Abstract
Haematopoietic stem cells (HSCs) arise in the developing aorta during embryogenesis. The number of HSC clones born has been estimated through transplantation, but experimental approaches to assess the absolute number of forming HSCs in a native setting have remained challenging. Here, we applied single-cell and clonal analysis of HSCs in zebrafish to quantify developing HSCs. Targeting creERT2 in developing cd41:eGFP+ HSCs enabled long-term assessment of their blood contribution. We also applied the Brainbow-based multicolour Zebrabow system with drl:creERT2 that is active in early haematopoiesis to induce heritable colour barcoding unique to each HSC and its progeny. Our findings reveal that approximately 21 HSC clones exist prior to HSC emergence and 30 clones are present during peak production from aortic endothelium. Our methods further reveal that stress haematopoiesis, including sublethal irradiation and transplantation, reduces clonal diversity. Our findings provide quantitative insights into the early clonal events that regulate haematopoietic development.
Blood homeostasis is maintained by proliferation and differentiation of clones of haematopoietic stem cells (HSCs). In vertebrates, clones arise in the haemogenic endothelium of the aortic floor, hereafter referred to as the ventral dorsal aorta (VDA) region, during the definitive wave of haematopoiesis1–3. From this stage on, blood production relies on the self-renewal and differentiation of existing clones rather than de novo generation of HSCs1. The number of clones generated in the VDA that sustain blood production for life remains unknown. Estimates for the number of HSCs that arise are based on genetic reporters, short-term fate mapping or transplantation4–7. Genetic reporters enable observation of putative HSCs, but cannot accurately quantify or evaluate their functional long-term output in vivo. Transplantation by limiting dilution is a standard and quantitative method for assessing HSC function and numbers, but multiple reports demonstrated altered HSC dynamics and clonal selection during transplantation, which do not reflect native haematopoiesis8–10. Alternative methods for studying endogenous HSCs may provide a more accurate view of the stem cell number.
Because their embryos are transparent and contain relatively few HSCs6,11, the zebrafish (Danio rerio) is a unique developmental system to observe native haematopoietic development in vivo. In Tg(cd41:eGFP) or Tg(Runx1+23:eGFP) zebrafish embryos, GFP+ haematopoietic stem and progenitor cells (HSPCs) appear in the VDA between 33 h post fertilization (hpf) and 54 hpf, with peak budding at 48 hpf6,12,13. Time-lapse imaging of these transgenic lines shows that, on average, about three cells per hour bud from the VDA and enter the circulation, suggesting that dozens of clones could give rise to definitive haematopoiesis5. By 80 hpf, on average five Runx1+23:eGFPhigh cells have engrafted the caudal haematopoietic tissue (CHT), whereas others remain in circulation. Transplantation by limiting dilution revealed that approximately one in three Runx1+23:eGFP+ cells have long-term repopulation potential6.
Here, we applied two independent fate mapping methods with clonal analysis in zebrafish to study contribution of embryonic HSCs to adult haematopoiesis. First, we established a single-cell gene induction method combined with a ligand-inducible creERT2 that induces permanent fluorophore expression in cd41:GFP+ cells and their progeny. Second, we employed the Zebrabow system with tissue-specific creERT2 expressed in haemogenic endothelium. These independent methods yield similar estimates for the number of HSC clones that are born in the VDA and survive to contribute to lifelong haematopoiesis. We show that the clonal contribution is altered after sublethal irradiation or transplant. Our study provides a framework for examining the clonality of blood in zebrafish and establishes that approximately 30 HSC clones arise during VDA development.
RESULTS
Laser-induced fate mapping of cd41+ cells measures the contribution of embryonic HSCs to adult blood
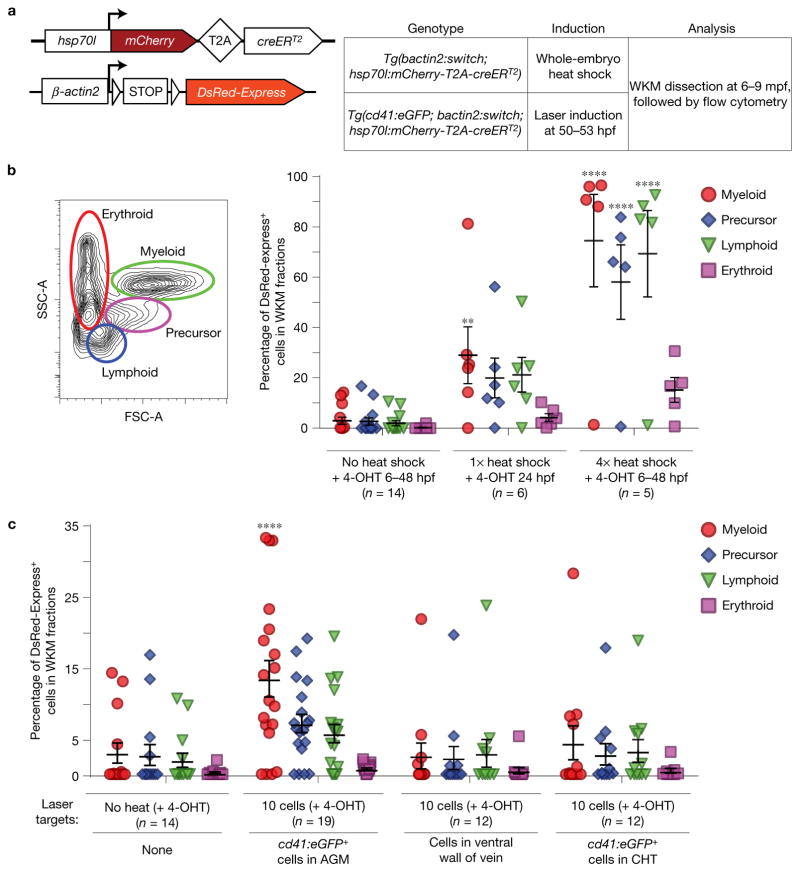
To evaluate the development of HSCs, we used the transgenic reporter line Tg(β-actin2:loxP-STOP-loxP-DsRed-Express)sd5, also called Tg(bactin2:switch)1, that drives widespread reporter expression from early development throughout adulthood (Fig. 1a). We crossed this reporter to the dual 4-hydroxytamoxifen- (4-OHT-) and heat-inducible creERT2 recombinase driver transgenic Tg(hsp70l:mCherry-T2A-creERT2)14. We heat-shocked double transgenic embryos and treated with 1 μM 4-OHT at 24 hpf, and observed high expression levels of DsRed-Express in all tissues at 5 days post fertilization (dpf) (Supplementary Fig. 1). To determine the efficiency of labelling adult haematopoietic cells, we isolated whole-kidney marrow (WKM) cells from 6–9 month old transgenic Tg(bactin2:switch; hsp70l:mCherry-T2A-creERT2) animals and analysed reporter expression by flow cytometry (Fig. 1b). WKM fractions from zebrafish that received no heat shock contained minimal numbers (<3%) of DsRed-Express+ cells. The myeloid, precursor and lymphoid fractions of zebrafish that received four heat-shock treatments (6 hpf, 24 hpf, 36 hpf and 48 hpf) contained 75% (±18.4%, s.e.m.), 58% (±14.8%), 69% (±17.2%) and 15% (±4.9%) DsRed-Express+ cells, respectively (Fig. 1b). Our data indicate that the majority of adult blood cells in the WKM are capable of being labelled with this system.
Figure 1.
Contribution of embryonic cd41:eGFP+ HSCs to adult kidney marrow. (a) Transgenes used for DsRed-Express labelling: bactin2:switch and hsp70l:mCherry-T2A-creERT2. mpf, months post-fertilization. (b) Quantification showing the percentage of DsRed-Express+ cells in each of the four WKM fractions after whole-embryo heat shock, with mean and s.e.m. (c) Quantification showing the percentage of DsRed-Express+ cells in each of the four WKM fractions after targeted laser induction, with mean and s.e.m. Each circle represents one animal. **P = 0.0067, ****P < 0.0001, with two-way analysis of variance (ANOVA).
