treated with tamoxifen to generate the cells described in Figure 6: Xbp1flox/flox × ER-Cre+ (XBP1 Δ), Xbp1flox/flox × Ern1flox/clox × ER-Cre+ (XBP1 Δ IRE1α Δ), or Xbp1flox/flox × ER-Cre- (WT). VSV-G-GFP was a kind gift of Dr. J Rose (Yale University, New Haven, CT) and Dr. A Geballe (Fred Hutchinson Cancer Research Center, Seattle, WA) and was maintained and titered in BHK cells. HSV-1-GFP (72) was a kind gift from Drs. P. Desai and S. Person (Johns Hopkins University, MD) and was maintained and titered in Vero cells. VSV-G pseudotyped lentivirus was made by harvesting culture supernatants of HEK-293T cells transfected with plasmids encoding VSV-G, GFP and Gag-Pol. Trans-packaged HCV replicons were prepared by transfecting the JFH/Gluc replicon into Huh-7.5[core-NS2] cells as previously described (50)
Infection and stimulation of cells
Cells were incubated for 1 h with VSV or HSV-1 in serum-free medium or pseudotyped virus in PBS 0.1% BSA, and then the inoculum was removed and incubation continued in complete media. Cells were incubated for 4 h with trans-packaged HCV, and then the inoculum was removed and incubation continued in complete media containing 60µM IRE1 Inhibitor II (Calbiochem). zVAD (Invivogen) was added at 20–100 µM after removal of the viral inoculum. Poly I:C (1 µg/mL) was delivered complexed to Lipofectamine 2000 (Invitrogen). Cells were treated with 0.1–1µM staurosporine (Enzo Life Sciences), 1µM gliotoxin (Sigma), 10µM ABT-737 (Santa Cruz Biotechnology), 50ng/ml TNF + 0.1µg/ml cycloheximide (Abcam), 2 µg/ml anti-mouse CD95 (BD Pharmingen) Fas activating antibody + 0.1µg/ml cycloheximide, 10–100µg/ml tunicamycin (Sigma), 1µM thapsigargin (Calbiochem) or 100µg/ml cycloheximide (high dose CHX). Cells were treated with IRE1 inhibitor 4µ8C (8-Formyl-7-hydroxy-4-methylcoumarin, Calbiochem) or the structurally similar compound AMC (7-Amino-4-methylcoumarin, Sigma) at 25µM for 3 days prior to infection or apoptosis induction, or 40µM IRE1 Inhibitor II (Calbiochem) for 24 h prior to apoptosis induction.
Plasmids
pFLAG.XBP1u.CMV2 (73) (Addgene plasmid # 21832, from David Ron); the empty vector control c-Flag pcDNA3 (74) (Addgene plasmid # 20011, from Stephen Smale); hIRE1a.pcD (Addgene plasmid # 21892, from Randal Kaufman); and hIRE1a wt (75) (Addgene plasmid # 20744, from Fumihiko Urano) were used. MEFs were transfected using TransIT-2020 (Mirus Bio). HCV NS4B was expressed with an authentic N-terminal Ala residue by fusing a human ubiquitin gene to a codon optimized NS4B gene (76): ubiquitin was amplified by using primers YO-0905 (TTA ATT AAC GAG GAT CCC GCC ACC ATG CAG ATC TTC GTG AAG AC) and YO-0928 (TCG ATC AGG GCT GCT CTG CTG GCT CCA CCG CGG AGA CGC AGC ACC); codon-optimized NS4B was amplified by using primers YO-0927 (GGT GCT GCG TCT CCG CGG TGG AGC CAG CAG AGC AGC CCT GAT CGA) and YO-0931 (GTT TAA ACT TAA CAA GGG ATG GGG CAG TCC T). Ubi-NS4B was then assembled in secondary PCRs with primers YO-0905 and YO-0931, cloned into pCR2.1 (Invitrogen) for sequencing, then subcloned into pIRES2-EGFP (Clontech) by using the common SacI and PstI restriction sites. H1-HeLa cells were transfected using TransIT-HeLa (Mirus Bio).
