Abstract
A host’s microbiota may increase, diminish, or have no effect at all on cancer susceptibility. Assigning causal roles in cancer to specific microbes and microbiotas, unraveling host-microbiota interactions with environmental factors in carcinogenesis, and exploiting such knowledge for cancer diagnosis and treatment are areas of intensive interest. This Review considers how microbes and the microbiota may amplify or mitigate carcinogenesis, responsiveness to cancer therapeutics, and cancer-associated complications.
The relationship between cancer and microbes is complex. Although cancer is generally considered to be a disease of host genetics and environmental factors, microorganisms are implicated in ~20% of human malignancies (1). Microbes present at mucosal sites can become part of the tumor microenvironment of aerodigestive tract malignancies, and intratumoral microbes can affect cancer growth and spread in many ways (2–6). In counterpoise, the gut microbiota also functions in detoxification of dietary components, reducing inflammation, and maintaining a balance in host cell growth and proliferation. The possibility of microbe-based cancer therapeutics has attracted interest for more than 100 years, from Coley’s toxins (one of the earliest forms of cancer bacteriotherapy) to the current era of synthetic biology’s designer microbes and microbiota transplants. Thus, interrogation of the roles of microbes and the microbiota in cancer requires a holistic perspective.
The ways in which microbes and the microbiota contribute to carcinogenesis, whether by enhancing or diminishing a host’s risk, fall into three broad categories: (i) altering the balance of host cell proliferation and death, (ii) guiding immune system function, and (iii) influencing metabolism of host-produced factors, ingested foodstuffs, and pharmaceuticals (Fig. 1). Assigning microbial communities, their members, and aggregate biomolecular activities into these categories will require a substantial research commitment. This Review discusses how microbes and the microbiota may contribute to cancer development and progression, responsiveness to cancer therapeutics, and cancer-associated complications.
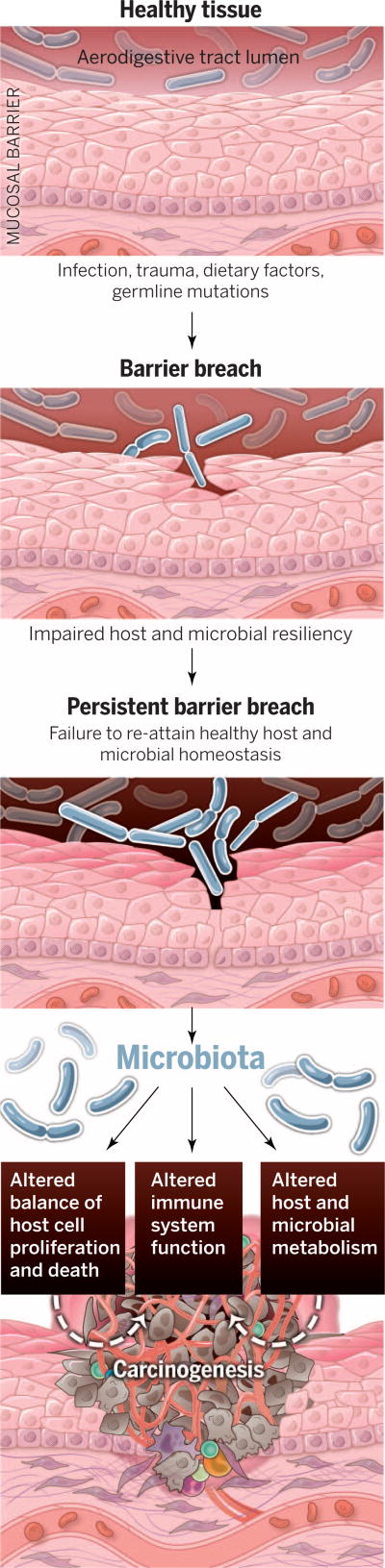
Fig. 1. The path from health to solid tumor malignancies at mucosal sites and the microbiota’s contribution.
Human body surfaces are subject to constant environmental insult and injury. Infections, trauma, dietary factors, and germline mutations can contribute to breach of the body’s mucosal barriers. In most individuals, barrier breaches are rapidly repaired and tissue homeostasis is restored. Impaired host or microbial resiliency contributes to persistent barrier breach and a failure to restore homeostasis. In these settings, the microbiota may influence carcinogenesis by (i) altering host cell proliferation and death, (ii) perturbing immune system function, and (iii) influencing metabolism within a host.
Microbial contributions to carcinogenesis
Of the estimated 3.7 × 1030 microbes living on Earth (7), only 10 are designated by the International Agency for Cancer Research (IACR) as carcinogenic to humans (1). Although most of these carcinogenic microbes colonize large percentages of the human population, only a subset of affected individuals develop cancer, because host and microbial genotypes influence cancer susceptibility.
Tumors arising at boundary surfaces, such as the skin, oropharynx, and respiratory, digestive, and urogenital tracts, harbor amicrobiota, which complicates cancer-microbe causality. Enrichment of a microbe at a tumor site does not connote that a microbe is directly associated, let alone causal, in disease. Rather, microbes may find a tumor’s oxygen tension or carbon sources permissive and take advantage of an underused nutritional niche. Decreased abundances of specific microbes may also place a host at enhanced risk for cancer development at sites local or distant from this microbial shift. Thus, rigorous frameworks for interpreting tumor-associated microbiota data are essential (2).
Oncomicrobes, shifting the balance of when to die and when to grow
Bona fide oncomicrobes—microbes that trigger transformation events in host cells—are rare. Beyond the 10 IACR-designated microbes, there are a handful of other microorganisms with robust but fewer aggregate data supporting their role in human carcinogenesis. As many of these and their carcinogenic mechanisms have been recently reviewed (2–6, 8), select activities representing common pathways by which microbes influence cancer will be highlighted.
Human oncoviruses can drive carcinogenesis by integrating oncogenes into host genomes. Human papillomaviruses (HPV) express oncoproteins such as E6 and E7. Data from recent genomic analyses of HPV+ cervical cancers suggest that viral integration also selectively triggers amplification of host genes in pathways with established roles in cancer (9).
