Abstract
Alpha1-antitrypsin deficiency (AATD) is a protein conformational disease with the most common cause being the Z-variant mutation in alpha1-antitrypsin (Z-AAT). The misfolded conformation triggered by the Z-variant challenges cellular proteostasis (protein folding) systems and fails to meet the ER export metrics, leading to decreased circulating AAT and deficient anti-protease activity in the plasma and lung. Here, we described the methods for measuring the secretion and neutrophil elastase (NE) inhibition activity of AAT/Z-AAT, as well as the response to histone deacetylase inhibitor (HDACi), a major proteostasis modifier that impacts the secretion and function of AATD from the liver to plasma. These methods provide a platform for further therapeutic development of proteostasis regulators for AATD.
1. Introduction
Alpha-1 antitrypsin deficiency (AATD) is a hereditary disorder due to mutations in the Alpha-1 antitrypsin (AAT) gene (1). AAT is the most abundant serine protease inhibitor (SERPIN) in the plasma that is synthesized in hepatocytes and plays a critical role in preventing the degradation of lung tissues by neutrophil elastase (NE). AAT is considered a metastable protein that is managed by a proteostasis program encompassing protein synthesis, folding, degradation, and membrane trafficking systems (2-5). This evolutionarily ancient system optimizes the ability of biology to synthesize, process and maintain the protein fold for function in response to cellular environments (6). AAT variants arising during normal evolution of the genome may exceed proteostasis capacitance and trigger disease phenotypes. The most common AAT variant causing AATD is the Z-variant (E342K). This variant has a slow folding rate and an increased tendency to form polymers through a ‘loop-sheet’ mechanism in the endoplasmic reticulum (ER) (7). Z-AAT polymer results in proteotoxic stress in the ER triggering liver diseases including neonatal hepatitis, cirrhosis, and hepatocellular carcinoma (8). On the other hand, the misfolded Z-AAT conformation decreases its trafficking efficacy along the secretory pathway and leads to a substantially reduced level of circulating AAT in the plasma required for anti-protease activity in the lung. Loss of functional AAT in the lung leads to emphysema and/or chronic obstructive pulmonary disease (COPD) (9). Thus, AATD is considered a ‘protein conformational disease’ (10, 11). Understanding and managing the balance between evolutionary diverse protein fold trajectories and the driving force of proteostasis system in managing folding with trafficking and function represents a largely untested mechanism for disease intervention through the development of proteostasis targeted therapeutics (12).
The acetylation and deacetylation status on protein lysine residue mediated by histone acetyltrasferases (HATs) and histone deacetylases (HDACs) has a major impact on the properties of both the protein fold and proteostasis system (13). It is well known that this modification on histone proteins controls the nucleosome structure and regulates gene expressions through epigenetic programs. Acetylation/deacetylation also impacts almost all proteostasis components and pathways, including the core chaperones responsible for assisting protein folding (e.g. Hsp90 (14), Hsp70 (15), Hsp40 (16), Bip (17)), the transcription factor HSF1 that is critical for stress response (18), Lys ubiquitination components (by direct competition) that are central in degradation processes (19) and cytoskeletal components responsible for membrane trafficking processes (20). The key role of HATs/HDACs in proteostasis is further supported by the beneficial effects of HDAC inhibitors (HDACi) in many protein misfolding diseases, such as cystic fibrosis (21), Gauchers disease (22), muscle atrophy (23), Niemann-Pick C (24) and neurodegenerative diseases (25).
We previously showed that the HDACi suberoylanilide hydroxamic acid (SAHA) improved the secreted functional pool of Z-AAT from less than 20% to ~50% of the level that observed for WT-AAT (26). Here, we describe the detailed methods for measuring the HDACi effect on both the trafficking efficacy and NE inhibition activity of AAT/Z-AAT. AAT has three N-linked glycosylation sites. The processing states of oligosaccharides can report the trafficking efficacy with immature, core-glycosylated isoform reflecting ER fraction and mature, complex-glycosylated isoform reflecting Golgi fraction (27). The mature isoform that is secreted from cell shows slower migration in SDS-PAGE than the immature isoform. To inhibit NE, AAT forms a covalent and irreversible complex with NE, which can be measured by SDS-PAGE as a readout for AAT function (28). Those assays can serve as a platform to test proteostasis compounds that have therapeutic potential for AATD such as HDACi described here.
