Abstract
The ovary’s primary function is to produce the mature female gamete, the oocyte that, following fertilization, can develop into an embryo, implant within the uterus and ultimately allow the mother’s genetic material to be passed along to subsequent generations. In addition to supporting the generation of the oocyte, the ovary and specific ephemeral tissues within it, follicles and corpora lutea, produce steroids that regulate all aspects of the reproductive system, including the hypothalamic/pituitary axis, the reproductive tract (uterus, oviduct, cervix), secondary sex characteristics all of which are also essential for pregnancy and subsequent nurturing of the offspring. To accomplish these critical roles, ovarian development and function are tightly regulated by a number of exogenous (hypothalamic/pituitary) and endogenous (intraovarian) hormones. Within ovarian cells, intricate signalling cascades and transcriptional and post-transcriptional gene regulatory networks respond to these hormonal influences to provide the exquisite control over all of the temporal and spatial events that must be synchronized to allow this organ to successfully complete its function. This book chapter will focus specifically on the role of non-coding RNAs, their identification and described functional roles within the ovary with respect to normal function and their possible involvement in diseases, which involve the ovary.
Keywords: Folliculogenesis, MicroRNA, Premature ovarian failure, Ovarian cancer, Endo-siRNA, Long non-coding RNA
5.1 Introduction
Cell signalling networks and transcriptional factors within ovarian tissues have been extensively studied, however, studies of regulatory non-coding RNAs, which includes small RNA species such as piRNA, microRNA, endogenous small interfering RNAs (endo-siRNA), snoRNA and snRNA, and long RNA species including long intergenic noncoding RNA (lincRNA), long non-coding (lncRNA), have only recently (mid-2000s) been studied in ovarian tissues (Carletti and Christenson 2009; Lau et al. 2009). Much of the early work that continues today relates to the identification of the small RNA species within the organ, tissues and specific cell types of the ovary. Indeed as of yet, no systematic analysis of lncRNA has been completed in ovarian tissue. Current research is now beginning to elucidate the functional consequences these regulatory RNA have within ovarian tissues. This chapter will cover both of these aspects in normal physiology as well as discuss the relevance of regulatory non-coding RNAs with respect to several prominent ovarian diseases including ovarian cancer, polycystic ovarian syndrome, premature ovarian failure and ovarian hyperstimulation. Lastly, it should be noted that the majority of the published work on non-coding RNAs is centred on microRNA (miRNA), due primarily to their earlier identification, their high conservation across species, and their described regulatory mechanisms, which has allowed scientists to more easily study them. Therefore, this chapter will have a major focus on this class of non-coding RNAs, and mention briefly the few studies that look at other regulatory non-coding RNAs in the ovary.
5.2 Non-coding RNA in Ovarian Development
Ovarian development begins during gestation with primordial germ cells and somatic cells coming together to form a cellular mass known as the primordial follicles. These immature follicles contain the oocyte (the germ cell) and a surrounding layer of squamous granulosa cells (somatic cells). Subsequent to primordial follicle formation and prior to puberty, local factors within the ovary induce follicular waves that lead to recruitment, development and growth of some the primordial follicle pool causing progression from primordial to pre-antral and antral stage follicles. Currently, the presence and role for lncRNA in developing ovarian tissues has yet to be reported, although it is expected that these regulatory RNA will play mechanistic and functionally important roles similar to what has been observed in other tissues (Kung et al. 2013). Conversely, microRNA have been widely described in developing ovaries and we summarize those studies below.
Gene expression analysis studies have shown that miRNA are widely expressed in the mammalian ovary (Tripurani et al. 2010; Zhao and Rajkovic 2008; Ahn et al. 2010; Ro et al. 2007; Choi et al. 2007), with estimates ranging from 373 to 679 different miRNA being present in whole ovarian tissues using cloning or sequencing based technology. These early studies, while confirming presence of miRNA, were limited in their scope, as most did not address the cell types or stages of the ovarian cycle during which the ovaries were collected. Developmental studies in rodent, bovine and ovine models also examined miRNA expression in whole ovaries of newborn and foetal animals (Tripurani et al. 2010; Zhao and Rajkovic 2008; Ahn et al. 2010; Choi et al. 2007; Torley et al. 2011; Veiga-Lopez et al. 2013; Luense et al. 2011). Using a microarray platform, Zhao et al. (Zhao and Rajkovic 2008) identified 173 microRNA that were significantly expressed in newborn mouse ovaries with members of the let-7 family and miR-199a being the most prevalent miRNA. Function and cell-specific localization of miR-199a and the let-7 family members within the developing ovary have yet to be determined. Furthermore, as few miRNA were known at the time of this study, only 373 different miRNA were evaluated. Cloning studies by Tripurani et al. (2010) and next generation sequencing studies by Ahn et al. (2010), which are not reliant on a priori identification of miRNA, indicated that newborn ovaries in cattle and mice have 679 and 398 expressed miRNA, respectively.