Expression analysis
RNA isolated using the RNeasy kit (Qiagen) was used to synthesize cDNA using the iScript cDNA synthesis kit (Bio-Rad) and qPCR was performed on a Stratagene MX3000P or Bio-Rad CFX Connect using SyberGreen (Bio-Rad) with primers as follows (all primers listed in 5’-3’ orientation): Ifna4 forward, CTG CTA CTT GGA ATG CAA CTC; Ifna4 reverse, CAG TCT TGC CAG CAA GTT GG; Ifnb1 forward, GCA CTG GGT GGA ATG AGA CTA TTG; Ifnb1 reverse, TTC TGA GGC ATC AAC TGA CAG GTC; Mx1 forward, AGT CCT TTC CAC AGG CAG AA; Mx1 reverse: CAT TGA GAG AAA CTC ACC TAA GAA C; Xbp1s forward, GAG TCC GCA GCA GGT; Xbp1s reverse, GTG TCA GAG TCC ATG GGA; Hspa5 (Bip) forward, TCA TCG GAC GCA CTT GGA; (Hspa5 Bip) reverse, CAA CCA CCT TGA ATG GCA AGA; Chop forward, GTC CCT AGC TTG GCT GAC AGA; Chop reverse, TGG AGA GCG AGG GCT TTG; Blos1 forward, CAA GGA GCT GCA GGA GAA GA; Blos1 reverse, GCC TGG TTG AAG TTC TCC AC; Pdgfrb forward, AAC CCC CTT ACA GCT GTC CT; Pdgfrb reverse, TAA TCC CGT CAG CAT CTT CC; human spliced XBP1 forward, TGC TGA GTC CGC AGC AGG TG; human spliced XBP1 reverse, GCT GGC AGG CTC TGG GGA AG. For analysis of Xbp1-splicing, primers flanking the spliced sequences in Xbp1 mRNA (forward, ACA CGC TTG GGA ATG GAC AC; reverse, CCA TGG GAA GAT GTT CTG GG) were used for PCR amplification, and products were separated by electrophoresis through a 3% agarose gel and visualized by ethidium bromide staining.
miRNA expression analysis
RNA isolated using the microRNeasy kit (Qiagen) was subjected to micoRNA profiling using the nCounter mouse miRNA expression assay v1.5 (Nanostring) according to the manufacturer’s protocol. Data were analyzed by using the nSolver software with normalization to the geometric mean of the top 100 miRNAs as recommended by the manufacturer. Quantitative RT-PCR with the miRCURY Universal RT microRNA qPCR system (Exiqon) was used to measure miR-125a-5p. Expression was calculated relative to the manufacturer’s suggested endogenous control (miR-103a-3p), with equivalent results also obtained relative to miR-16-5p.
Assessment of cell death
Cells were stained with the LIVE/DEAD Fixable Far Red Dead Cell Stain Kit (Molecular Probes) and analyzed by flow cytometry on a BD FACSCalibur or BD LSRFortessa. Viability was also assessed by measuring MTS reduction using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega). Cells were stained with the Caspase-3, Active Form, Apoptosis Kit (BD Pharmingen) and analyzed by flow cytometry on a BD FACSCalibur. Caspase-3/7 activity was measured using the SensoLyte Homogeneous Rh110 Caspase-3/7 Assay Kit (AnaSpec).
Western blotting
Cell pellets were lysed in SDS sample buffer (Cell Signaling Technology). Proteins were separated by SDS/PAGE, transferred to PVDF membranes, and interrogated with antibodies against Mcl-1 (BioLegend-613601), Bcl-XL (Cell Signaling Technology-54H6) or β-actin (Cell Signaling Technology-13E5).
Intracellular staining
Cells were fixed in Cytofix/Cytoperm solution (BD) on ice, washed with Perm/Wash buffer (BD) and stained with DyLight 550 conjugated antibody against Beclin 1 (Novus Biologicals NB110-87318R) or an equal concentration of isotype control antibody. Washed cells were analyzed by flow cytometry on a BD LSRFortessa.
siRNA, miR mimetics, and inhibitors
Gene specific siGENOME siRNA or siGENOME Non-Targeting siRNA #4 (which targets firefly luciferase mRNA and has at least 4 mismatches to all mouse genes) obtained from Dharmacon (Thermo Fisher Scientific) was delivered complexed to Lipofectamine RNAiMAX (Invitrogen). After incubation for 48h, cells were replated at equal density prior to infection or apoptosis induction. miR mimetics, miRIDIAN microRNA hairpin inhibitor negative control #1, and miRIDIAN miR-125a hairpin inhibitor were obtained from Dharmacon (Thermo Fisher Scientific) and delivered complexed to Lipofectamine RNAiMAX (Invitrogen).