2. Materials
2.1 Cellular sample preparation
Cell lines: Although AAT is mainly synthesized and secreted from hepatocytes, the large amount of endogenous WT-AAT limits the use of liver cell line to study the biology of exogenously-expressed Z-variant AAT. Hence we used a human epithelial colorectal carcinoma cell line (HCT116) with undetectable endogenous AAT to generate a stable cell line that expresses the exogenous WT-AAT or Z-AAT with FLAG and HA tags at the N-terminus, as described previously (26).
Culture medium: DMEM (Dulbecco’s Modified Eagle Medium) and Fetal Bovine Serum (FBS).
SAHA (Cayman Chemical, Ann Arbour, MI, USA) was dissolved in DMSO to prepare a stock solution of 50 mM and stored in aliquots at −20°C.
Cell culture supplements and equipment: Bio-safety hood, 70% ethanol, incubator maintained at 37°C and 5% CO2, water bath, microscope, 12-well cell culture dish, pipettes, pipetman,, 1× PBS buffer (137 mM NaCl, 2.7 mM KCl, 1.8 mM KH2PO4 and 15.2 mM Na2HPO4/7H2O). All the above need to be in aseptic environment or sterilized to avoid contamination.
Cell lysis buffer: 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% (v/v) Triton X-100 containing protease inhibitors cocktail (Roche, Mannheim, Germany) at 2 mg/ml.
Centrifuge capable of holding 1.5 ml tubes and reaching centrifugal force of 20,000× g at 4°C.
2.2 Immunoblotting AAT/Z-AAT
Bradford protein assay kit (Thermo Scientific, Rockford, IL, USA).
SDS-PAGE materials: 8% and 10% SDS-PAGE gels, 6× SDS-PAGE loading buffer (30% glycerol, 120 mM Tris pH 7.0, 6% SDS and 0.6% Bromophenol Blue) with 6% β-mercaptoethanol, heating block, electrode/running tank, 1× running buffer (25 mM Tris, 192 mM glycine and 0.1% SDS), Seeblue pre-stained protein standard (Life technologies, Grand Island, NY, USA).
Protein transfer materials: 1× transfer buffer (25 mM Tris and 192 mM glycine), nitrocellulose membrane, wet-electroblotting systems (transfer tank, cassette, blot paper, glass tube, fiber pad, ice packs, glass dish, and stir bar).
Antibodies: AAT antibody (Immunology Consultants Laboratory, Inc. Portland, OR, USA), Hsp90 antibody (Enzo Life Sciences, Plymouth Meeting, PA, USA.), Peroxidase conjugated secondary antibody (Mouse anti-Goat IgG for AAT primary antibody, and goat anti-rabbit IgG for Hsp90 antibody).
1× TBST buffer (25 mM Tris, 150 mM NaCl, 2 mM KCl, pH 7.4, 0.1% Tween-20).
Non-fat dry milk (LabScientific, Highlands, NJ, USA).
ECL solution: Solution-1 (2.5 mM luminol, 0.45 mM p-Coumaric Acid and 0.1 M Tris pH 8.8). Solution-2 (0.02% Hydrogen Peroxide and 0.1 M Tris pH 8.8).
Film, developing cassette, dark room and developer.
2.3 Detecting the complex of AAT/Z-AAT and NE
Human Neutrophil Elastase (Innovative research, MI, USA).
Phenylmethanesulfonylfluoride (PMSF) was dissolved in isopropanol to a concentration of 20 mM (20×) stock solution and stored at −20°C.
3. Methods
3.1 Cell culture, compound treatment and sample collection
Plate 10×104 WT- and Z- HCT116 cells in 12-well tissue culture dish with DMEM containing 10% FBS and let them grow to confluent state (usually 2-3 days).
When the cells are confluent, renew the culture medium (DMEM containing 10% FBS).
Add SAHA to a final concentration of 5 μM for compound treated wells and equal volume of DMSO for control wells.
Incubate the cells at 37°C for 24 hours.
At the end of the treatment, remove and discard culture medium. Wash the cells with 1× PBS twice.
Add 350 μM serum free medium (only DMEM) to collect the secreted AAT/Z-AAT.
Incubate the cells at 37°C for 2 hours.
After 2 hours collection, harvest the culture medium that contains the secreted AAT/Z-AAT. Samples can be aliquoted and stored at −80°C.
Put the culture dish on ice, and wash the dish with cold 1× PBS twice.
Add 50 μl lysis buffer to each well.
Lyse for 30 minutes on ice and rock the dish every 10 minutes.
Scrap the wells once the 30 minutes is complete and transfer the lysate to 1.5 ml centrifuge tubes.
Spin the lysate at 20,000× g for 20 minutes at 4 °C.