Tg(cd41:eGFP) has been used to track the migration of HSCs from the VDA to the CHT, thymus and pronephros5,13. Tg(cd41:eGFP;bactin2:switch;hsp70l;mCherry-T2A-creERT2) embryos were treated with 4-OHT and subjected to whole-embryo heat-shock induction between 50 hpf and 53 hpf (Supplementary Fig. 2a). Confocal microscopy of primordial thymuses at 75 hpf revealed abundant double-positive cd41:eGFP+;DsRed-Express+ cells, whereas in the absence of heat only six double-positive cells per thymic lobe were detected on average (Supplementary Fig. 2d). To more specifically label individual HSPCs, we adapted an hsp70l laser induction method that was used to trace microglial precursors in zebrafish15. At 48 hpf, there are only 10–15 cd41:eGFP+ cells along the VDA1,16. When we specifically targeted ten cd41:eGFP+ cells in the VDA with laser induction, we observed approximately 13 double-positive cells per thymic lobe, a significant doubling relative to the negative control (Supplementary Fig. 2b–d). These results demonstrate that we can specifically target nascent cd41:eGFP-marked HSCs as they emerge. We then analysed their contribution to the adult marrow. Consisting mainly of neutrophils, the myeloid fraction represents the current, steady-state HSC contribution to the marrow due to its restricted lifespan of a few days17. Between 11% and 16% (13.4% ± 2.5% s.e.m.) of this fraction were DsRed-Express+ at 6–9 months post induction (Fig. 1c). Without considering the precise fate of each HSC and with an approximate targeting efficiency of 45%, we conservatively estimate that there are between 25 and 36 HSCs born from the VDA (see Methods for estimation). To assess the accuracy of laser induction, we targeted ten cd41:eGFP− cells (which we would not expect to contribute to haematopoiesis) in the neighbouring ventral wall of the cardinal vein. WKM fractions from these animals resembled those of negative controls (Fig. 1c). Altogether, our observations suggest that approximately 30 HSC clones are in the VDA around 50 hpf, but there is appreciable background in this system. Our conclusion also assumes that targeted cd41:eGFP+ cells contribute to the marrow.
Optimized Zebrabow labelling using draculin:creERT2 induces colour labelling of embryonic haematopoiesis
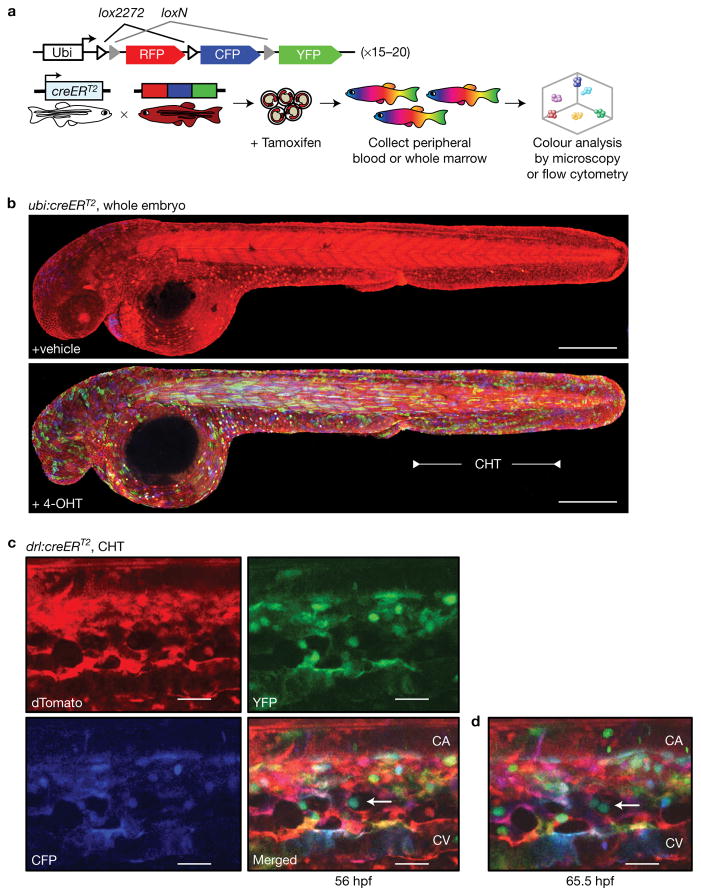
To independently confirm and corroborate our findings, we sought to genetically label individual emerging HSC clones with the Zebrabow system18–20. Three fluorescent proteins (dTomato (RFP), CFP, and YFP) are driven by a ubiquitous promoter in a single cassette and flanked by unique lox site variants (Fig. 2a). In its default state, the Zebrabow transgene expresses dTomato. Upon stochastic recombination by creERT2, either CFP or YFP is exclusively expressed. Multiple independent integrations of the Zebrabow construct in the genetic background lead to different expression levels of the fluorophores after label induction. The resulting unique colour barcodes are inherited by all descendants of the labelled cell. This system has been used for clonal tracking and fate mapping in a variety of localized tissues, including heart, brain, large intestine and cornea19,21–23. While a recent study used a similar system in tracking mouse T-cell clones24, multicolour fate mapping has not been applied to tracking of HSCs and their descendants.
Figure 2.
Zebrabow label induction with drl:creERT2 induces mosaic colour barcoding in blood and vasculature during embryogenesis. (a) The Zebrabow transgene and strategy to induce clonal labelling during early blood development. (b) Tg(Zebrabow-M;ubi:creERT2) embryos were treated with vehicle control (top) or 4-OHT (bottom) at 24 hpf and imaged by confocal microscopy at 48 hpf to measure mosaic label induction (the blue heart of the vehicle control is eGFP signal from a heart-specific cmlc2:eGFP marker on the ubi:creERT2 construct; scale bar, 300 μm). (c) Tg(Zebrabow-M;drl:creERT2) embryos were treated at 24 hpf and the CHT region was imaged by confocal microscopy at 48 hpf. Merging images of the three fluorescent proteins gives the blood and vasculature a variety of hues (CA, caudal artery; CV, caudal vein; scale bar, 25 μm). (d) Time-lapse confocal microscopy of these embryos reveals division of putative stem cells (white arrow) in perivascular niches that yield progeny with identical colours. (scale bar, 25 μm)
For clonal labelling of HSCs, we crossed Tg(Zebrabow-M) transgen-ics (~20 transgene insertions) to Tg(ubi:creERT2)25 and Tg(draculin (drl):creERT2)26. The drl regulatory elements are active during early development in all lineages derived from the lateral plate mesoderm and subsequently refine to cardiovascular lineages including HSCs, thus covering all haematopoietic cells (Supplementary Video 1). To extend our previous genetic lineage tracing26 and to demonstrate that the drl promoter is active in HSPCs, we crossed Tg(drl:eGFP) with Tg(Runx1+23:mCherry)6 reporter zebrafish that mark HSPCs and analysed co-expression by time-lapse confocal microscopy of the VDA and CHT (Supplementary Fig. 3a–c). By flow cytometry, we found that the drl promoter is active in more than 95% of Runx1+23:mCherry+ cells throughout development (Supplementary Fig. 3d–e). Transplantation of sorted drl:eGFP+ubi:mCherry+ cells into irradiated recipients demonstrated multilineage engraftment after 3 months, corroborating the view that drl reporters are active in functional HSCs (Supplementary Fig. 3f–h). Tg(Zebrabow-M;ubi:creERT2) and Tg(Zebrabow-M;drl:creERT2) embryos were treated with 10–15 μM 4-OHT at 24 hpf, immediately prior to HSC emergence. Confocal microscopy of whole embryos and the CHT showed stochastic colour expression in multiple cell types at 48 hpf (Fig. 2b,c). We used time-lapse imaging to identify putative stem cells, characterized by persistence in perivascular niches and a slow division rate (Fig. 2c and Supplementary Video 2)6. Observation of these cells revealed that they give rise to progeny of identical colour (Fig. 2d).