Luciferase assay
Conditioned cell culture medium was collected at 24h or 48h after infection, clarified by centrifugation, mixed with 1/4 volume 5× lysis buffer (New England Biolabs) and assayed for luciferase activity (New England Biolabs).
Liver specimens
Liver samples from percutaneous biopsies of liver transplant recipients chronically infected with HCV or from HCV negative control liver transplant recipients were obtained with the approval of the University of Washington Institutional Review Board. Tissue was archived in RNALater and stored at −80 °C. RNA was isolated using the microRNeasy kit (Qiagen).
Statistical analyses
Sample size was chosen according to previous experience in similar experiments; all samples were included in analysis. Unpaired student's t-test or Mann-Whitney test was used for comparisons between two groups. P values of less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank H. Dong and K. Hayashi for technical support. We thank B. Cookson and N. Kaminski for sharing laboratory facilities and equipment. We thank L. Glimcher, J. Rose, A. Geballe, P. Desai, S. Person, D. Ron, S. Smale, R. Kaufman and F. Urano for providing us with various reagents. Funding: S.L.F. was supported by NIH award T32 HL007974. This work was supported by funding from the Howard Hughes Medical Institute and NIH R01 AI054359 and AI064705 (to A.I.) and K08 AI119142 (to S.L.F.). B.D.L. was funded by R01 AI087925.
Footnotes
Author contributions: S.L.F., T.J.R., R.D.M., T.I., C.S.L., B.D.L. and A.I. designed the experiments. S.L.F., T.R.J., R.D.M., and B.D.L. performed the experiments. S.L.F., T.J.R., R.D.M., and A.I. analyzed the data. S.L.F and A.I. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The Xbp1+/+ and Xbp1−/− MEFs and Xbp1flox/flox mice require a material transfer agreement from Harvard University. The Ern1flox/flox mice require a material transfer agreement from RIKEN BRC. The following plasmids require a material transfer agreement from Addgene: pFLAG.XBP1u.CMV2, c-Flag pcDNA3, hIRE1a.pcD and hIRE1a wt. The JFH-1 HCV replicon requires material transfer agreements from Rockefeller University and Apath LLC (in the US) or from the Tokyo Metropolitan Organization for Medical Research and Toray Industries (elsewhere). The replicon was packaged by using the HCV strain J6 structural genes, which was obtained under a material transfer agreement with the US National Institutes of Health. The Huh-7.5 cell line requires a material transfer agreement from Washington University and Apath LLC.
Supplementary Materials
Fig. S1. Increased interferon and ISG expression during VSV infection in Xbp1 deficient MEFs.
Fig. S2. Xbp1 siRNA knockdown mimics Xbp1 deficiency and Xbp1 reconstitution reverses resistance to cell death.
Fig. S3. Xbp1 deficiency does not increase susceptibility to a noncytotoxic virus and mimics BCL2 overexpression.
Fig. S4. Resistance to virally induced apoptosis in Xpb1 deficient cells is independent of Beclin 1.
Fig. S5. Death of VSV and HSV infected cells does not require Chop.
Fig. S6. VSV infection does not activate the unfolded protein response.
Fig. S7. The resistance of Xbp1-deficient cells to apoptosis results from the activation of IRE1α.
Fig. S8. miR-125a regulates resistance to apoptosis.
