Abstract
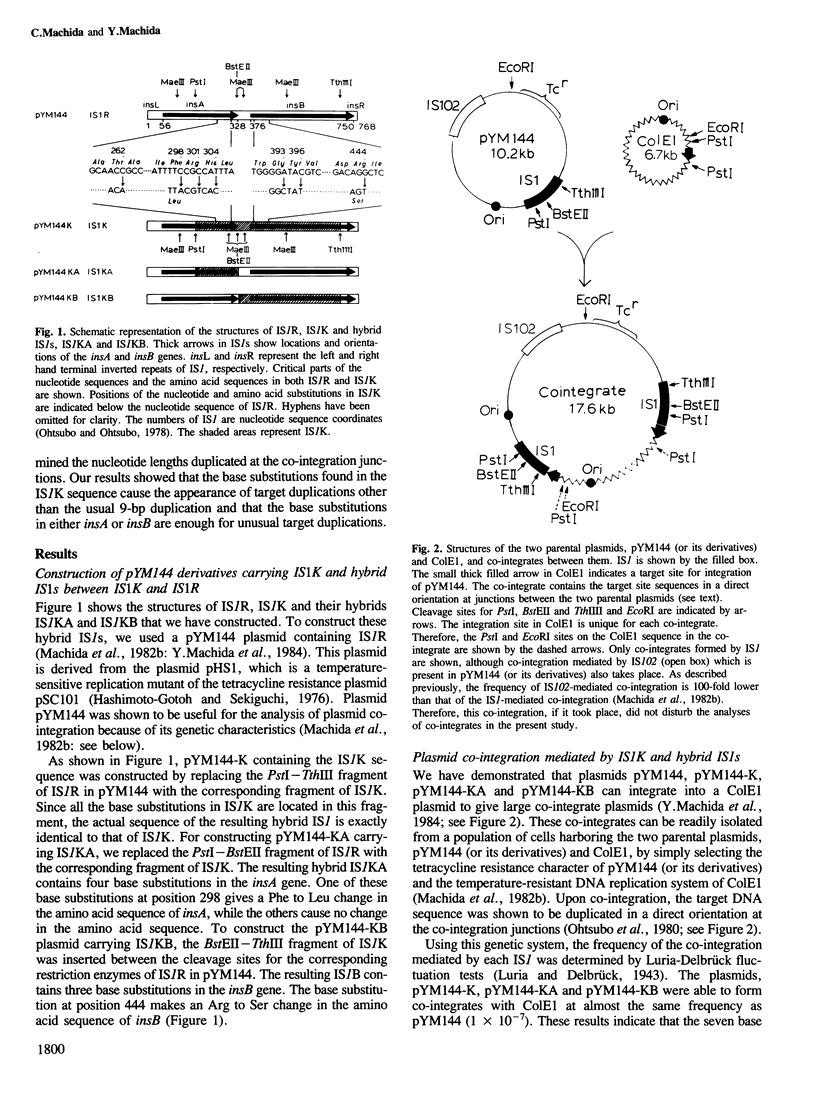
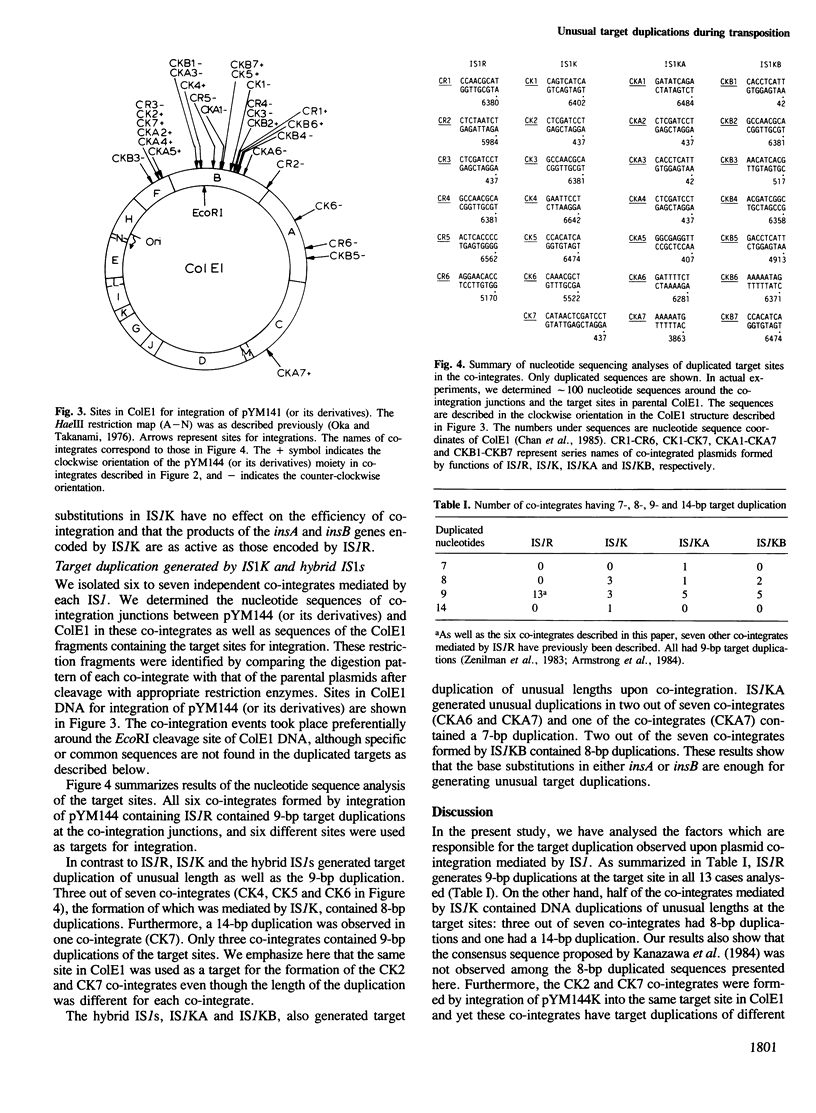
We demonstrate that base substitutions in the IS1 sequence affect the length of the nucleotide sequence which is duplicated during IS1-mediated co-integration. IS1K, an IS1 variant present in the Escherichia coli chromosome, has seven base substitutions in its sequence as compared with that of IS1R derived from the plasmid R100. All substitutions are located in the internal region of IS1K. We have constructed plasmids containing IS1R, IS1K and hybrids between them: one contains four base substitutions causing an amino acid substitution in the insA gene and the other has three substitutions producing an amino acid substitution in the insB gene. We have isolated co-integrate plasmids formed by each IS1 and analysed nucleotide sequences of the target sites duplicated at the co-integration junctions. The results show that IS1K generates duplications of 8 or 14 bp as well as 9 bp, while IS1R exclusively generates the 9-bp duplications. Both hybrid IS1s also create 8- or 7-bp target duplications in addition to 9-bp duplications. These results indicate that the base substitutions in either insA or insB are sufficient for the occurrence of unusual target duplications, suggesting that both genes are involved in the target duplication.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong K. A., Acosta R., Ledner E., Machida Y., Pancotto M., McCormick M., Ohtsubo H., Ohtsubo E. A 37 X 10(3) molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J Mol Biol. 1984 May 25;175(3):331–348. doi: 10.1016/0022-2836(84)90352-8. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Johnsrud L., Miller J. H. DNA sequence at the integration sites of the insertion element IS1. Cell. 1978 Mar;13(3):411–418. doi: 10.1016/0092-8674(78)90315-x. [DOI] [PubMed] [Google Scholar]
- Chan P. T., Ohmori H., Tomizawa J., Lebowitz J. Nucleotide sequence and gene organization of ColE1 DNA. J Biol Chem. 1985 Jul 25;260(15):8925–8935. [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Faelen M., Toussaint A., De Lafonteyne J. Model for the enchancement of lambde-gal integration into partially induced Mu-1 lysogens. J Bacteriol. 1975 Mar;121(3):873–882. doi: 10.1128/jb.121.3.873-882.