Zebrabow labelling in adults marks multilineage, polyclonal HSPCs
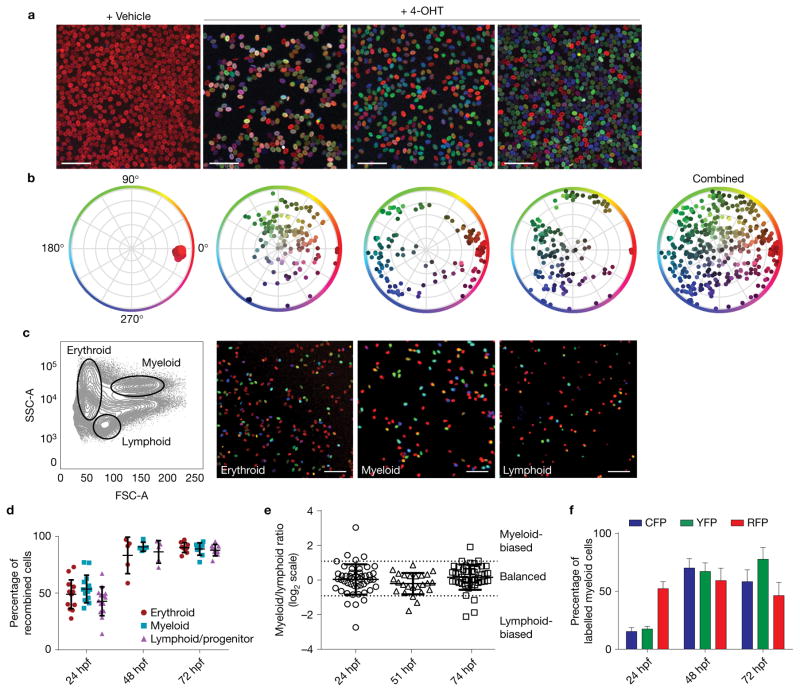
To evaluate HSC clonality, we raised 4-OHT-treated Tg(Zebrabow-M;drl:creERT2) embryos for 2.5–3 months before analysing their mature blood. This time frame allows for growth of the kidney, and the long chase period ensures that all colour recombination in the mature blood pool is HSC derived. Each treated zebrafish displayed a unique combination of hues in the peripheral blood, representing a polyclonal state (Fig. 3a,b). Control zebrafish treated with vehicle or lacking the creERT2 transgene showed no detectable recombination (Fig. 3a,b, n> 75 control animals tested). To demonstrate labelling of multipotent HSCs, we sorted blood lineages by forward and side scatter (FSC and SSC) characteristics from WKM of 4-OHT-treated zebrafish and imaged cells at 3 months post induction; we found that all lineages contained labelled cells (Fig. 3c). Percentages of recombined cells, measured by flow cytometry, were similar across multiple lineages, which is consistent with labelling of multipotent HSCs (Fig. 3d and Supplementary Fig. 4a–c). Analysis of myeloid and lymphoid lineages revealed similar colour clusters in each population (Supplementary Fig. 4d). We quantified the lineage output of HSC clones from adults by measuring the ratio of myeloid to lymphoid contribution from each coloured clone (Fig. 3e). Our data show that the majority of HSC clones (n = 104/119) from 24 hpf to 74 hpf have a balanced lineage output (myeloid/lymphoid ratio between 0.5 and 2). A few colour clones (n= 15/119) seemed to have biased output (myeloid/lymphoid ratio below 0.5 or above 2), but these were rare and generally small clones. We found that labelled cells constituted more than 95% of the marrow, indicating the entire haematopoietic system could be fate mapped (Fig. 3d). To rule out colour bias in the system, we quantified the percentage of recombination for each fluorophore in mature granulocytes. We found no significant difference between percentages of cells expressing CFP or YFP, suggesting that neither recombination event is preferred or provides a competitive advantage to cells (Fig. 3f). These data show that early embryonic label induction can mark endogenous, multipotent HSCs with heritable colour barcodes.
Figure 3.
Label induction in Zebrabow embryos leads to multilineage, polyclonal colour labelling in adult blood and marrow. (a) Tg(ubi:Zebrabow-M;drl:creERT2) embryos were treated with 4-OHT during development and grown to adulthood. Peripheral blood smears were imaged by confocal microscopy and show multicoloured blood in treated fish (scale bar, 50 μm). (b) Multiple, unique colours are evident by measurement of hue and saturation. (c) Mature blood lineages were sorted by fluorescence-activated cell sorting (FACS) and imaged, demonstrating labelling of multipotent HSCs (scale bar, 50 μm). (d) Quantification of labelling efficiency in the mature blood populations at multiple treatment times, measured by flow cytometry (n = 14, 5 and 9 fish for 24, 48 and 72 hpf, respectively). (e) Quantification of lineage contribution at multiple treatment times by flow cytometry. Each marker corresponds to an individual colour clone with the ratio of its myeloid/lymphoid contribution on a log2 scale. Thresholds for lineage bias are defined as ratios below 0.5 or above 2 (mean with s.d., n = 48, 25 and 46 clones for 24, 51 and 74 hpf, respectively). (f) Quantification of fluorophore-positive cells in multiple fish at multiple treatment times. Comparison of CFP+ and YFP+ percentages by two-tailed t-test showed no statistically significant differences at any time (P = 0.594, 0.792 and 0.192 and n = 12, 5 and 10 fish for 24, 48 and 72 hpf, respectively). Error bars show mean and s.e.m. (d,f).
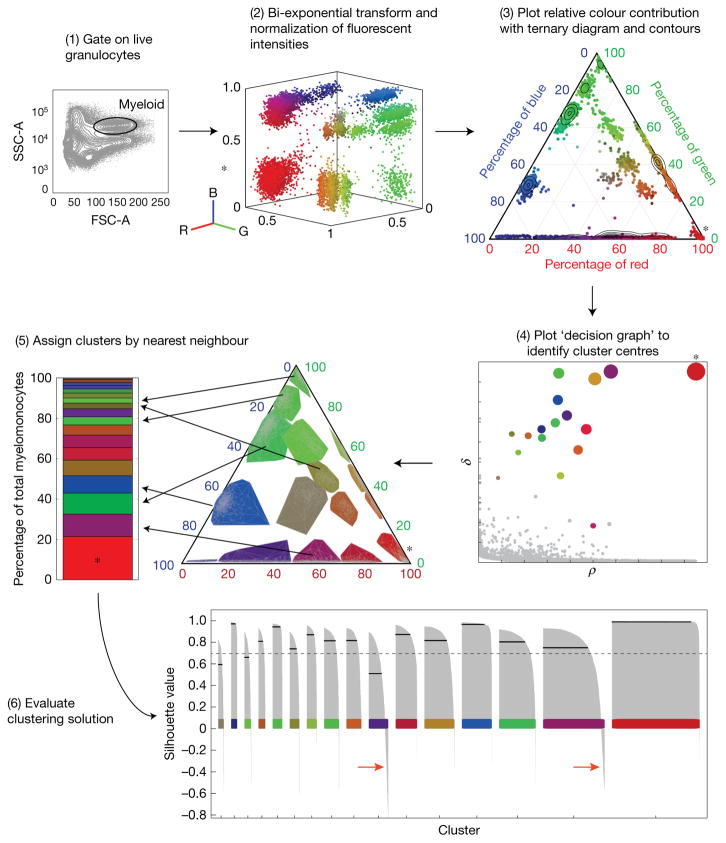
To quantify colour barcodes, we developed a custom MATLAB graphical user interface to cluster flow cytometry data (Fig. 4). With flow cytometry, clusters that contributed less than 0.5% of the granulocyte population could be detected, demonstrating a low threshold of detection in this system (Supplementary Fig. 5). To evaluate the potential total number of colours in the Zebrabow system, we carried recombination through the germline (Supplementary Fig. 6a). Tg(Zebrabow-M;ubi:creERT2) embryos were treated at 50% epiboly with 4-OHT to induce mosaic recombination in germ cells. Embryos generated by in-crossing of these parents were grown to adulthood. Flow cytometric analysis of WKM showed single-hue blood: a result of the combination of random recombination events in the parental transgenes (Supplementary Fig. 6b,c). By mapping these single colours into the total available colour space, we estimated that the Zebrabow system is capable of producing about 40 (25–59, 95% CI, n= 13) distinguishable colours in WKM, which is highly dependent on transgene copy number (Supplementary Fig. 6d–f).