Microbes also drive transformation by affecting genomic stability, resistance to cell death, and proliferative signaling. Many bacteria have evolved mechanisms to damage DNA, so as to kill competitors and survive in the microbial world. Unfortunately, these bacterial defensive factors can lead to mutational events that contribute to carcinogenesis (Fig. 2). Examples include colibactin encoded by the pks locus [expressed by B2 group Escherichia coli (10) as well as by other Enterobacteriaceae (11)], Bacteroides fragilis toxin (Bft) produced by enterotoxigenic B. fragilis, and cytolethal distending toxin (CDT) produced by several ε- and γ-proteobacteria. Colibactin has emerged as a molecule of interest in colorectal carcinogenesis, given the detection of pks+ E. coli in human colorectal cancers and the ability of colibactin-expressing E. coli to potentiate intestinal tumorigenesis in mice (12, 13). Accumulating data also support a role for enterotoxigenic B. fragilis in both human and animal models of colon tumors (14–17). Both colibactin and CDT can cause double-stranded DNA damage in mammalian cells (18). In contrast, Bft acts indirectly by eliciting high levels of reactive oxygen species (ROS), which in turn damage host DNA (19). Chronically high ROS levels can outpace a host’s DNA repair mechanisms, leading to DNA damage and mutations (Fig. 2).
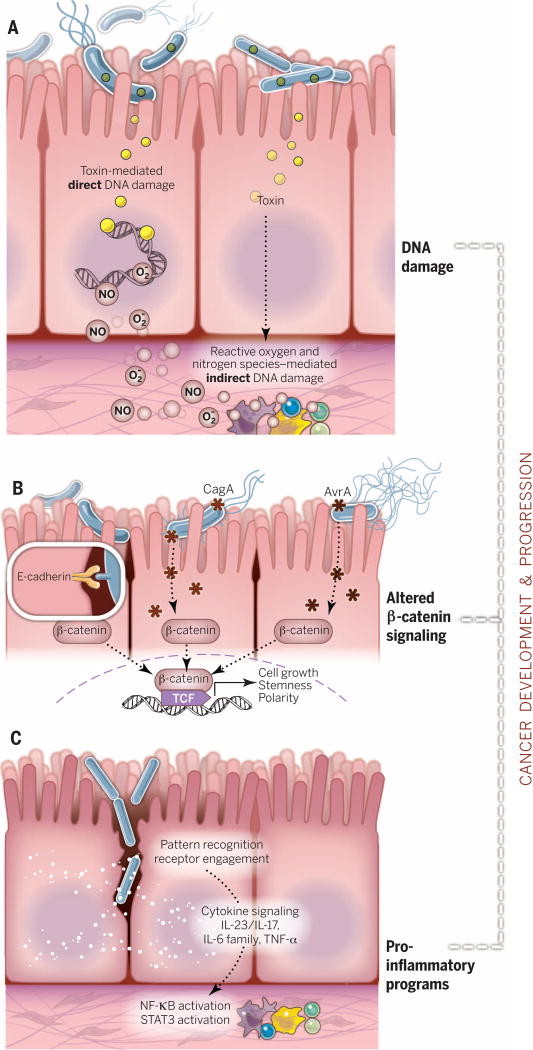
Fig. 2. Mechanisms by which microbes influence cancer development and progression.
(A) Bacterial toxins can directly damage host DNA. Bacteria also damage DNA indirectly via host-produced reactive oxygen and nitrogen species. When DNA damage exceeds host cell repair capacity, cell death or cancer-enabling mutations occur. (B) β-Catenin signaling alterations are a frequent target of cancer-associated microbes. Some microbes bind E-cadherin on colonic epithelial cells, with altered polarity or within a disrupted barrier, and trigger β-catenin activation. Other microbes inject effectors (e.g., CagA or AvrA) that activate β-catenin signaling, resulting in dysregulated cell growth, acquisition of stem cell–like qualities, and loss of cell polarity. (C) Proinflammatory pathways are engaged upon mucosal barrier breach in an evolving tumor. Loss of boundaries between host and microbe engages pattern recognition receptors and their signaling cascades. Feedforward loops of chronic inflammation mediated by NF-κB and STAT3 signaling fuel carcinogenesis within both transforming and nonneoplastic cells within the tumors.
Beyond damaging DNA, several microbes possess proteins that engage host pathways involved in carcinogenesis. The Wnt/β-catenin signaling pathway, which regulates cell stemness, polarity, and growth (20), is one example and is altered in many malignancies. Several cancer-associated bacteria also can influence β-catenin signaling (Fig. 2). Oncogenic type 1 strains of Helicobacter pylori express a protein called CagA, which is injected directly into the cytoplasm of host cells and aberrantly modulates β-catenin to drive gastric cancer (8). CagA-mediated β-catenin activation leads to up-regulation of genes involved in cellular proliferation, survival, and migration, as well as angiogenesis—all processes central to carcinogenesis. Fusobacterium nucleatum is a member of the oral microbiota and is associated with human colorectal adenomas and adenocarcinomas and amplified intestinal tumorigenesis in mice (21–24). F. nucleatum expresses FadA, a bacterial cell surface adhesion component that binds host E-cadherin, leading to β-catenin activation (25). Enterotoxigenic B. fragilis, which is enriched in some human colorectal cancers (14), can stimulate E-cadherin cleavage via Btf, leading to β-catenin activation (26). Salmonella typhi strains that maintain chronic infections secrete AvrA, which can activate epithelial β-catenin signaling (27, 28), and are associated with hepatobiliary cancers (29–31).
This phenomenon of activating β-catenin signaling reflects an interesting convergence of evolution, as several of these bacteria are normal constituents of the human microbiota. Although microbial engagement of β-catenin signaling may reflect a drive to establish a niche in a new tissue site, the presence of these cancer-potentiating microbes and their access to E-cadherin in evolving tumors demonstrate that a loss of appropriate boundaries and barrier maintenance between host and microbe is a critical step in the development of some tumors (Figs. 1 and 2).
The immune system, microbes, microbiota, and cancer
Mucosal surface barriers permit host-microbial symbiosis (32); they are susceptible to constant environmental insult and must rapidly repair to reestablish homeostasis. Compromised resiliency of the host or microbiota can place tissues on a path to malignancy. Cancer and inflammatory disorders can arise when barriers break down and microbes and immune systems find themselves in geographies and assemblages for which they have not coevolved. Once barriers are breached, microbes can further influence immune responses in evolving tumor microenvironments by eliciting proinflammatory or immunosuppressive programs (Fig. 2).