Collect the supernatant of the lysate to a new set of tubes. Samples can be stored at −80°C.
3.2 Measuring the secretion of AAT/Z-AAT in response to SAHA
Determine protein concentration of the supernatant of cell lysate by using Bradford protein assay kit.
Prepare loading samples by adding SDS-PAGE loading buffer and heating the sample at 95°C for 5 minutes.
Load 15 μg lysate supernatant samples to 10% SDS-PAGE gel for probing the immature and mature AAT/Z-AAT. Load 20 μl collected culture medium samples to another 10% SDS-PAGE gel for probing the secreted AAT/Z-AAT. Load the pre-stained protein standard ladder on both gels.
Run gel at 30 mA/gel and 200 voltage until the 50 kDa marker band nears the bottom (AAT is 52 kDa).
Stop the gel and perform protein transfer to nitrocellulose membrane at 500 mA of current, 100 V of voltage for 1.5 hour.
Here we use Hsp90 as loading control. The immature, mature and secreted bands of AAT/Z-AAT (52 kDa of molecular weight) are between the markers of 50 kDa and 64 kDa (Seeblue pre-stained protein standard). Cut the blot above 64 kDa to separate the intracellular AAT/Z-AAT from Hsp90 (See instructions note 6 and 7 when considering other proteins as loading control).
Block the blots in 5% milk (diluted in TBST) at room temperature for 1 hour.
Dilute the AAT antibody and Hsp90 antibody in 1% milk with 1:2000 and 1:25000 ratio respectively. Incubate the blots in primary antibody at 4°C on the rocker platform overnight.
Next day, discard the primary antibody and wash the blots with TBST buffer 10 minutes for three times.
Then incubate the blots in peroxidase -conjugated secondary antibody (1:10,000 diluted in 1% milk) for 1 hour at room temperature.
Discard the secondary antibody and wash the blots with TBST buffer 10 minutes for three times.
After the wash, dab the blots on filter paper; place the blots on plastic wrap; add the ECL mixture (mix equal volume of solution 1 and 2) 2 ml per blot and incubate for 1 minute. Then dab excess liquid off and put blots in the developing cassette with transparent sheet protector.
Film exposure and developing in the dark room.
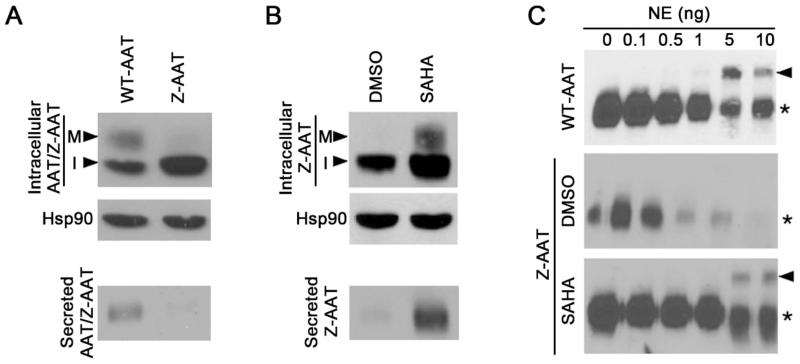
As shown in Fig.1A, the mature fraction (M) of WT-AAT migrates slower than the immature fraction (I). Compared to WT-AAT, the mature and secreted fraction of Z-AAT is significantly less, while the immature fraction is more, indicating Z-AAT is largely retained in the ER. SAHA significantly increases both the mature and secreted fraction (Figure 1B), indicating the correction effect of HDACi on the deficient secretion of Z-AAT.
Figure 1.
Western blot shows the secretion and NE inhibition activity of Z-AAT with or without SAHA treatment. (A) Secretion of WT-AAT or Z-AAT. M, mature fraction; I, immature fraction. (B) Secretion of Z-AAT under DMSO or SAHA condition. (C) NE inhibition activity of WT-AAT and Z-AAT with or without SAHA treatment. The band indicated by arrow is the AAT-NE complex. The band labeled by asterisk is the unbounded AAT. This figure is derived from reference (26).
3.3 Measuring the anti-protease of AAT/Z-AAT in response to SAHA
Prepare NE solution in PBS buffer with a serial of different concentration as 0.1, 0.5, 1, 5 and 10 ng/μl.
Add 1 μl each of the above NE solution to 20 μl of the culture medium collected in each experiment that contains secreted AAT or Z-AAT.
Tap the tubes several times to mix the solution, and then shortly spin down the mixture.
Incubate tubes at 37°C for 30 minutes to perform the binding reaction.