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill R., Heffron F., Dougan G., Falkow S. Analysis of sequences transposed by complementation of two classes of transposition-deficient mutants of Tn3. J Bacteriol. 1978 Nov;136(2):742–756. doi: 10.1128/jb.136.2.742-756.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D. IS1 insertion generates duplication of a nine base pair sequence at its target site. Cell. 1978 Mar;13(3):419–426. doi: 10.1016/0092-8674(78)90316-1. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Sherratt D. J. Sequence analysis at IS1 insertion sites: models for transposition. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1257–1261. doi: 10.1101/sqb.1979.043.01.142. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Gotoh T., Sekiguchi M. Mutations of temperature sensitivity in R plasmid pSC101. J Bacteriol. 1977 Aug;131(2):405–412. doi: 10.1128/jb.131.2.405-412.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S., Hiestand-Nauer R., Arber W. Transposable element IS1 intrinsically generates target duplications of variable length. Proc Natl Acad Sci U S A. 1985 Feb;82(3):839–843. doi: 10.1073/pnas.82.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S., Hiestand-Nauer R. Insertion element IS1 can generate a 10-base pair target duplication. Gene. 1986;45(2):233–235. doi: 10.1016/0378-1119(86)90260-x. [DOI] [PubMed] [Google Scholar]
- Iida S., Marcoli R., Bickle T. A. Variant insertion element IS1 generates 8-base pair duplications of the target sequence. Nature. 1981 Nov 26;294(5839):374–376. doi: 10.1038/294374a0. [DOI] [PubMed] [Google Scholar]
- Johnsrud L. DNA sequence of the transposable element IS1. Mol Gen Genet. 1979 Jan 31;169(2):213–218. doi: 10.1007/BF00271673. [DOI] [PubMed] [Google Scholar]
- Kanazawa H., Kiyasu T., Noumi T., Futai M., Yamaguchi K. Insertions of transposable elements in the promoter proximal region of the gene cluster for Escherichia coli H+-ATPase: 8 base pair repeat generated by insertion of IS1. Mol Gen Genet. 1984;194(1-2):179–187. doi: 10.1007/BF00383514. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida C., Machida Y., Ohtsubo E. Both inverted repeat sequences located at the ends of IS1 provide promoter functions. J Mol Biol. 1984 Aug 5;177(2):247–267. doi: 10.1016/0022-2836(84)90455-8. [DOI] [PubMed] [Google Scholar]