Proinflammatory responses can be procarcinogenic
Both the chronic, high-grade inflammation of inflammatory disorders (e.g., inflammatory bowel disease) and the lower-grade smoldering inflammation of malignancies and obesity drive a tumor-permissive milieu. Inflammatory factors such as reactive oxygen and nitrogen species, cytokines, and chemokines can contribute to tumor growth and spread (Fig. 2). Data from human tissues and animal models show that tumors can up-regulate and activate many pattern recognition receptors, including Toll-like receptors (3, 8). Activation of these receptors results in feedforward loops of activation of NF-κΒ, a master regulator of cancer-associated inflammation (33) (Fig. 2). Numerous cancer-associated microbes appear to activate NF-κΒ signaling within the tumor microenvironment [e.g., the colon cancer–associated F. nucleatum (23)]. The activation of NF-κΒ by F. nucleatum may be the result of pattern recognition receptor engagement (10, 34–37) or FadA engagement of E-cadherin (25). Other pattern recognition receptors, such as the nucleotide-binding oligomerization domain–like receptor (NLR) family members NOD-2, NLRP3, NLRP6, and NLRP12, may play a role in mediating colorectal cancer; mice deficient in these NLRs display an enhanced susceptibility to colitis-associated colorectal cancer (caCRC) (38–44).
Engagement of the immune system within the tumor microenvironment is not restricted to the innate immune system. Once barriers are breached and the innate immune system is activated, subsequent adaptive immune responses ensue, often with deleterious consequence for tumor progression. The interleukin-23 (IL-23)–IL-17 axis (45), tumor necrosis factor–α (TNF-α)–TNF receptor signaling (3, 5, 6, 46), IL-6–IL-6 family member signaling (46, 47), and STAT3 activation (48, 49)—an output of these cytokine-mediated signaling pathways—all represent innate and adaptive pathways contributing to tumor progression and growth (Fig. 2).
The microbiota is responsive and adapts to changes in its host, such as inflammation. Adaptation to new selective pressures may result in a microbiota at a tissue site that is not well suited for barrier repair, immune homeostasis, or maintenance of traditional host and microbe boundaries. Mouse models of caCRC furnish insight in this regard. One such model uses azoxymethane, a genotoxin, and dextran sodium sulfate, a colon barrier–disrupting agent. Either agent alone results in colon tumors in susceptible mouse strains; using them together accelerates tumorigenesis. Although this model does not recapitulate the molecular and environmental events that lead to caCRC, it provides an opportunity to study the convergence of an environmental genotoxin, barrier disruption, and severe chronic inflammation on cancer development.
Microbiota transfer studies in caCRC models support the idea that perturbations to a host immune system, either by genetic deletion or genotoxin coupled with inflammatory stimulus, may select for microbiotas enriched for bacterial clades adept at attaching to host surfaces, invading host tissue, or triggering host inflammatory mediators (21, 22, 40, 50, 51). Fecal microbiota from Nod2- or Nlrp6-deficient mice acquire features that enhance the susceptibility of wild-type mice to caCRC (40, 44). In mice, the gut microbiota modulate colon tumorigenesis, independent of genetic deficiencies. When germ-free mice were colonized with microbes from donors with or without caCRC, followed by treatments that induced caCRC, those recipients that received gut microbiomes from caCRC-bearing mice developed more tumors (51). Similar mouse experiments using fecal transfers from humans with colon cancer suggest that there are microbiome structures, both protective and risk-elevating, that influence tumorigenesis (52).
Inflammation also results in the generation of respiratory electron acceptors such as nitrate, ethanolamine, and tetrathionate, which some bacterial clades can use for their own fitness advantage (53–59). Several bacteria (e.g., E. coli and Salmonella spp.) can use these electron acceptors and also possess the key features that reinforce the chronic inflammatory programs that can enhance cancer growth and spread. However, it remains to be determined whether bacterial use of these electron acceptors enhances cancer growth.
Immune-dampening responses can be cancer-permissive
Microbes not only trigger and reinforce proinflammatory immune circuits but also exploit or elicit immunosuppressive responses. A microbe may take advantage of preexisting immunosuppression or elicit immune-dampening responses to avoid destruction. Chronic systemic immunosuppression, as seen with advanced HIV infection, increases the risk for many cancers, especially virally associated malignancies. Microbial-elicited immunosuppression can also contribute to impaired antitumor immunity. Most current cancer-directed immunotherapies are focused on rousing immune responsiveness to tumors (60). The colon cancer–associated bacterium F. nucleatum may directly inhibit antitumor immunity by engaging TIGIT, a receptor with immunoglobulin and ITIM domains expressed on some T cells and natural killer cells, and blocking its ability to kill tumor cells (61). Whether microbes contribute to immunotherapeutic resistance in other cancers remains to be investigated.
Interrogating the role of microbes and microbiotas in cancer with new and old technologies
Microbiota studies in cancer remain at an early stage. Information gathering and descriptive studies are still necessary, and many critical questions remain. What other mechanisms might microbes use to influence tumorigenesis? If single microbes can compromise antitumor immunity or enhance susceptibility to oncomicrobes, are there configurations of the microbiota that do this, too (or are protective)? Are there microbes or microbiotas that enhance responsiveness to immunotherapies or other therapeutic interventions? To answer these questions, it is important to identify the key next steps in understanding how the human microbiota affects tumor growth and spread.
Sequencing-based technologies are a boon to both cancer biology and microbiology. Cancer genomes and their functional analyses have led to the implementation of precision medicine approaches to cancer care. Efforts to sequence individual microbes and human microbiomes are providing insight into how they influence human health and disease. Computational tools that identify microbial data within human sequencing data sets are welcome new additions to the armamentarium of cancer microbe hunters (62, 63).
Despite the affordable price of sequencing, advances in culture techniques (64–66), and high-throughput analysis pipelines, the path of cancer microbiome discovery is fraught with pitfalls. Cancers may develop over decades, and different microbes and microbiotas may participate at distinct stages of the neoplastic process. For many malignancies, by the time a cancer is detected, the window of opportunity for identifying the inciting microbial agent(s) may have passed, allowing these organisms to remain elusive. However, the microbiota should remain a focus of study in locally advanced and metastatic cancer, as microbes may contribute to an established cancer’s continued growth and spread.