After the reaction, add SDS-PAGE loading buffer and heat the samples at 95°C for 5 minutes. (See Note 8 for experiment improvement).
Run the samples in 8% SDS-PAGE and follow the same western-blot procedure described above (Use AAT antibody to detect the AAT-NE complex).
As shown in Fig.1C, the covalent complex between WT-AAT and NE is observed when 5 ng of NE was added in the solution (the band indicated by arrow). No complex is observed for Z-AAT. In contrast, after SAHA treatment, we observed the complex between Z-AAT and NE, which indicates SAHA increase the anti-protease activity of secreted Z-AAT.
Acknowledgments
This work was supported by grants from National Institute of Health HL095524 for WEB. CW is supported by Postdoctoral Research fellowship from Alpha-1 Foundation.
Footnotes
For HCT116 cell maintenance, the medium (DMEM containing 10% FBS) needs to be renewed every 2 to 3 days, and a sub-cultivation ratio of 1:3 to 1:8 is recommended. The culture environment is 37°C with 95% air and 5% carbon dioxide.
A dose-dependent increase effect of SAHA was observed in our previous work (26). 0.5 μM of SAHA starts to show correction effect and 5 μM SAHA gives the maximal correction effect. Higher dose (>5 μM) is toxic to the cell.
After collecting the culture medium that contains secreted AAT/Z-AAT, it is better to add fresh medium back to the cell to prevent drying, which gives more time to prepare the next step.
The pellet of cell lysate can be kept for the analysis of aggregates of AAT/Z-AAT as described in (29).
The loading volume of collected culture medium can be normalized according to the protein concentration of cell lysate.
Actin (43 kDa for either alpha or beta subunit) and tubulin (55 kDa for either alpha or beta subunit) are not recommended for loading control since they have similar molecular weight with AAT (52 kDa) and can’t be separated from AAT in SDS-PAGE.
GAPDH (36 kDa) can be used as loading control for intracellular AAT. If GAPDH is used as loading control, make sure that (a) GAPDH doesn’t run out of the gel, and (b) Cut the membrane below the 50 kDa marker band carefully to separate AAT and GAPDH. Ponceau staining of the membrane can give a general view about how the samples run on the SDS-PAGE, which helps the proper cutting.
It is recommended to add 1mM PMSF to stop the NE binding reaction before adding SDS-PAGE loading buffer. After forming the covalent complex with NE, AAT undergoes large conformational changes and becomes disorder in some region, which makes the complex very susceptive to be digested by protease including NE. By adding PMSF immediately after the reaction, we can detect very weak Z-AAT and NE complex (data not shown).
Reference
- 1.Ghouse R, Chu A, Wang Y, Perlmutter DH. Mysteries of alpha1-antitrypsin deficiency: emerging therapeutic strategies for a challenging disease. Disease models & mechanisms. 2014;7:411–419. doi: 10.1242/dmm.014092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 3.Bouchecareilh M, Conkright JJ, Balch WE. Proteostasis strategies for restoring alpha1-antitrypsin deficiency. Proceedings of the American Thoracic Society. 2010;7:415–422. doi: 10.1513/pats.201001-016AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchecareilh M, Balch WE. Proteostasis: a new therapeutic paradigm for pulmonary disease. Proceedings of the American Thoracic Society. 2011;8:189–195. doi: 10.1513/pats.201008-055MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouchecareilh M, Balch WE. Proteostasis, an emerging therapeutic paradigm for managing inflammatory airway stress disease. Current molecular medicine. 2012;12:815–826. doi: 10.2174/156652412801318782. [DOI] [PubMed] [Google Scholar]
- 6.Powers ET, Balch WE. Diversity in the origins of proteostasis networks--a driver for protein function in evolution. Nature reviews. Molecular cell biology. 2013;14:237–248. doi: 10.1038/nrm3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gooptu B, Dickens JA, Lomas DA. The molecular and cellular pathology of alpha(1)-antitrypsin deficiency. Trends in molecular medicine. 2014;20:116–127. doi: 10.1016/j.molmed.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Teckman JH. Liver disease in alpha-1 antitrypsin deficiency: current understanding and future therapy. Copd. 2013;10(Suppl 1):35–43. doi: 10.3109/15412555.2013.765839. [DOI] [PubMed] [Google Scholar]
- 9.Brebner JA, Stockley RA. Recent advances in alpha-1-antitrypsin deficiency-related lung disease. Expert review of respiratory medicine. 2013;7:213–229. doi: 10.1586/ers.13.20. quiz 230. [DOI] [PubMed] [Google Scholar]