Figure 4.
Quantification of colour barcodes in mature granulocytes from Zebrabow-labelled marrow. The intensity levels of the three fluorescent Zebrabow-derived proteins can be analysed by flow cytometry. (1) Myeloid cells are isolated by FSC and SSC characteristics. (2) Their intensity values are transformed and normalized. R, red; B, blue; G, green. (3) Ternary plots show the relative contribution of each fluorescent protein to the colour of the cell. (4) Colour barcodes are determined by supervised, density-based clustering to identify cluster centres. ρ, local density; δ, distance from points of higher density. (5) Clusters can be pseudo-coloured by mean colour of the cluster, and bar graphs show quantification of the clonal contribution to the entirety of the myeloid population. (6) The quality of clustering is evaluated by calculating a silhouette value (s-value) for each cell with a given clustering solution. s-values are plotted for each cell belonging to each colour cluster (grey bars), where the total average (solid black line) of s-values for a given cluster measures the quality of that cluster (dashed black line, threshold for high quality clusters). Cells with s-values below zero most probably do not belong to that cluster (red arrows). The asterisk indicates polyclonal RFP+-only cells derived from unrecombined clones.
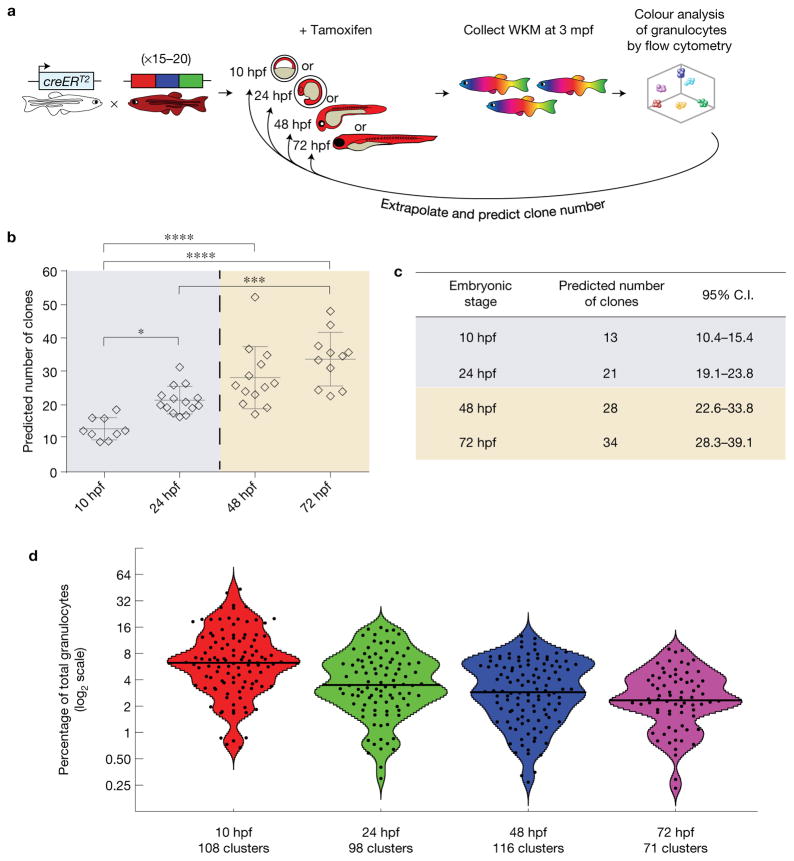
Fate mapping with clonal analysis at distinct developmental times defines the number of nascent clones
We hypothesized that early induction of clonal Zebrabow labelling in HSCs with long-term fate mapping, followed by quantification of colour barcodes in mature granulocytes, would enable us to map the number of functional HSC clones born at the time of label induction. Tg(Zebrabow-M;drl:creERT2) embryos were treated with 15 μM 4-OHT overnight (16–24 h) at distinct developmental times. We grew embryos to adulthood (2.5–3 months) and analysed their WKM by flow cytometry (Fig. 5a and Supplementary Fig. 7a–c). Quantification of colour barcodes in mature, short-lived granulocytes enabled us to estimate the number of contributing clones that existed during the time of label induction (Fig. 5b,c, Methods). This analysis predicted that, at the beginning of somitogenesis, around 13 clones (10.4–15.4, 95% CI, n = 9) are responsible for generating the haemogenic endothelium. A few hours before definitive haematopoiesis begins (24 hpf), on average 21 (19.1–23.8, 95% CI, n= 14) HSC clones initiate the entire adult haematopoietic system. At 48 hpf, we found that on average 28 (22.6–33.8, 95% CI, n= 13) HSC clones were present, which is in close agreement with findings from our single-cell laser induction approach, yet is substantially higher than the number of HSCs observed through reporter gene expression in zebrafish5,6. To verify that overnight treatment with 4-OHT does not over- or underestimate the number of HSC clones, we repeated treatment of Tg(Zebrabow-M;drl:creERT2) embryos at defined 4 h intervals (Supplementary Fig. 8a). Analysis of mature granulocytes in adult marrow of these fish predicted 20 clones (16.1–23.4, 95% CI, n = 8) present at 24–28 hpf and 26 clones (22.9–29.5, n = 5) at 51–55 hpf, which agrees with our estimates from overnight treatment (Supplementary Fig. 8b,c). Plotting the contribution of all clones revealed non-normal distributions of clonal output (Fig. 5d). Our data provide a quantitative assessment of endogenous HSC clone number in the developing aorta.
Figure 5.
Clonal analysis with fate mapping predicts the number of stem cells born during development. (a) Tg(ubi:Zebrabow-M;drl:creERT2) embryos were treated at distinct developmental times and grown to adulthood. (b) Colour barcodes in granulocytes were quantified and extrapolated to predict the number of clones present during label induction that contribute to adult blood (the dashed line marks the beginning of definitive haematopoiesis; n = 9, 14, 13 and 11 fish for 10, 24, 48 and 72 hpf, respectively). Each marker represents one animal. Error bars show mean and s.e.m. (*P = 0.026, ***P = 0.0003, ****P < 0.0001, one-way ANOVA). (c) Table showing quantification of these predicted values with 95% confidence intervals (C.I.). (d) Violin plots showing the distribution of all colour clones from all analysed zebrafish. The x axis shows the treatment time, while the y axis shows the log2 percentage contribution of each clone (black dots) to the total granulocyte pool. At later treatment times, the mean clonal contribution (black line) decreases as the number of clones increases (n = 108, 98, 116 and 71 colour clusters for 10, 24, 48 and 72 hpf, respectively). Cluster data are available in Supplementary Table 1.
Longitudinal analysis of the erythroid population reveals little HSPC turnover
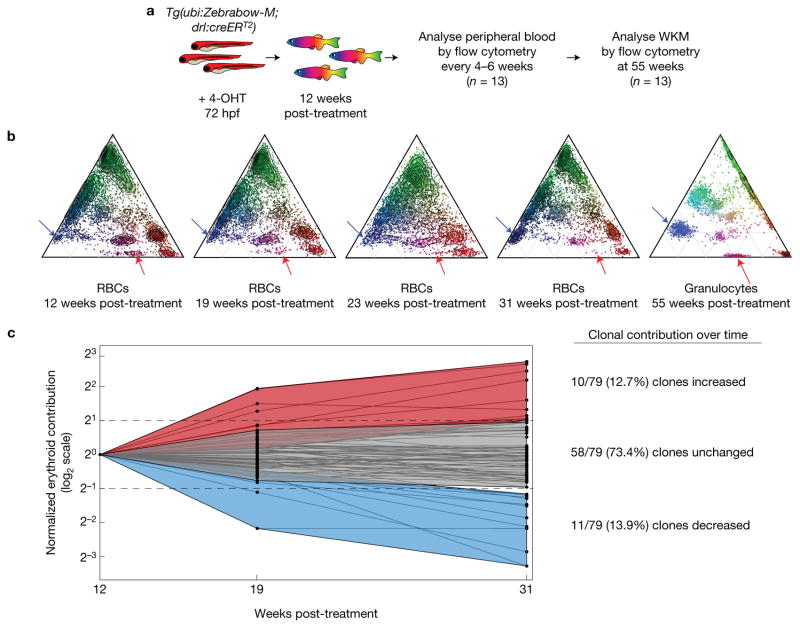
After defining the number of nascent HSPCs in development, we investigated clonal dynamics in the adult. We treated Tg(Zebrabow-M;drl:creERT2) embryos with 4-OHT at 72 hpf and grew the animals to adulthood. Every 4–6 weeks for 5 months, we obtained peripheral blood samples by retro-orbital bleeding (Fig. 6a, n = 13), and collected a terminal marrow sample at 11 months. We found that Zebrabow transgene expression remained stable in the erythrocyte population for at least 5 months (Fig. 6b). We quantified the contribution of 79 total clones from 13 zebrafish at three times (Fig. 6c). We did not observe any evidence of new colour barcodes appearing over time. The majority (73.4%) of clones maintained a stable contribution to the peripheral blood after 19 weeks. These findings are consistent with reports that clonal dynamics and competition occur at the progenitor level8,27.