References
- 1.Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annual review of immunology. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, Wang A. Virus-induced ER stress and the unfolded protein response. Frontiers in plant science. 2012;3:293. doi: 10.3389/fpls.2012.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nature reviews. Molecular cell biology. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 6.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annual review of biochemistry. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 7.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 8.Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiological reviews. 2011;91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- 9.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. The Journal of cell biology. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 11.Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nature immunology. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu F, Yu X, Wang H, Zuo D, Guo C, Yi H, Tirosh B, Subjeck JR, Qiu X, Wang XY. ER stress and its regulator X-box-binding protein-1 enhance polyIC-induced innate immune response in dendritic cells. European journal of immunology. 2011;41:1086–1097. doi: 10.1002/eji.201040831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith JA, Turner MJ, DeLay ML, Klenk EI, Sowders DP, Colbert RA. Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-beta induction via X-box binding protein 1. European journal of immunology. 2008;38:1194–1203. doi: 10.1002/eji.200737882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho JA, Lee AH, Platzer B, Cross BC, Gardner BM, De Luca H, Luong P, Harding HP, Glimcher LH, Walter P, Fiebiger E, Ron D, Kagan JC, Lencer WI. The unfolded protein response element IRE1alpha senses bacterial proteins invading the ER to activate RIG-I and innate immune signaling. Cell host & microbe. 2013;13:558–569. doi: 10.1016/j.chom.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Eckard SC, Rice GI, Fabre A, Badens C, Gray EE, Hartley JL, Crow YJ, Stetson DB. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nature immunology. 2014;15:839–845. doi: 10.1038/ni.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hur KY, So JS, Ruda V, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V, Iwawaki T, Glimcher LH, Lee AH. IRE1alpha activation protects mice against acetaminophen-induced hepatotoxicity. The Journal of experimental medicine. 2012;209:307–318. doi: 10.1084/jem.20111298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, Blumberg RS. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Y, Gao B, Ye L, Kong L, Jing W, Yang X, Wu Z, Ye L. Hepatitis C virus non-structural protein NS4B can modulate an unfolded protein response. Journal of microbiology. 2005;43:529–536. [PubMed] [Google Scholar]
- 19.Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH. An essential role in liver development for transcription factor XBP-1. Genes & development. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- 20.Hetz C, Lee AH, Gonzalez-Romero D, Thielen P, Castilla J, Soto C, Glimcher LH. Unfolded protein response transcription factor XBP-1 does not influence prion replication or pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:757–762. doi: 10.1073/pnas.0711094105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hobbs JA, Schloemer RH, Hommel-Berrey G, Brahmi Z. Caspase-3-like proteases are activated by infection but are not required for replication of vesicular stomatitis virus. Virus research. 2001;80:53–65. doi: 10.1016/s0168-1702(01)00350-1. [DOI] [PubMed] [Google Scholar]
- 22.Hobbs JA, Hommel-Berrey G, Brahmi Z. Requirement of caspase-3 for efficient apoptosis induction and caspase-7 activation but not viral replication or cell rounding in cells infected with vesicular stomatitis virus. Human immunology. 2003;64:82–92. doi: 10.1016/s0198-8859(02)00702-4. [DOI] [PubMed] [Google Scholar]
- 23.Thomson BJ. Viruses and apoptosis. International journal of experimental pathology. 2001;82:65–76. doi: 10.1111/j.1365-2613.2001.iep0082-0065-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoyer-Hansen M, Jaattela M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell death and differentiation. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 25.Margariti A, Li H, Chen T, Martin D, Vizcay-Barrena G, Alam S, Karamariti E, Xiao Q, Zampetaki A, Zhang Z, Wang W, Jiang Z, Gao C, Ma B, Chen YG, Cockerill G, Hu Y, Xu Q, Zeng L. XBP1 mRNA splicing triggers an autophagic response in endothelial cells through BECLIN-1 transcriptional activation. The Journal of biological chemistry. 2013;288:859–872. doi: 10.1074/jbc.M112.412783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Molecular and cellular biology. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baltzis D, Qu LK, Papadopoulou S, Blais JD, Bell JC, Sonenberg N, Koromilas AE. Resistance to vesicular stomatitis virus infection requires a functional cross talk between the eukaryotic translation initiation factor 2alpha kinases PERK and PKR. Journal of virology. 