Beyond sequencing, microscopy and flow cytometry–based approaches are useful tools to detect and study tumor-associated microbiotas. Human colon tumors may harbor specific consortia of bacteria that assemble themselves into biofilms (17). These biofilms appear to be specific to certain biogeographies within the gastrointestinal tract and have members that have been associated with colorectal adenomas and adenocarcinomas in human and mouse studies (e.g., enterotoxigenic B. fragilis and F. nucleatum). Microbiological studies of the oral cavity have shed light on microbial biofilms and their roles in human health and disease (67, 68). Within biofilms, microbial cross-feeding and co-metabolism occur (69). Consortia of tumor-associated microbes have the potential to generate metabolites that require collective microbial metabolism, and these co-metabolites may contribute to or halt carcinogenesis. The role of microbial metabolism in host physiology is an exciting area, with several recent studies reexamining the role of microbial metabolites in cancer (4, 70).
Microbes, metabolism, and cancer
In 1956, Warburg put forth the hypothesis that altered cellular metabolism is the root cause of carcinogenesis (71), and cancer cell metabolism is currently a promising therapeutic target (72). Microbes participate in a range of host metabolic activities. Microbial metabolites or co-metabolites (generated with contributions from both host and microbe) can contribute to inflammatory tone and can influence the balance of proliferation and cell death in tissues (4). Consideration of the effects of a microbiota’s metabolism, and specifically microbial metabolites generated within the tumor microenvironment, on cancer growth and spread adds another therapeutic and diagnostic angle for targeting cancers through metabolic alterations.
A meal fit/unfit for a tumor: Fiber and fats
What defines a microbial oncometabolite (73), and how are such metabolites generated? Both the host and its microbes affect the metabolism of dietary fiber, fats, ethanol, and phytoestrogens. As with microbes, metabolites can affect immune cell function, barrier function, and cell proliferation and death. Metabolites generated from dietary fiber and fats that have an established effect on cancer are considered below, along with recent insights.
Intestinal fermentation of dietary fiber by members of the colonic microbiota results in the generation of several short-chain fatty acids (SCFAs) including acetic, propionic, and butyric acids. These SCFAs have a range of effects on many cell types, including anti-inflammatory effects on myeloid cells (74) and colonic regulatory T cells (75–77), with consequences for intratumoral inflammation. SCFA’s effects may be tuned by the receptors that they bind (e.g., Niacr1/Gpr109a, Gpr43, Gpr41, or Olfr78). Gpr109a is a receptor for niacin and butyrate. It plays an important role in mediating the effects of dietary fiber and the microbiota in the colon, where it is expressed by both colonic epithelial cells and intestinal myeloid cells. Activation of Gpr109a by butyrate results in anti-inflammatory host responses in myeloid cells that lead to regulatory T cell generation, and loss of Gpr109a increases susceptibility to caCRC (78).
SCFAs also affect host gene expression patterns, cell proliferation, and cell death via both receptor-mediated and receptor-independent mechanisms. SCFAs and their activation of Gpr43 reduce the proliferation rate of leukemia cells (79). In a study of ~70 human colon adenocarcinomas, GPR43 expression was reduced in cancer versus healthy tissue; restoration of GPR43 in a human colon cancer line increased apoptotic cell death upon SCFA exposure (80).
SCFAs’ effects on host cellular processes vary according to concentration and host genotype. Two recent mouse studies, which arrived at different conclusions regarding the relationship of dietary fiber, the microbiota, and butyrate to colorectal tumorigenesis, reflect this heterogeneous response to SCFAs (Fig. 3). Dietary fiber and butyrate-producing bacteria suppressed tumors in mice that harbored strictly defined microbial communities, received specialized diets, and were treated with azoxymethane and dextran sodium sulfate (81). This study’s data supported a model wherein the glycolytic metabolism of cancer cells resulted in reduced metabolism of butyrate and enhanced butyrate nuclear accumulation. High intranuclear butyrate levels increased histone acetylation and led to increased apoptosis and reduced cellular proliferation. In a mouse model of intestinal tumorigenesis driven by mutations in both the Apc gene and the mismatch repair gene Msh2, the microbiota and butyrate had tumor-promoting effects (82). Butyrate’s principal effect in this model system was to drive a hyperproliferative response in Msh2-deficient epithelial cells. Cancer genetics and butyrate concentrations were critical factors in SCFAs’ disparate effects on tumorigenesis between these studies. These studies underscore the challenges of translating microbiome, diet, and cancer basic science data into consensus guidelines for dietary interventions to reduce cancer risk. Given that a single microbial metabolite can mediate a range of effects in tumor models, investigators will require additional experimental systems to unravel the effects of the human-microbial meta-metabolome for health and cancer susceptibility.
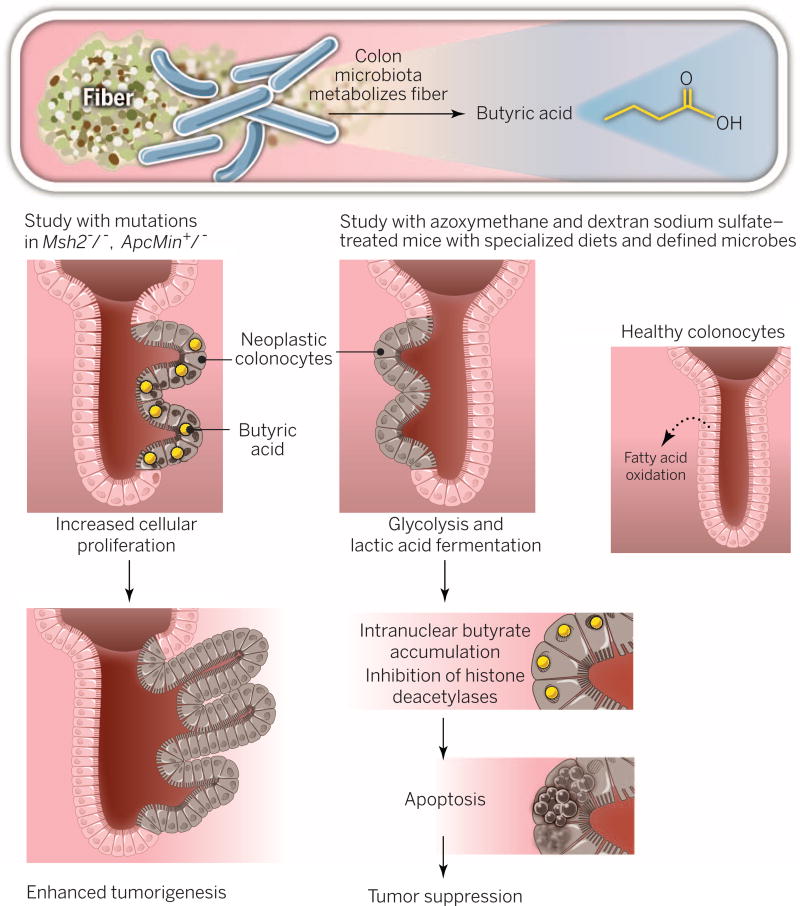
Fig. 3. Dietary fiber, microbiota, butyrate, and tumorigenesis.