Figure 6.
Longitudinal dynamics of labelled erythrocytes. (a) Strategy to evaluate clonal dynamics in the erythroid lineage. (b) Example ternary diagrams that show colour barcode distribution measured by flow cytometry from a single fish over time. The last ternary plot shows colours in granulocytes at a final time (red arrows, clone with decreased contribution over time in erythrocytes (RBCs); blue arrows, clone with increased contribution). (c) Quantification of individual clonal contribution over three analysis times. The contributions of individual colour clones (black dots) were compared across multiple times (log2 scale). Data were normalized to the contribution at the first time. The majority of clones maintain a stable contribution to the blood pool, while a few clones increase (>2-fold) or decrease (>2-fold) in contribution over time relative to the initial time.
Stress haematopoiesis leads to clonal selection
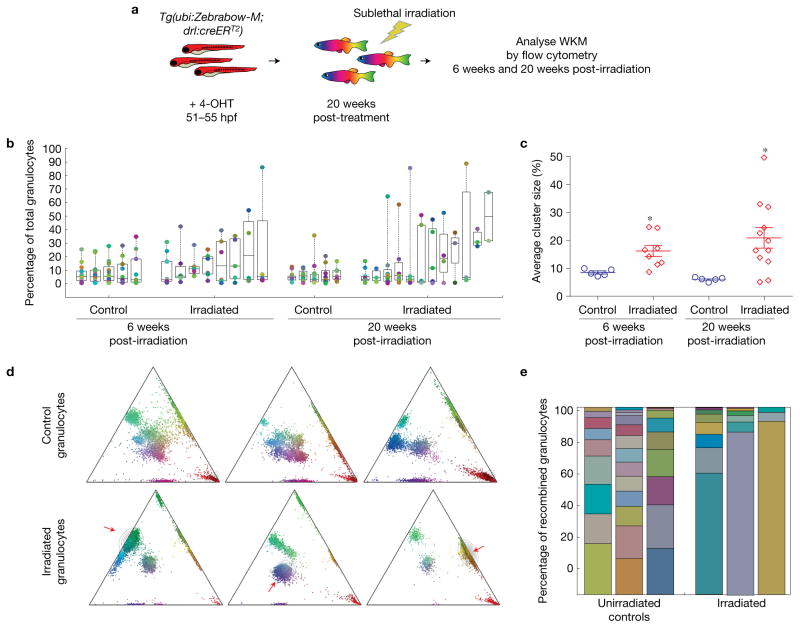
To investigate effects of perturbations on the clonality of haematopoiesis, we sublethally irradiated 5 month old Tg(Zebrabow-M;drl:creERT2) fish that had been treated with 4-OHT at 51 hpf and analysed their marrow 6 weeks and 20 weeks post irradiation (Fig. 7a). In non-irradiated fish, we found a large number of granulocyte clones (8–15 clusters), with a large distribution of colour barcode and clonal contribution (0.25–35.7% contribution per cluster). In contrast, sublethally irradiated marrow displayed fewer colour barcodes in the granulocyte population (2–10 clusters, Fig. 7b). These clusters constituted a higher percentage of the myeloid population (0.3–88.9%), suggesting that some clones expanded in the marrow due to stress induced by irradiation (6 weeks post-treatment (wpt), average clonal contribution 8.5% control versus 16.2% irradiated, P = 0.017; 20 wpt, 6.1% control versus 21% irradiated, P = 0.021). In the total pool of irradiated zebrafish, we found three examples where a single cluster constituted more than 80% of the myeloid population, indicating a severe decrease in clonal diversity (Fig. 7d,e). Our findings demonstrate that sublethal irradiation leads to increased selection amongst clones.
Figure 7.
Sublethal irradiation of Zebrabow-labelled marrow. (a) Strategy to measure the effects of sublethal irradiation on clonal diversity in Zebrabow-labelled marrow. (b) Quantification of colour barcodes in mature granulocytes as a percentage of total cells (log2 scale) in control or irradiated animals. Each column of circles corresponds to an individual animal. Box plots show the median and interquartile range (n = 6 and 8 fish for control and irradiated conditions at 6 weeks post irradiation, n = 5 and 12 fish for control and irradiated conditions at 20 weeks post irradiation). (c) Summary of sublethal irradiation data, showing mean cluster sizes in control and irradiated animals (error bars, s.e.m.). Each marker corresponds to an individual animal (n values are identical to those in (b), *P < 0.05, P = 0.017 at 6 weeks, P = 0.021 at 20 weeks, two-tailed t-test). (d) Example ternary plots from control and irradiated adult zebrafish demonstrating severe reduction of clonal diversity. The red arrows indicate an expanded clone. (e) Stacked bar graphs showing the clonal distribution of control and irradiated adult zebrafish.
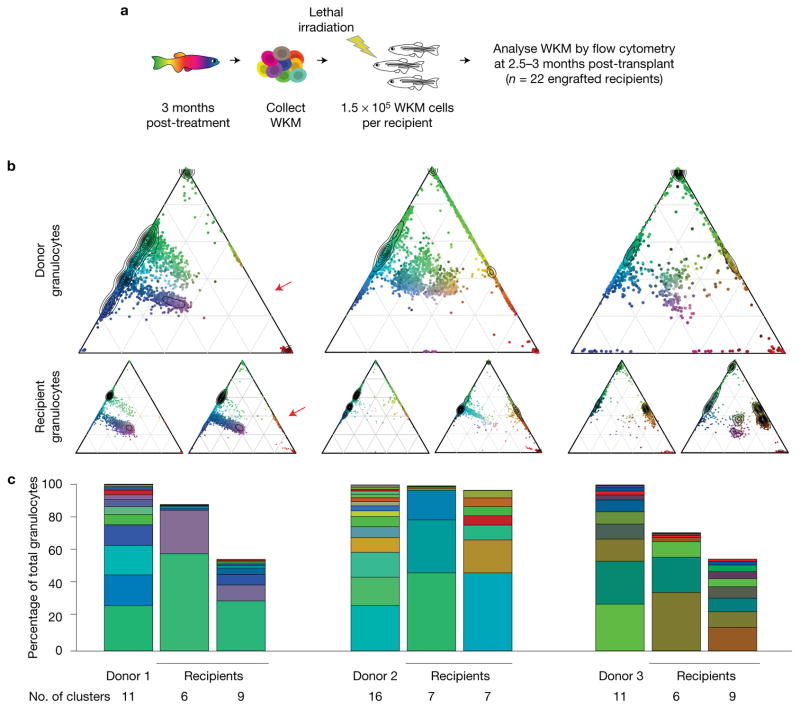
We transplanted Zebrabow-labelled WKM into irradiated recipients to investigate differences between native and transplant haematopoiesis (Fig. 8a). Tg(Zebrabow-M;drl:creERT2) donors treated at 48 hpf or 72 hpf with 4-OHT were grown to adulthood. At 3–6 months, marrow was isolated from these donors and transplanted into lethally irradiated recipients. At 2.5–3 months post transplant, recipient marrow analysed by flow cytometry revealed long-term, multilineage reconstitution (Fig. 8b), confirming that barcoded cells were derived from HSCs. Engraftment was generally polyclonal, but clonal diversity was reduced in the majority of recipients when compared with donors, suggesting that clonal selection occurs (Fig. 8b). There was only one example where a new colour barcode emerged in recipients, which possibly indicates activation of quiescent HSCs following transplantation (Fig. 8b, red arrow). Clonal contribution did not correlate with contribution in the donors, suggesting that clonal dynamics were altered upon transplant. Analysis of multiple recipients that received cells from the same marrow revealed that the clonal contribution is not consistent across different recipients (Fig. 8c). These results suggest that clonal diversity is reduced during stress haematopoiesis, and our data suggest that clonal engraftment after transplantation does not correlate with prior contribution to the mature blood pool.