2004;78:12747–12761. doi: 10.1128/JVI.78.23.12747-12761.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burnett HF, Audas TE, Liang G, Lu RR. Herpes simplex virus-1 disarms the unfolded protein response in the early stages of infection. Cell stress & chaperones. 2012;17:473–483. doi: 10.1007/s12192-012-0324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulvey M, Arias C, Mohr I. Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. Journal of virology. 2007;81:3377–3390. doi: 10.1128/JVI.02191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganguly A, Basu S, Chakraborty P, Chatterjee S, Sarkar A, Chatterjee M, Choudhuri SK. Targeting mitochondrial cell death pathway to overcome drug resistance with a newly developed iron chelate. PloS one. 2010;5:e11253. doi: 10.1371/journal.pone.0011253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho AT, Li QH, Hakem R, Mak TW, Zacksenhaus E. Coupling of caspase-9 to Apaf1 in response to loss of pRb or cytotoxic drugs is cell-type-specific. The EMBO journal. 2004;23:460–472. doi: 10.1038/sj.emboj.7600039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen VT, Lee JS, Qian ZJ, Li YX, Kim KN, Heo SJ, Jeon YJ, Park WS, Choi IW, Je JY, Jung WK. Gliotoxin isolated from marine fungus Aspergillus sp. induces apoptosis of human cervical cancer and chondrosarcoma cells. Marine drugs. 2014;12:69–87. doi: 10.3390/md12010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pardo J, Urban C, Galvez EM, Ekert PG, Muller U, Kwon-Chung J, Lobigs M, Mullbacher A, Wallich R, Borner C, Simon MM. The mitochondrial protein Bak is pivotal for gliotoxin-induced apoptosis and a critical host factor of Aspergillus fumigatus virulence in mice. The Journal of cell biology. 2006;174:509–519. doi: 10.1083/jcb.200604044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varfolomeev EE, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann JS, Mett IL, Rebrikov D, Brodianski VM, Kemper OC, Kollet O, Lapidot T, Soffer D, Sobe T, Avraham KB, Goncharov T, Holtmann H, Lonai P, Wallach D. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity. 1998;9:267–276. doi: 10.1016/s1074-7613(00)80609-3. [DOI] [PubMed] [Google Scholar]
- 35.Byrd AE, Aragon IV, Brewer JW. MicroRNA-30c-2* limits expression of proadaptive factor XBP1 in the unfolded protein response. The Journal of cell biology. 2012;196:689–698. doi: 10.1083/jcb.201201077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romero-Ramirez L, Cao H, Nelson D, Hammond E, Lee AH, Yoshida H, Mori K, Glimcher LH, Denko NC, Giaccia AJ, Le QT, Koong AC. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer research. 2004;64:5943–5947. doi: 10.1158/0008-5472.CAN-04-1606. [DOI] [PubMed] [Google Scholar]
- 37.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM, O'Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 39.Cubillos-Ruiz JR, Silberman PC, Rutkowski MR, Chopra S, Perales-Puchalt A, Song M, Zhang S, Bettigole SE, Gupta D, Holcomb K, Ellenson LH, Caputo T, Lee AH, Conejo-Garcia JR, Glimcher LH. ER Stress Sensor XBP1 Controls Anti-tumor Immunity by Disrupting Dendritic Cell Homeostasis. Cell. 2015;161:1527–1538. doi: 10.1016/j.cell.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cross BC, Bond PJ, Sadowski PG, Jha BK, Zak J, Goodman JM, Silverman RH, Neubert TA, Baxendale IR, Ron D, Harding HP. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E869–878. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Molecular cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 42.Coelho DS, Domingos PM. Physiological roles of regulated Ire1 dependent decay. Frontiers in genetics. 2014;5:76. doi: 10.3389/fgene.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maurel M, Chevet E. Endoplasmic reticulum stress signaling: the microRNA connection. American journal of physiology. Cell physiology. 2013;304:C1117–1126. doi: 10.1152/ajpcell.00061.2013. [DOI] [PubMed] [Google Scholar]
- 44.Tong Z, Liu N, Lin L, Guo X, Yang D, Zhang Q. miR-125a-5p inhibits cell proliferation and induces apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2015;75:129–136. doi: 10.1016/j.biopha.2015.07.036. [DOI] [PubMed] [Google Scholar]
- 45.Svensson D, Gidlof O, Turczynska KM, Erlinge D, Albinsson S, Nilsson BO. Inhibition of microRNA-125a promotes human endothelial cell proliferation and viability through an antiapoptotic mechanism. Journal of vascular research. 2014;51:239–245. doi: 10.1159/000365551. [DOI] [PubMed] [Google Scholar]
- 46.Balakrishnan A, Stearns AT, Park PJ, Dreyfuss JM, Ashley SW, Rhoads DB, Tavakkolizadeh A. Upregulation of proapoptotic microRNA mir-125a after massive small bowel resection in rats. Annals of surgery. 2012;255:747–753. doi: 10.1097/SLA.0b013e31824b485a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Machida K, Tsukiyama-Kohara K, Seike E, Tone S, Shibasaki F, Shimizu M, Takahashi H, Hayashi Y, Funata N, Taya C, Yonekawa H, Kohara M. Inhibition of cytochrome c release in Fas-mediated signaling pathway in transgenic mice induced to express hepatitis C viral proteins. The Journal of biological chemistry. 2001;276:12140–12146. doi: 10.1074/jbc.M010137200. [DOI] [PubMed] [Google Scholar]