Metabolism of fiber by colonic microbes results in generation of butyric acid. When genetic mutations in Msh2 and Apc are present, butyrate increases cell proliferation and enhances tumorigenesis. Data from another model of colorectal carcinogenesis indicate the opposite outcome: Neoplastic colonocytes engage in glycolysis for cellular energy, unlike healthy colonocytes (which favor fatty acid oxidation). As a result, butyrate accumulates in the nucleus of neoplastic cells, engaging tumor-suppressive pathways and apoptosis.
In contrast with the conflicting basic science and epidemiological data surrounding dietary fiber (83), there is consensus that high saturated fat intake heightens cancer risk. Debate surrounding a high-fat diet (HFD) focuses on several mechanisms that may act alone or in combination, involving obesity, the microbiome, bile acids, and inflammation. There are a myriad of studies exploring the interconnection between obesity and malignancy (84–86). Obesity is now regarded as an inflammatory state (87), and we are learning more about the gut microbiome’s contribution to obese and lean states (88, 89). Data support the idea that inflammation, the microbiota, and obesity constitute an inseparable trio that fuels cancer. However, a recent study suggests otherwise. In a mouse model of duodenal hyperplasia, adenomas, and invasive cancer driven by k-ras mutation, HFD and microbial dysbiosis amplified tumor growth and spread in the absence of obesity or the development of a robust proinflammatory response (90);mutated k-ras modulated Paneth cell antimicrobial expression and HFD affected intestinal mucin expression, thereby altering the intestinal microbiota. The fecal microbiota of HFD k-ras mutant mice was sufficient to transmit the cancer-potentiating effects of the HFD when transferred to antibiotic-treated k-ras mutant mice.
Another mechanism by which HFD influences cancer risk is via bile acids that are produced to solubilize and digest the consumed fats—specifically, the microbially generated secondary bile acids. The role of secondary bile acids in increased or decreased cancer risk has been studied for decades (2). One recent study provided new insight into deoxycholic acid’s prooncogenic mechanisms in liver cancer: HFD or genetic susceptibility to obesity can increase deoxycholic acid–mediated activation of a mitogenic and proinflammatory response program in hepatic stellate cells, thereby potentiating liver cancer in mice (91). These studies reinforce the importance of gene-environment interactions in carcinogenesis and underscore the need to consider how dietary patterns influence the genomes and genomic outputs of both host and microbiome in mitigating or amplifying cancer risk.
Drugs, bugs, and cancer
The gut microbiota function in drug metabolism, influencing toxicity and efficacy (92, 93). Because chemotherapeutic agents have a narrow therapeutic window, there is interest in the microbiota’s modulation of chemotherapy toxicity and efficacy (Fig. 4). Irinotecan is a topoisomerase-1 inhibitor that is used in combination with other chemotherapies to treat several cancers. A common side effect is diarrhea. For some patients, the severity of the diarrhea requires hospitalization. Microbial-produced β-glucuronidases regulate levels of irinotecan’s bioactive form within the intestinal lumen and thus influence irinotecan’s toxicity (94). Oral bacterial β-glucuronidase inhibitors blunt the dose-limiting toxicities of irinotecan in mice and do not harm host cells or kill bacteria, which suggests that microbial metabolism is a plausible target in cancer care (95).
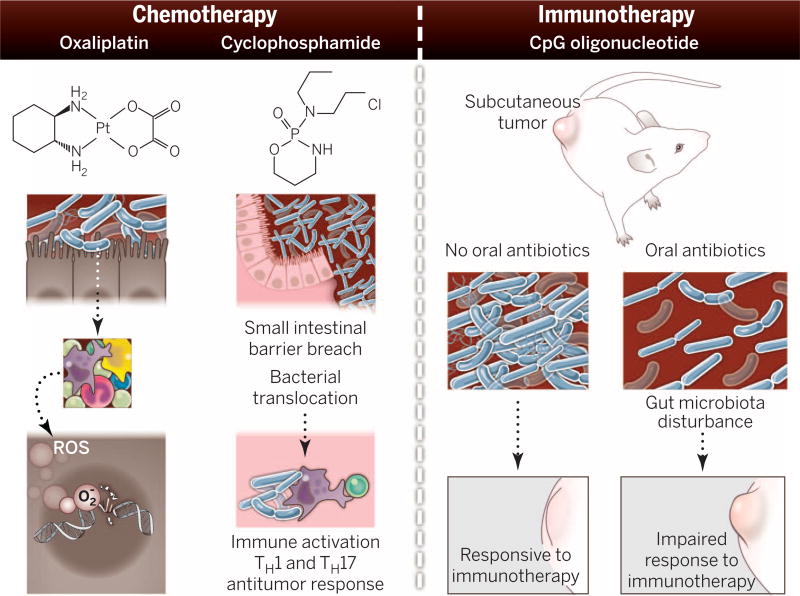
Fig. 4. How the microbiota modulate chemotherapy and immunotherapy efficacy in mouse models.
The gut microbiota stimulate immune cells to produce reactive oxygen species (ROS). ROS enhance DNA damage caused by oxaliplatin, blocking DNA replication and transcription and resulting in cell death. Cyclophosphamide can cause small intestinal barrier breach. This barrier disruption results in bacterial translocation that potentiates antitumor TH1 and TH17 responses. CpG oligonucleotides are a microbial-associated molecular pattern and are used in immunotherapy. Antibiotic disruption of the gut microbiota in mice compromised the efficacy of CpG in a mouse subcutaneous tumor model.
The gut microbiota also affect the efficacy of chemotherapy. Oxaliplatin is a platinum-based chemotherapy used to treat several gastrointestinal malignancies. Together, the microbiota and immune system contribute to oxaliplatin’s efficacy (96). The gut microbiota prime myeloid cells for high-level ROS production. The resultant intratumoral oxidative stress augments oxaliplatin-associated DNA damage, triggering cancer cell death (96). Cyclophosphamide, an alkylating agent used in hematologic malignancies and solid tumors, can injure the small intestinal epithelium. The ensuing barrier breach results in gut microbiota–dependent, T helper (TH) cell–mediated antitumor responses (97). Delineating the roles of gut microbiota in response to chemotherapy in model systems and undertaking epidemiologic studies with microbiome analysis in patients with and at risk for cancer will be critical for realizing the microbiota as an adjuvant therapy that enhances efficacy or attenuates toxicity of chemotherapies.