Figure 8.
Transplant of Zebrabow-labelled marrow into lethally irradiated recipients. (a) Transplantation strategy for labelled marrow. (b) Example ternary diagrams showing colour distribution in granulocytes for three donors with two recipients per donor (red arrow, new colour barcode in recipient). (c) Bar graphs show colour barcode distributions in granulocytes of donors and recipients depicted in b. Each column corresponds to a single animal.
DISCUSSION
Our study establishes two independent fate mapping methods to quantify emergence of HSCs during development. Both methods reveal similar estimates for the number of nascent HSCs born during embryogenesis. During development of the dorsal aorta, we find that approximately 30 clones are responsible for generating the entire haematopoietic system, which will sustain the animal through its life. We and others have previously reported that the CHT, equivalent to the mammalian fetal liver, contains fewer than ten Runx1+23:eGFP+ or cd41:eGFPlow HSPCs after 50 hpf5,6. The discrepancy between these numbers and our fate mapping prediction could be explained by transgene variegation of previously used transgenic insertions, stages of stem cell maturation over multiple days, or presence of circulating HSCs that are challenging to quantify.
Our estimates are similar to quantification of embryonic HSC pools in both human28 and mouse7,29 systems. Analysis of X-chromosome inactivation in human samples estimates that approximately 16 clones are responsible for initiating the haematopoietic system28. Limiting dilution analysis in mouse embryonic tissues revealed that approximately three transplantable HSCs exist at the time of aorta–gonad–mesonephros (AGM) budding (E10-E11), but this number expands to approximately 66 definitive HSCs at E127. Since this large expansion cannot be explained by cell division alone, it is likely that many more than three pre-definitive HSCs are maturing at this stage7. A major advantage of long-term fate mapping with genetic means to evaluate HSC output is the ability to capture such immature HSCs. Our studies predict that the relative number of HSCs in the zebrafish AGM equivalent (~30) would be in the same range as in the mouse. It is thus likely that transplant efficiencies of mouse AGM cells into adult hosts greatly underestimate the actual frequency of murine HSCs29.
Laser-induced fate mapping of cd41:eGFP+ HSCs or Zebrabow labelling using the drl regulatory elements labels the vast majority of adult marrow and in many instances more than 95% of mature blood cells. When we observed longitudinal dynamics of erythropoiesis with Zebrabow, there was no evidence of new colour barcodes arising up to 8 months post label induction. Granulocytes displayed similar colour barcodes one year after label induction. These data suggest that the majority of definitive HSCs are active during embryogenesis but show limited dynamics in adulthood. This conclusion is consistent with recent fate mapping efforts in mouse, which demonstrated that the bulk of HSCs are highly active in producing progenitor and mature compartments during embryogenesis27. Labelling of HSCs during embryogenesis with an HSC-specific Tie2:iCre reached equilibrium with label in the peripheral system within one week27. In contrast, induction of labelling in adult mouse revealed that the HSC contribution is not observed until 16–20 weeks later. Another report using inducible transposon barcodes in mouse haematopoiesis also found that blood production in the adult is mainly driven by successive waves of progenitor populations as opposed to contribution from HSCs8. Our findings in light of these data suggest that HSCs are most productive during early clonal events in the embryo.
Pertaining to stress haematopoiesis, sublethal irradiation and transplantation influences clonal diversity of the marrow. Sublethal irradiation led to increased production of mature blood from fewer clones. One possible mechanism is that irradiation eliminates some HSPC clones, and their empty niches are taken over by descendants of surviving HSCs. Transplantation of labelled marrow also reduced clonality in recipients, which may be selective or induced by environmental triggers. We found no observable correlation between clonal distribution in donor and recipient marrows. This suggests that clonal activity in the donor does not reflect or determine the dynamics of marrow reconstitution. The reduction in clonality is consistent with other reports comparing barcoded native and transplant haematopoiesis8,30. Our data combined with these studies demonstrate that repopulation dynamics of HSC clones after transplant are highly unpredictable.
The fate mapping systems introduced in our study will be useful to evaluate the genetics of clonal diversity in both native and malignant settings. The production of blood by one or a few stem cell clones with somatic mutations renders patients susceptible to multiple haematopoietic malignancies31,32. Our fate mapping systems combined with mosaic gene knockout technologies33 in zebrafish could be instrumental to study the genetics of clonality.
METHODS
Methods, including statements of data availability and any associated accession codes and references, are available in the online version of this paper.
Note: Supplementary Information is available in the online version of the paper
METHODS
Zebrafish stocks and embryos
Zebrafish are bred and maintained as described34 and in accordance with University of California, San Diego and Boston Children’s Hospital Institutional Animal Care and Use Committee protocols, and according to Animal Resources at Children’s Hospital (ARCH) guidelines. Tg(cd41:eGFP)35, Tg(bactin2:loxP-STOP-loxP-DsRed-Express)sd51, Tg(hsp70l:mCherry-T2A-creERT2)14, Tg(ubi:Zebrabow-M)19, Tg(ubi:mCherry)25, Tg(ubi:creERT2)25, Tg(drl:eGFP)26, Tg(drl:creERT2)26 and Tg(Runx1+23:mCherry)6 have been previously described. Embryos are collected and kept in embryo buffer (5 mM NaCl, 0.17 mM KCl, 0.25 mM CaCl2 and 0.15 mM MgSO4). At 24 hpf, embryos are dechorionated with 50 mg ml−1 pronase (Roche) and then placed in embryo buffer supplemented with 0.003% 1-phenyl-2-thiourea (Sigma) to prevent pigmentation for optimal visualization (for imaging studies). Embryos were grown at 28 °C in an incubator. Both sexes were used for the studies, ranging in age from early embryo to 1.5 years old.
Whole-embryo heat shock and tamoxifen treatments
Embryos are transferred to a 50 ml conical tube, and the buffer is replaced with embryo buffer that has been heated to 38 °C. The tube containing the embryos is immediately placed in a 38 °C water bath for the duration of the heat shock. Upon completion of heat shock, embryos are transferred to a 10 cm petri dish, buffer is replaced with embryo buffer containing 1 μM 4-OHT (Sigma H7904)25, and embryos are kept in a 28 °C incubator. After 12 h of incubation with 4-OHT, embryos are placed in fresh embryo buffer. For adult blood analysis, embryos are transferred to a 28 °C aquatic housing system (Aquaneering) at 6 dpf. Embryos lacking both heat and 4-OHT treatment did not show any DsRed-Express signal. We observed mCherry expression in the presence of heat shock, regardless of 4-OHT treatment. To restrict experimental variables to heat induction, all experiments were conducted in the presence of 4-OHT. Percentages of DsRed-Express+ erythrocytes were low, probably due to the reduced activity of the β-actin2 promoter upon erythroid differentiation1,36.
For Zebrabow labelling, embryos are dechorionated following the procedure described above and transferred to six-well plates at a density of 25–30 embryos per well. Embryo buffer is replaced with embryo buffer containing 10–15 μM 4-OHT (Sigma, H7904), and embryos are kept in the dark overnight (16 h treatments) or for 4 h in a 28 °C incubator. After treatment, embryos are washed with fresh embryo buffer and transferred to new embryo buffer in a 28 °C incubator.