- 49.Kamegaya Y, Hiasa Y, Zukerberg L, Fowler N, Blackard JT, Lin W, Choe WH, Schmidt EV, Chung RT. Hepatitis C virus acts as a tumor accelerator by blocking apoptosis in a mouse model of hepatocarcinogenesis. Hepatology. 2005;41:660–667. doi: 10.1002/hep.20621. [DOI] [PubMed] [Google Scholar]
- 50.Phan T, Kohlway A, Dimberu P, Pyle AM, Lindenbach BD. The acidic domain of hepatitis C virus NS4A contributes to RNA replication and virus particle assembly. Journal of virology. 2011;85:1193–1204. doi: 10.1128/JVI.01889-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rebsamen M, Meylan E, Curran J, Tschopp J. The antiviral adaptor proteins Cardif and Trif are processed and inactivated by caspases. Cell death and differentiation. 2008;15:1804–1811. doi: 10.1038/cdd.2008.119. [DOI] [PubMed] [Google Scholar]
- 52.Scott I, Norris KL. The mitochondrial antiviral signaling protein, MAVS, is cleaved during apoptosis. Biochemical and biophysical research communications. 2008;375:101–106. doi: 10.1016/j.bbrc.2008.07.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, van Delft MF, Bedoui S, Lessene G, Ritchie ME, Huang DC, Kile BT. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rongvaux A, Jackson R, Harman CC, Li T, West AP, de Zoete MR, Wu Y, Yordy B, Lakhani SA, Kuan CY, Taniguchi T, Shadel GS, Chen ZJ, Iwasaki A, Flavell RA. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159:1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y, Brandizzi F. IRE1: ER stress sensor and cell fate executor. Trends in cell biology. 2013;23:547–555. doi: 10.1016/j.tcb.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PloS one. 2009;4:e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, Paton AW, Paton JC, Walter P, Ashkenazi A. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science. 2014;345:98–101. doi: 10.1126/science.1254312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L, Truitt M, McManus MT, Ruggero D, Goga A, Papa FR, Oakes SA. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338:818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lerner AG, Upton JP, Praveen PV, Ghosh R, Nakagawa Y, Igbaria A, Shen S, Nguyen V, Backes BJ, Heiman M, Heintz N, Greengard P, Hui S, Tang Q, Trusina A, Oakes SA, Papa FR. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell metabolism. 2012;16:250–264. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sandow JJ, Dorstyn L, O'Reilly LA, Tailler M, Kumar S, Strasser A, Ekert PG. ER stress does not cause upregulation and activation of caspase-2 to initiate apoptosis. Cell death and differentiation. 2014;21:475–480. doi: 10.1038/cdd.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K, Hori S, Kakizuka A, Ichijo H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes & development. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 66.Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nature reviews. Drug discovery. 2013;12:703–719. doi: 10.1038/nrd3976. [DOI] [PubMed] [Google Scholar]
- 67.Fung TS, Liao Y, Liu DX. The endoplasmic reticulum stress sensor IRE1alpha protects cells from apoptosis induced by the coronavirus infectious bronchitis virus. Journal of virology. 2014;88:12752–12764. doi: 10.1128/JVI.02138-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hassan IH, Zhang MS, Powers LS, Shao JQ, Baltrusaitis J, Rutkowski DT, Legge K, Monick MM. Influenza A viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IRE1) stress pathway. The Journal of biological chemistry. 2012;287:4679–4689. doi: 10.1074/jbc.M111.284695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Foxman EF, Storer JA, Vanaja K, Levchenko A, Iwasaki A. Two interferon-independent double-stranded RNA-induced host defense strategies suppress the common cold virus at warm temperature. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:8496–8501. doi: 10.1073/pnas.1601942113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iwawaki T, Akai R, Yamanaka S, Kohno K. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16657–16662. doi: 10.1073/pnas.0903775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. The Journal of experimental medicine. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. Journal of virology. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 74.Sanjabi S, Williams KJ, Saccani S, Zhou L, Hoffmann A, Ghosh G, Gerondakis S, Natoli G, Smale ST. A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes & development. 2005;19:2138–2151. doi: 10.1101/gad.1329805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lipson KL, Ghosh R, Urano F. The role of IRE1alpha in the degradation of insulin mRNA in pancreatic beta-cells. PloS one. 2008;3:e1648. doi: 10.1371/journal.pone.0001648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kazakov T, Yang F, Ramanathan HN, Kohlway A, Diamond MS, Lindenbach BD. Hepatitis C virus RNA replication depends on specific cis- and trans-acting activities of viral nonstructural proteins. PLoS pathogens. 2015;11:e1004817. doi: 10.1371/journal.ppat.1004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.