The microbiota and immunotherapy: Friend or foe?
The success of immunotherapy (in the form of cytokine therapy, targeting immune checkpoint blockade, and vaccine therapy) has been one of the most exciting developments in cancer care over the past decade (98). Given the intertwined nature of the microbiota and the immune system, it is plausible that the microbiota influence a host’s responsiveness to immunotherapy. In support of this idea, antibiotic-mediated disruption of the microbiota in mice bearing subcutaneous tumors impaired the effectiveness of CpG oligonucleotide immunotherapy (Fig. 4) (96). Observations that immunotherapies are showing efficacy in melanoma and bladder, renal, and lung cancer but not in cancer of the colon (which is densely populated by bacteria) fuel interest in how the microbiota contributes to immunotherapy’s efficacy. Furthermore, given the severe colitis observed in some patients receiving immunotherapies (99) (e.g., antibodies to CTLA4 and PD-L1) and the role of gut microbes in colitis, it is possible that the gut microbiota influences this toxicity. As patient populations expand, investigators will hopefully interrogate whether there are microbiota that are predictive for colitis and other toxicities. Examining the microbiota and its effects on immunotherapy efficacy and toxicity in preclinical models and patients is a critical next step.
Hematopoietic transplants, complications, and the microbiota
Allogeneic hematopoietic stem cell transplant (allo-HSCT), a mainstay in hematologic malignancy treatment, is a challenge to both host and microbiota. An individual’s microbiota is confronted with a new host within its host as well as chemotherapy, radiation, oral and gastrointestinal barrier breach, and broad-spectrum antibiotics. Studies have begun to examine perturbations to the gut microbiota and clinical outcomes during allo-HSCT (100).
Bacteremia, Clostridium difficile infection, and graft-versus-host disease (GVHD) are common events in allo-HSCT patients. Bacteremias with vancomycin-resistant Enterococcus (VRE) are a grave concern. Two preclinical studies examining how antibiotics perturb the gut microbiota to enable VRE displacement of a healthy microbiota (101) and how the anaerobic bacteria Barnesiella spp. may confer resistance to VRE (102) have provided mechanistic insight into these bloodstream infections. These studies set the stage for a clinical study showing that enterococcal gut microbiota domination was associated with a factor of 9 higher risk of VRE bacteremia in allo-HSCT patients (103). Hospitalized patients and allo-HSCT patients both confront toxigenic C. difficile infection. Using mouse models, microbiome analysis, and allo-HSCT patient populations, researchers identified a microbe that can restore bile acid–mediated resistance to C. difficile (104). The workflows of this precision medicine–based study are applicable to many diseases associated with altered microbiotas.
Allo-HSCT patients can experience gastrointestinal, pulmonary, and skin complications after transplant; some of these are idiopathic clinical syndromes while others are GVHD manifestations. Using shotgun DNA sequencing of colon tissue and the PathSeq pipeline, investigators found that Bradyrhizobium enterica was enriched in affected colonic tissue from patients with idiopathic colitis after receiving a cord blood transplant (105), providing insight and a potential treatment. Using samples from mice and humans that had undergone allogeneic bone marrow transplants, investigators characterized the gut microbiota changes in active intestinal GVHD (106). In mice, depletion of lactobacilli exacerbated GVHD-associated intestinal inflammation and their reintroduction attenuated inflammation (106). The challenge intrinsic to these studies, and realized in (104), is to use our evolving knowledge of the microbiome and microbes to identify bacteriotherapy for cancer and its complications.
Back to the future: Perspectives and directions for cancer bacteriotherapy
The genesis of immunotherapy came from an appreciation for the co-adaptation between host and microbe. Exploiting this knowledge and using bacteria to trigger the immune system to attack and destroy cancers dates back to the 1850s, when several German physicians noticed that some cancer patients with active infections showed signs of tumor regression. This led Coley to test bacterial extracts in patients with bone cancers around 1900. Heat-killed cultures of Streptococcus pyogenes and Serratiamarcescens, or Coley’s toxins, were one of earliest forms of immunotherapy (60). Since this seminal work, one bacterium has entered the mainstream of cancer treatment. For the past three to four decades, Bacillus Calmette-Guerin (BCG) has been used to treat non–muscle-invasive bladder cancer. The live bacteria, which are delivered directly into the bladder, elicit inflammation that triggers an antitumor immune response (107). Much still remains to be learned about the immune response to BCG and antitumor immunity, and why BCG loses efficacy once the cancer is more invasive (108).
Over the past 30 years, several bacterial-based approaches to cancer therapy have emerged. Bacterial-based vaccines that express tumor antigens have shown efficacy in preclinical studies, and recombinant Listeria monocytogenes–based vaccines showed tremendous promise in mice (109). Interest remains in using bacteria as a delivery vehicle for plant toxins, such as ricin and saporin, or pseudomonal exotoxins that can block protein synthesis and induce apoptosis in cancer cells (110). Bacteria have evolved elegant systems to communicate with each other, to kill one another (111), and to deliver their effectors into host cells (112). The extension and application of these secretion systems, which have been honed by millennia of evolution, seems like a therapeutic slam dunk but has been challenging in practice. A recent study in dogs (113) has breathed new life into the concept of bacteriotherapy with Clostridium novyii, which emerged as a promising concept in preclinical models almost 15 years ago (114); however, balancing toxicity with efficacy remains difficult.
Synthetic biology approaches to cancer care hold enormous potential, especially those that make use of bacteria. These efforts involve the reengineering of bacterial cells for the delivery of biomolecules under tunable networks and on/off toggle switches triggered by host responses (115). The goals are simple: to target cancers and minimize damage to healthy tissues via genetic network designs informed by engineering principles. Proof of concept that designer microbes can invade cancer cells (116) to target and perturb key cancer pathways has been established (117). Evaluation in robust preclinical models will be the next step. Application and design for cancer care will need to focus on maximizing anticancer responses while minimizing toxicities and infectious complications.