Single-cell laser induction
Embryos are anaesthetized with 0.02% tricaine (ethyl-3-aminobenzoate) (Sigma, E10521) in embryo buffer. Embryos are transferred onto a 40 mm glass-bottom dish with 0.17 mm thickness (WillCo Wells, GWST 5040). The dish is placed on the platform of a Leica DMI6000 inverted microscope. Individual cells are targeted with a 440 nm wavelength laser dye (5 mM coumarin 440 in methanol, with peak λ ~ 435 nm) with a MicroPoint system (Photonic Instruments) routed through the epifluorescence port and ×20 objective. Each cell is targeted with a 10 Hz pulse for 2 min. Embryos are transferred to a 10 cm petri dish containing 1 μM 4-OHT in embryo buffer and left to recover for 12 h in a 28 °C incubator. Buffer is then replaced with fresh embryo buffer. For adult blood analysis, embryos are transferred to a 28 °C aquatic housing system (Aquaneering) at 6 dpf.
Imaging and microscopy
During imaging, embryos are anaesthetized with 0.02% tricaine in embryo buffer. Embryos are imaged using a Leica DMI6000 inverted microscope, Leica SP5 inverted confocal microscope (Wetzlar) or Leica SP8X inverted confocal microscope (for Zebrabow images). Images are analysed with Imaris (Bitplane), Volocity Acquisition and Restoration software (Improvision) and ImageJ/Fiji. With the SP5, DsRed-Express and mCherry are excited by 543 and 594 nm laser lines, respectively; eGFP is excited by a 488 nm laser line. For Zebrabow colour detection, CFP, YFP and dTomato are excited with 447 nm, 515 nm and 555 nm laser lines, respectively. Individual channels were uniformly adjusted with Photoshop CS6 (Adobe) for display purposes. All quantification and data analysis were conducted on raw images. Imaging experiments were repeated at least three times.
Cell collection and flow cytometry
Adult fish between the ages of three and nine months post fertilization (3 mpf to 9 mpf) are anaesthetized with 0.02% tricaine in embryo buffer and dissected under a Leica MZ75 light microscope to collect the WKM. A single-cell suspension is mechanically prepared by pipetting the WKM up and down 50 times in cold 1× PBS (or 0.9× PBS with 2% FBS) and then passed through a 40 μm nylon mesh. Samples are subjected to 5 nM SYTOX Red dead cell stain (Invitrogen, S34859) or 3 nM DRAQ-7 dead cell stain (Abcam, ab109202) for Zebrabow analysis. Flow cytometry was done with a BD LSR II machine or BD FACS Aria IIIu Cell Sorter (Becton Dickinson) and analysed with FloJo software (Tree Star). For optimized Zebrabow colour detection, a 445 nm laser was used for excitation of CFP.
Adult–adult HSPC transplantation and sublethal irradiation
drl:GFP transplant
WKM from 3 month Tg(drl:GFP;ubi:mCherry) zebrafish was isolated and sorted by FACS. Cells were specifically enriched from the lymphoid/progenitor and precursor gates. Double-positive cells (6 × 104) were transplanted into irradiated casper recipients (n = 30 recipients). At 3 months post-transplant, WKM and peripheral blood from recipient fish was collected and analysed by flow cytometry to detect chimaerism levels of mCherry and eGFP-positive cells. We confirmed multilineage reconstitution by observing differentiated eGFP-positive cells in multiple cell populations, as determined by forward and side-scatter profiles.
Zebrabow transplant
WKM from 3 month Tg(ubi:Zebrabow-M;drl:creERT2) fish that were treated at either 48 hpf or 72 hpf with 4-OHT was isolated according to the procedures described above. WKM cells were transplanted into irradiated casper recipients (11 donors, average of eight recipients/donor) at 1.5× 105 non-erythroid cells per recipient. At 2.5–3 months post transplant, WKM and peripheral blood from recipient fish was collected and analysed as described above.
Sublethal irradiation
Tg(ubi:Zebrabow-M;drl:creERT2) fish that were treated at 51 hpf with 4-OHT were grown to 5 months. They were then subjected to split dose 30 Gy irradiation. WKM was isolated from irradiated fish or non-irradiated controls at 6 and 20 weeks post irradiation and analysed by flow cytometry.
Zebrabow longitudinal analysis
Zebrafish were anaesthetized with 0.02% tricaine in fish water and bled retro-orbitally with a 10 μl bevelled Hamilton syringe. Blood was collected in 0.9× PBS with 2% FBS and heparin (1 U). Because there are insufficient numbers of granulocytes in the zebrafish peripheral blood, we were only able to observe clonal dynamics of the erythrocyte population (Fig. 6b). Erythrocytes are relatively dim in Zebrabow expression, so we chose the three times where the detection by flow cytometry was optimal. We used a custom MATLAB graphical user interface with manual gating in ratiometric ternary colour space to quantify individual clones longitudinally. The contribution of each clone was normalized to the first time of collection. We define a stable contribution to the peripheral blood as a contribution between 0.5- and twofold relative to the first time.
Zebrabow cluster analysis
A custom MATLAB graphical user interface was developed to quantify colour barcodes using flow cytometry data exported from FloJo software (Tree Star). Live granulocytes, which are mainly composed of neutrophils (~80%), were isolated by FSC/SSC characteristics (step 1). These cells express the Zebrabow transgenes at the highest level in the marrow and are most useful for studying the clonal contribution from stem cells due to their short lifespan. We transformed (logicle) and normalized the intensities of the three Zebrabow fluorescent proteins in these cells and plotted them in 2D with ternary plots (step 2). This method revealed visual clusters that correspond to unique colour barcodes (step 3), where dTomato+CFP−YFP− cells denote polyclonal cells derived from HSCs that did not recombine. These red cells were ignored for analysis. To more accurately quantify the colour clusters, we employed a recently described clustering method that measures the density (ρ) of each cell in colour space (RGB) or ratiometric ternary space and the distance that each cell is from cells with higher density (δ)37. The cells with high ρ and δ values are most likely to be cluster centres. Plotting these metrics revealed the cluster centres (step 4), and the remainder of the cells were assigned to clusters by a nearest neighbour algorithm (step 5). The quality of clustering solutions was evaluated by the silhouette criterion, which measures the similarity between points in a cluster when compared with points in other clusters38 (Fig. 4 and Supplementary Fig. 5).
In cases of low labelling density, where the majority of the mature blood cells derived from HSCs that did not recombine, we could be more confident that non-red clusters are clonal in origin. In cases of high label density, where the majority of the cells have recombined, we observed some clusters that constituted significant percentages of the mature blood (>20%). It is possible that large HSC clones were present, but it is more likely that multiple, smaller clones expressed the same colour barcode. To accurately predict the clonal contribution in the blood, we ignored these large clusters (see examples in Supplementary Fig. 5 and Supplementary Fig. 7). High quality clusters (average silhouette value > 0.6) that contributed less than 0.5% of the granulocyte population could be detected. Clusters that contained fewer than 25 cells (out of 10,000 sampled, 0.25%) were also ignored for analysis. With treatment after 72 hpf, it is likely that we have saturated the number of colours possible for the Zebrabow system, which obscures clonal analysis.
Calculation of HSC number
Laser-induced fate mapping
Heat shock by targeted laser induction is not 100% efficient. To test for efficiency with our system, we targeted mesenchymal cells in the medial fin fold with laser induction. We found that approximately 45% of targeted cells express the DsRed-Express reporter. Out of ten cd41:eGFP+ cells, we estimate that ~4.5 cells on average actually recombine to turn on the reporter. In targeted adults, we found that 13.4% of granulocytes were labelled on average. 13.4% of labelled granulocytes/4.5 HSCs = ~ 3% granulocytes/HSC. Extrapolating to the whole population, 100%/3% = 33 HSCs.