- 10.Kopito RR, Ron D. Conformational disease. Nature cell biology. 2000;2:E207–209. doi: 10.1038/35041139. [DOI] [PubMed] [Google Scholar]
- 11.Nyon MP, Gooptu B. Therapeutic targeting of misfolding and conformational change in alpha1-antitrypsin deficiency. Future medicinal chemistry. 2014;6:1047–1065. doi: 10.4155/fmc.14.58. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, Balch WE. Managing the adaptive proteostatic Landscape: restoring resilience in alpha-1 antitrypsin deficiency. In: Sandhaus RA, Wanner A, editors. Alpha-1 antitrypsin: role in health and disease. Springer; 2015. in press. [Google Scholar]
- 13.Hutt DM, Balch WE. Expanding proteostasis by membrane trafficking networks. Cold Spring Harbor perspectives in medicine. 2013;3:1–21. doi: 10.1101/cshperspect.a013383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Molecular cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 15.Marinova Z, Ren M, Wendland JR, Leng Y, Liang MH, Yasuda S, Leeds P, Chuang DM. Valproic acid induces functional heat-shock protein 70 via Class I histone deacetylase inhibition in cortical neurons: a potential role of Sp1 acetylation. Journal of neurochemistry. 2009;111:976–987. doi: 10.1111/j.1471-4159.2009.06385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hageman J, Rujano MA, van Waarde MA, Kakkar V, Dirks RP, Govorukhina N, Oosterveld-Hut HM, Lubsen NH, Kampinga HH. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Molecular cell. 2010;37:355–369. doi: 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Rao R, Nalluri S, Kolhe R, Yang Y, Fiskus W, Chen J, Ha K, Buckley KM, Balusu R, Coothankandaswamy V, Joshi A, Atadja P, Bhalla KN. Treatment with panobinostat induces glucose-regulated protein 78 acetylation and endoplasmic reticulum stress in breast cancer cells. Molecular cancer therapeutics. 2010;9:942–952. doi: 10.1158/1535-7163.MCT-09-0988. [DOI] [PubMed] [Google Scholar]
- 18.Westerheide SD, Anckar J, Stevens SM, Jr., Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Molecular & cellular proteomics: MCP. 2011;10:M111 013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nature reviews. Molecular cell biology. 2011;12:773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- 21.Hutt DM, Herman D, Rodrigues AP, Noel S, Pilewski JM, Matteson J, Hoch B, Kellner W, Kelly JW, Schmidt A, Thomas PJ, Matsumura Y, Skach WR, Gentzsch M, Riordan JR, Sorscher EJ, Okiyoneda T, Yates JR, 3rd, Lukacs GL, Frizzell RA, Manning G, Gottesfeld JM, Balch WE. Reduced histone deacetylase 7 activity restores function to misfolded CFTR in cystic fibrosis. Nature chemical biology. 2010;6:25–33. doi: 10.1038/nchembio.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, Yang C, Chen M, Ye DY, Lonser RR, Brady RO, Zhuang Z. Histone deacetylase inhibitors prevent the degradation and restore the activity of glucocerebrosidase in Gaucher disease. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21200–21205. doi: 10.1073/pnas.1119181109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moresi V, Williams AH, Meadows E, Flynn JM, Potthoff MJ, McAnally J, Shelton JM, Backs J, Klein WH, Richardson JA, Bassel-Duby R, Olson EN. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pipalia NH, Cosner CC, Huang A, Chatterjee A, Bourbon P, Farley N, Helquist P, Wiest O, Maxfield FR. Histone deacetylase inhibitor treatment dramatically reduces cholesterol accumulation in Niemann-Pick type C1 mutant human fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5620–5625. doi: 10.1073/pnas.1014890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coppede F. The potential of epigenetic therapies in neurodegenerative diseases. Frontiers in genetics. 2014;5:220. doi: 10.3389/fgene.2014.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouchecareilh M, Hutt DM, Szajner P, Flotte TR, Balch WE. Histone deacetylase inhibitor (HDACi) suberoylanilide hydroxamic acid (SAHA)-mediated correction of alpha1-antitrypsin deficiency. The Journal of biological chemistry. 2012;287:38265–38278. doi: 10.1074/jbc.M112.404707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hebert DN, Lamriben L, Powers ET, Kelly JW. The intrinsic and extrinsic effects of N-linked glycans on glycoproteostasis. Nature chemical biology. 2014;10:902–910. doi: 10.1038/nchembio.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gettins PG. Serpin structure, mechanism, and function. Chemical reviews. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 29.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, Michalopoulos G, Perlmutter DH. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]