Like synthetic biology, microbiome studies have emerged as a promising area of investigation for cancer care over the past decade. The microbiome may afford many answers to several looming questions in cancer biology: What are the critical gene-environmental interactions in cancer susceptibility? Why do certain foods or dietary patterns confer increased or decreased risk in certain populations and individuals? Why do chemotherapies, immunotherapies, and preventive agents fail or succeed for patients, irrespective of host germline or cancer genotype? The microbiome seems to provide many potential answers in the forms of select clades, consortia, metabolites, and enzymatic activities, but it remains unclear whether and how these will translate from preclinical models to humans. One opportunity for the microbiota in the near term is as a biomarker for diagnosis (118), prognostication, or identifying those most at risk for treatment-related complications. Although there may be dissent about the best next steps, there is consensus that therapeutic consideration of cancer and the microbiota requires a multidisciplinary approach and more intensive investigation.
Acknowledgments
W.S.G. thanks C. Brennan, C. Gallini, G. Hold, C. Lesser, and M. Howitt for critical reading of the manuscript. Supported by NIH grant R01CA154426, a Burroughs Wellcome Career in Medical Sciences Award, a Searle Scholars Award, a Cancer Research Institute Investigator Award, and a research grant from Hoffman-LaRoche. W.S.G. is a Synlogic SAB member.
References
- 1.de Martel C, et al. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 2.Sears CL, Garrett WS. Cell Host Microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwabe RF, Jobin C. Nat. Rev. Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Louis P, Hold GL, Flint HJ. Nat. Rev. Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 5.Elinav E, et al. Nat. Rev. Cancer. 2013;13:759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 6.Irrazábal T, Belcheva A, Girardin SE, Martin A, Philpott DJ. Mol. Cell. 2014;54:309–320. doi: 10.1016/j.molcel.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 7.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D’Hondt S. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abreu MT, Peek RM., Jr Gastroenterology. 2014;146:1534–1546. e3. doi: 10.1053/j.gastro.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ojesina AI, et al. Nature. 2014;506:371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nougayrède J-P, et al. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 11.Putze J, et al. Infect. Immun. 2009;77:4696–4703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arthur JC, et al. Nat. Commun. 2014;5:4724. doi: 10.1038/ncomms5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arthur JC, et al. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boleij A, et al. Clin. Infect. Dis. 2015;60:208–215. doi: 10.1093/cid/ciu787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sears CL, et al. Clin. Infect. Dis. 2008;47:797–803. doi: 10.1086/591130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S, et al. Nat. Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dejea CM, et al. Proc. Natl. Acad. Sci. U.S.A. 2014;111:18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerra L, Guidi R, Frisan T. FEBS J. 2011;278:4577–4588. doi: 10.1111/j.1742-4658.2011.08125.x. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin AC, et al. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clevers H, Nusse R. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 21.Castellarin M, et al. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kostic AD, et al. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kostic AD, et al. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCoy AN, et al. PLOS ONE. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubinstein MR, et al. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sears CL. Clin. Microbiol. Rev. 2009;22:349–369. doi: 10.1128/CMR.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu R, et al. Oncogenesis. 2014;3:e105. doi: 10.1038/oncsis.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu R, et al. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;303:G1113–G1125. doi: 10.1152/ajpgi.00453.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dutta U, Garg PK, Kumar R, Tandon RK. Am. J. Gastroenterol. 2000;95:784–787. doi: 10.1111/j.1572-0241.2000.01860.x. [DOI] [PubMed] [Google Scholar]
- 30.Lazcano-Ponce EC, et al. CA Cancer J. Clin. 2001;51:349–364. doi: 10.3322/canjclin.51.6.349. [DOI] [PubMed] [Google Scholar]
- 31.Wistuba II, Gazdar AF. Nat. Rev. Cancer. 2004;4:695–706. doi: 10.1038/nrc1429. [DOI] [PubMed] [Google Scholar]
- 32.Hooper LV, Littman DR, Macpherson AJ. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DiDonato JA, Mercurio F, Karin M. Immunol. Rev. 2012;246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang G, Chen R, Rudney JD. J. Periodontal Res. 2011;46:558–567. doi: 10.1111/j.1600-0765.2011.01373.x. [DOI] [PubMed] [Google Scholar]
- 35.Milward MR, et al. Clin. Exp. Immunol. 2007;148:307–324. doi: 10.1111/j.1365-2249.2007.03342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen-Vercoe E, Strauss J, Chadee K. Gut Microbes. 2011;2:294–298. doi: 10.4161/gmic.2.5.18603. [DOI] [PubMed] [Google Scholar]
- 37.Park S-R, et al. Infect. Immun. 2014;82:1914–1920. doi: 10.1128/IAI.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen IC, et al. J. Exp. Med. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen GY, Liu M, Wang F, Bertin J, Núñez G. J. Immunol. 2011;186:7187–7194. doi: 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu B, et al. Proc. Natl. Acad. Sci. U.S.A. 2013;110:9862–9867. doi: 10.1073/pnas.1307575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu B, et al. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21635–21640. doi: 10.1073/pnas.1016814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaki MH, Vogel P, Body-Malapel M, Lamkanfi M, Kanneganti T-D. J. Immunol. 2010;185:4912–4920. doi: 10.4049/jimmunol.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaki MH, et al. Cancer Cell. 2011;20:649–660. doi: 10.1016/j.ccr.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Couturier-Maillard A, et al. J. Clin. Invest. 2013;123:700–711. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grivennikov SI, et al. Nature. 2012;491:254–258. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grivennikov SI, Karin M. Ann. Rheum. Dis. 2011;70(suppl. 1):i104–i108. doi: 10.1136/ard.2010.140145. [DOI] [PubMed] [Google Scholar]