Zebrabow clonal labeling
Colour barcodes were quantified according to the method described above in granulocytes from adult zebrafish that had been treated with 4-OHT at distinct developmental times. Clustering solutions for each adult marrow were evaluated by the silhouette value criterion, and clusters with silhouette values less than 0.75 were excluded from analysis. Outlier clusters, defined as more than 1.5 multiplied by the interquartile range of the clusters, were also excluded from analysis. These large outlier clusters are more likely to be polyclonal populations, where multiple stem cell clones express identical colour barcodes. All of the clusters that fit this criteria are shown in the violin plots of Fig. 5d. For each zebrafish, we obtained the average contribution of the clones that fitted these criteria. The average clonal contribution was then used to extrapolate to the entirety of the granulocyte pool (for example, an average clonal contribution of 5% extrapolates to 100%/5% = 20 clones). Averaging the data over multiple adult zebrafish led to the extrapolated clone numbers provided in Fig. 5b,c.
Our estimates of predicted clones rely on two main assumptions. First, we assume that rare colour barcodes in the mature granulocytes are more likely to be clonal populations. Second, we assume that the distribution of these rare barcodes reflects the distribution of clones in the remainder of the mature blood. It is also important to note that these predicted clones may not be functional HSCs at the time of label induction. In early treatments, we are most probably labelling a combination of functional HSCs, immature HSCs and haemogenic endothelium that will give rise to functional HSCs.
Statistics and reproducibility
Graphs and statistical analysis are done with Excel (Microsoft) and Prism (GraphPad Software). Descriptive statistics provide the mean and s.e.m. or s.d. P-values are obtained with two-tailed t-tests or one-way or two-way ANOVA, as denoted in figure legends. Confidence intervals were 95%. No statistical method was used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments or outcome assessment.
Experiments for Figs 1, 3e, 5, 6 and 7 and Supplementary Figs 3d–h, 6c–e and 8 were conducted once with the biological replicates noted in figure legends. Experiments for the remaining figures, especially those showing representative images (Figs 2 and 3 and Supplementary Figs 1, 2, 3 and 6), were repeated independently with similar results at least twice with the biological replicates noted in the figure legends.
Code availability
All computational code (written in the MATLAB environment) used to analyse the data is available from the corresponding author upon reasonable request.
Data availability
All data supporting the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank the following for their support and contributions: M. Distel, E. Butko, J. Bertrand and D. Stachura for critical reading of the manuscript; K. Richter for laboratory management; J. Abkowitz for comments and suggestions; A. Schier and A. Pan for the Zebrabow-M line and guidance; L. Ding and D. Tom at the HMS Enhanced Neuroimaging Core for help with confocal microscopy. This work was supported by HHMI and NIH F31HL126338 (J.H.), NIH R01-DK074482 (D.T.), NIH F32DK752433 (B.S.), the Deutsche Forschungsgemeinschaft (SFB655), the TU Dresden and the European Union (Zf Health), NIH R01-HL04880, NIH P01-HL032262, NIH 5P30-DK49216, NIH 5R01-DK53298, NIH 5U01-HL10001, NIH R24-DK09276 and an MPN Research Foundation Grant. C.M. received funding from an EMBO long-term fellowship, an HSFP long-term fellowship, an SNF advanced fellowship and an SNF professorship.
Footnotes
AUTHOR CONTRIBUTIONS
J.H. and B.S. designed and carried out experiments, analysed the data, and wrote the manuscript. B.S. and S.H. designed and carried out laser fate mapping experiments. E.D. and J.M. carried out experiments. C.M., M.B., D.T. and L.Z. designed experiments, edited the manuscript and supervised the project.
COMPETING FINANCIAL INTERESTS
L.Z. is a founder and stock holder of Fate, Inc., and Scholar Rock.
References
- 1.Bertrand JY, et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zovein AC, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boisset JC, et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- 5.Kissa K, et al. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood. 2008;111:1147–1156. doi: 10.1182/blood-2007-07-099499. [DOI] [PubMed] [Google Scholar]
- 6.Tamplin OJ, et al. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell. 2015;160:241–252. doi: 10.1016/j.cell.2014.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumaravelu P, et al. Quantitative developmental anatomy of definitive haematopoietic stem cells/long-term repopulating units (HSC/RUs): role of the aortagonad–mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver. Development. 2002;129:4891–4899. doi: 10.1242/dev.129.21.4891. [DOI] [PubMed] [Google Scholar]
- 8.Sun J, et al. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S, et al. Dynamics of HSPC repopulation in nonhuman primates revealed by a decade-long clonal-tracking study. Cell Stem Cell. 2014;14:473–485. doi: 10.1016/j.stem.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan CT, Lemischka IR. Clonal and systemic analysis of long-term hematopoiesis in the mouse. Genes Dev. 1990;4:220–232. doi: 10.1101/gad.4.2.220. [DOI] [PubMed] [Google Scholar]
- 11.Hess I, Iwanami N, Schorpp M, Boehm T. Zebrafish model for allogeneic hematopoietic cell transplantation not requiring preconditioning. Proc Natl Acad Sci USA. 2013;110:4327–4332. doi: 10.1073/pnas.1219847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma D, Zhang J, Lin H, Italiano J, Handin RI. The identification and characterization of zebrafish hematopoietic stem cells. Blood. 2011;118:289–297. doi: 10.1182/blood-2010-12-327403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertrand JY, Kim AD, Teng S, Traver D. CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development. 2008;135:1853–1862. doi: 10.1242/dev.015297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hans S, et al. Generation of a non-leaky heat shock-inducible Cre line for conditional Cre/lox strategies in zebrafish. Dev Dynam. 2011;240:108–115. doi: 10.1002/dvdy.22497. [DOI] [PubMed] [Google Scholar]
- 15.Xu J, et al. Temporal-spatial resolution fate mapping reveals distinct origins for embryonic and adult microglia in zebrafish. Dev Cell. 2015;34:632–641. doi: 10.1016/j.devcel.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- 17.Dixon G, Elks PM, Loynes CA, Whyte MKB, Renshaw SA. A method for the in vivo measurement of zebrafish tissue neutrophil lifespan. ISRN Hematol. 2012;2012:1–6. doi: 10.5402/2012/915868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 19.Pan YA, et al. Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development. 2013;140:2835–2846. doi: 10.1242/dev.094631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weissman TA, Pan YA. Brainbow: new resources and emerging biological applications for multicolor genetic labeling and analysis. Genetics. 2015;199:293–306. doi: 10.1534/genetics.114.172510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta V, Poss KD. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature. 2012;484:479–484. doi: 10.1038/nature11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loulier K, et al. Multiplex cell and lineage tracking with combinatorial labels. Neuron. 2014;81:505–520. doi: 10.1016/j.neuron.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 23.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 24.Martinez RJ, Neeld DK, Evavold BD. Identification of T cell clones without the need for sequencing. J Immunol Methods. 2015;424:28–31. doi: 10.1016/j.jim.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosimann C, et al. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Werdich AA, et al. Chamber identity programs drive early functional partitioning of the heart. Nat Commun. 2015;6:8146. doi: 10.1038/ncomms9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Busch K, et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- 28.Catlin S, Busque L, Gale R. The replication rate of human hematopoietic stem cells in vivo. Blood. 2011;117:4460–4466. doi: 10.1182/blood-2010-08-303537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arora N, et al. Effect of developmental stage of HSC and recipient on transplant outcomes. Dev Cell. 2014;29:621–628. doi: 10.1016/j.devcel.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu R, Neff N, Quake S, Weissman I. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol. 2011;29:928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shlush LI, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steensma DP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ablain J, Durand EM, Yang S, Zhou Y, Zon LI. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev Cell. 2015;32:756–764. doi: 10.1016/j.devcel.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 5. Univ. Oregon Press; 2007. [Google Scholar]
- 35.Lin HF, et al. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood. 2005;106:3803–3810. doi: 10.1182/blood-2005-01-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Traver D, et al. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez A, Laio A. Clustering by fast search and find of density peaks. Science. 2014;344:1492–1496. doi: 10.1126/science.1242072. [DOI] [PubMed] [Google Scholar]
- 38.Kaufman L, Rousseeuw PJ. Finding Groups in Ordinal Data. An Introduction to Cluster Analysis. Wiley-Interscience; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available from the corresponding author upon reasonable request.