- Machida C., Machida Y., Wang H. C., Ishizaki K., Ohtsubo E. Repression of cointegration ability of insertion element IS1 by transcriptional readthrough from flanking regions. Cell. 1983 Aug;34(1):135–142. doi: 10.1016/0092-8674(83)90143-5. [DOI] [PubMed] [Google Scholar]
- Machida Y., Machida C., Ohtsubo E. A novel type of transposon generated by insertion element IS102 present in a pSC101 derivative. Cell. 1982 Aug;30(1):29–36. doi: 10.1016/0092-8674(82)90008-3. [DOI] [PubMed] [Google Scholar]
- Machida Y., Machida C., Ohtsubo E. Insertion element IS1 encodes two structural genes required for its transposition. J Mol Biol. 1984 Aug 5;177(2):229–245. doi: 10.1016/0022-2836(84)90454-6. [DOI] [PubMed] [Google Scholar]
- Machida Y., Machida C., Ohtsubo H., Ohtsubo E. Factors determining frequency of plasmid cointegration mediated by insertion sequence IS1. Proc Natl Acad Sci U S A. 1982 Jan;79(2):277–281. doi: 10.1073/pnas.79.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyman K., Nakamura K., Ohtsubo H., Ohtsubo E. Distribution of the insertion sequence IS1 in gram-negative bacteria. Nature. 1981 Feb 12;289(5798):609–612. doi: 10.1038/289609a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H., McCormick M., Machida C., Machida Y. Mechanism of insertion and cointegration mediated by IS1 and Tn3. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):283–295. doi: 10.1101/sqb.1981.045.01.041. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H. Plasmids containing insertion elements are potential transposons. Proc Natl Acad Sci U S A. 1980 Feb;77(2):750–754. doi: 10.1073/pnas.77.2.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo H., Nyman K., Doroszkiewicz W., Ohtsubo E. Multiple copies of iso-insertion sequences of IS1 in Shigella dysenteriae chromosome. Nature. 1981 Aug 13;292(5824):640–643. doi: 10.1038/292640a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Takanami M. Cleavage map of colicin E1 plasmid. Nature. 1976 Nov 11;264(5582):193–196. doi: 10.1038/264193a0. [DOI] [PubMed] [Google Scholar]
- Sengstag C., Iida S., Hiestand-Nauer R., Arber W. Terminal inverted repeats of prokaryotic transposable element IS186 which can generate duplications of variable length at an identical target sequence. Gene. 1986;49(1):153–156. doi: 10.1016/0378-1119(86)90395-1. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]