- 47.Grivennikov SI. Cancer Cell. 2013;24:145–147. doi: 10.1016/j.ccr.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu H, Pardoll D, Jove R. Nat. Rev. Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li N, Grivennikov SI, Karin M. Cancer Cell. 2011;19:429–431. doi: 10.1016/j.ccr.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warren RL, et al. Microbiome. 2013;1:16. doi: 10.1186/2049-2618-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zackular JP, et al. MBio. 2013;4:e00692–13. doi: 10.1128/mBio.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baxter NT, Zackular JP, Chen GY, Schloss PD. Microbiome. 2014;2:20. doi: 10.1186/2049-2618-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faber F, Bäumler AJ. Immunol. Lett. 2014;162:48–53. doi: 10.1016/j.imlet.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rivera-Chávez F, et al. PLOS Pathog. 2013;9:e1003267. doi: 10.1371/journal.ppat.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winter SE, Bäumler AJ. Gut Microbes. 2014;5:71–73. doi: 10.4161/gmic.27129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nuccio S-P, Bäumler AJ. MBio. 2014;5:e00929–14. doi: 10.1128/mBio.00929-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winter SE, et al. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thiennimitr P, et al. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winter SE, et al. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mellman I, Coukos G, Dranoff G. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gur C, et al. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kostic AD, et al. Nat. Biotechnol. 2011;29:393–396. doi: 10.1038/nbt.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Riley DR, et al. PLOS Comput. Biol. 2013;9:e1003107. doi: 10.1371/journal.pcbi.1003107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goodman AL, et al. Proc. Natl. Acad. Sci. U.S.A. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vanwonterghem I, Jensen PD, Ho DP, Batstone DJ, Tyson GW. Curr. Opin. Biotechnol. 2014;27:55–64. doi: 10.1016/j.copbio.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 66.Sizova MV, et al. Appl. Environ. Microbiol. 2012;78:194–203. doi: 10.1128/AEM.06813-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wright CJ, et al. Mol. Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Curtis MA, Zenobia C, Darveau RP. Cell Host Microbe. 2011;10:302–306. doi: 10.1016/j.chom.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boyle KE, Heilmann S, van Ditmarsch D, Xavier JB. Curr. Opin. Microbiol. 2013;16:207–212. doi: 10.1016/j.mib.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Medzhitov R, Schneider DS, Soares MP. Science. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Warburg O. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 72.Vander Heiden MG. Nat. Rev. Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 73.Bultman SJ, Jobin C. Cell Host Microbe. 2014;16:143–145. doi: 10.1016/j.chom.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maslowski KM, et al. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith PM, et al. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arpaia N, et al. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Furusawa Y, et al. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 78.Singh N, et al. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bindels LB, et al. Br. J. Cancer. 2012;107:1337–1344. doi: 10.1038/bjc.2012.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tang Y, Chen Y, Jiang H, Robbins GT, Nie D. Int. J. Cancer. 2011;128:847–856. doi: 10.1002/ijc.25638. [DOI] [PubMed] [Google Scholar]
- 81.Donohoe DR, et al. Cancer Discov. 2014 doi: 10.1158/2159-8290.CD-14-0501. [DOI] [Google Scholar]
- 82.Belcheva A, et al. Cell. 2014;158:288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 83.Song M, Garrett WS, Chan AT. Gastroenterology. 2015 doi: 10.1053/j.gastro.2014.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.O’Rourke RW. Surg. Obes. Relat. Dis. 2014;10:1208–1219. doi: 10.1016/j.soard.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Park J, Morley TS, Kim M, Clegg DJ, Scherer PE. Nat. Rev. Endocrinol. 2014;10:455–465. doi: 10.1038/nrendo.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Berger NA. Ann. N.Y. Acad. Sci. 2014;1311:57–76. doi: 10.1111/nyas.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gregor MF, Hotamisligil GS. Annu. Rev. Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 88.Ley RE. Curr. Opin. Gastroenterol. 2010;26:5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- 89.Turnbaugh PJ, et al. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schulz MD, et al. Nature. 2014;514:508–512. doi: 10.1038/nature13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoshimoto S, et al. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 92.Patterson AD, Turnbaugh PJ. Cell Metab. 2014;20:761–768. doi: 10.1016/j.cmet.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Carmody RN, Turnbaugh PJ. J. Clin. Invest. 2014;124:4173–4181. doi: 10.1172/JCI72335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roberts AB, Wallace BD, Venkatesh MK, Mani S, Redinbo MR. Mol. Pharmacol. 2013;84:208–217. doi: 10.1124/mol.113.085852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wallace BD, et al. Science. 2010;330:831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iida N, et al. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Viaud S, et al. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Couzin-Frankel J. Science. 2013;342:1432–1433. doi: 10.1126/science.342.6165.1432. [DOI] [PubMed] [Google Scholar]
- 99.Gedye C, van der Westhuizen A, John T. Intern. Med. J. 2014 doi: 10.1111/imj.12653. [DOI] [PubMed] [Google Scholar]
- 100.Taur Y, et al. Blood. 2014;124:1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ubeda C, et al. J. Clin. Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ubeda C, et al. Infect. Immun. 2013;81:965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Taur Y, et al. Clin. Infect. Dis. 2012;55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Buffie CG, et al. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bhatt AS, et al. N. Engl. J. Med. 2013;369:517–528. doi: 10.1056/NEJMoa1211115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jenq RR, et al. J. Exp. Med. 2012;209:903–911. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ingersoll MA, Albert ML. Mucosal Immunol. 2013;6:1041–1053. doi: 10.1038/mi.2013.72. [DOI] [PubMed] [Google Scholar]
- 108.Redelman-Sidi G, Glickman MS, Bochner BH. Nat. Rev. Urol. 2014;11:153–162. doi: 10.1038/nrurol.2014.15. [DOI] [PubMed] [Google Scholar]
- 109.Pan ZK, Ikonomidis G, Lazenby A, Pardoll D, Paterson Y. Nat. Med. 1995;1:471–477. doi: 10.1038/nm0595-471. [DOI] [PubMed] [Google Scholar]
- 110.Nair N, Kasai T, Seno M. Anticancer Res. 2014;34:6289–6296. [PubMed] [Google Scholar]
- 111.Ho BT, Dong TG, Mekalanos JJ. Cell Host Microbe. 2014;15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Galán JE, Wolf-Watz H. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 113.Roberts NJ, et al. Sci. Transl. Med. 2014;6:249ra111. doi: 10.1126/scitranslmed.3008982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dang LH, Bettegowda C, Huso DL, Kinzler KW, Vogelstein B. Proc. Natl. Acad. Sci. U.S.A. 2001;98:15155–15160. doi: 10.1073/pnas.251543698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ruder WC, Lu T, Collins JJ. Science. 2011;333:1248–1252. doi: 10.1126/science.1206843. [DOI] [PubMed] [Google Scholar]
- 116.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. J. Mol. Biol. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 117.Xiang S, Fruehauf J, Li CJ. Nat. Biotechnol. 2006;24:697–702. doi: 10.1038/nbt1211. [DOI] [PubMed] [Google Scholar]
- 118.Zackular JP, Rogers MAM, Ruffin MT, 4th, Schloss PD. Cancer Prev. Res. 2014;7:1112–1121. doi: 10.1158/1940-6207.CAPR-14-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]