In the bovine foetal ovary, 8 miRNA of the 679 miRNA present (bta-miR-99a, bta-miR-10b, bta-miR-199a-3p, bta-miR-199a-5p, bta-miR-424, bta-miR-100, bta-miR-455 and bta-miR-214) had more than tenfold greater relative abundance in the foetal ovary when compared to other somatic tissues (brain, hypothalamus, spleen, heart, lung, kidney, small intestine and muscle), four of these miRNA (bolded) exhibited higher expression in human ovaries too (Tripurani et al. 2010; Liang et al. 2007). In their studies, characterizing newborn mouse ovaries, Ahn et al. (2010) found that miRNA were the most abundant (over 25 %) form of all small RNA species. Through quantifying the total number of reads, miR-320, let-7i and let-7d were identified as the most highly expressed miRNA in the newborn ovaries (Ahn et al. 2010). Comparison of ovarian tissue to brain, lung, heart, stomach, liver, muscle, kidney and uterus indicated that miR-202 and miR-298 were also enriched in mouse gonadal tissues (Ahn et al. 2010). Again as previously mentioned, isolation, expression and most importantly functional analysis of these miRNAs seen in whole tissue from specific ovarian cell populations are needed to determine whether these miRNA are critical players in ovarian development and function.
In two time course studies in foetal sheep, Torley et al. (2011) and Veiga-Lopez et al. (2013) examined miRNA expression during gonadal differentiation. These two studies compared the relative expression patterns of 128 and 742 different miRNA in foetal ovaries between gestational day (GD) 42 and 75 or GD 65 and 90, respectively. The time points span the period of meiotic resumption (GD 55) and primordial germ cell formation (GD 75) in the first study and are prior to primordial follicle assembly (GD 65) and towards the completion of the dramatic tissue remodelling associated with the ovigerous cord formation (GD 90) in the latter study. Interestingly, Torley et al. (2011) observed that 62 of 128 miRNA were differentially expressed with equal distribution over time (one-half being increased at GD 42 and the other half induced at GD 75), while Veiga-Lopez et al. (2013) observed that only 21 of the 742 miRNA examined in their study were differentially expressed with 13 being increased at GD 65 and 8 increased at GD 90. It is noteworthy that GD 65 and GD 90 are both after the onset of meiotic resumption, and thus, may present a more homogenous set of tissues. In addition to the developmental analysis, this latter study also examined the effect of maternal exposure of the environmental disruptor, bisphenol A (BPA), on foetal ovarian miRNA expression (Veiga-Lopez et al. 2013). Maternal exposure to BPA induced a dramatic decrease in 45 different miRNA in ovaries of GD 65 foetuses (Veiga-Lopez et al. 2013) and 11 miRNAs at GD 90, when compared to same age controls. Interestingly, BPA only caused miRNA to decline in foetal sheep ovaries, the authors evaluated several of the enzymatic genes (Drosha, Dicer and DiGeorge syndrome critical region gene 8) as a possible mechanism and found no differences (Veiga-Lopez et al. 2013). An attractive hypothesis includes the possibility that these miRNA have oestrogen response elements within their promoters that could interact with the weak estrogenic BPA molecule to thereby influence their expression. Together all of these studies provide evidence that miRNA change during ovarian development, yet the role of specific miRNA and their functional impact on ovarian development remains to be determined.
5.3 Noncoding RNAs Involved in Ovarian Physiology
5.3.1 MicroRNA and Endo-siRNA on Oocyte Development
During the early stages of folliculogenesis, Dicer1 the RNase III enzyme that functions in the biogenic pathways of both endo-siRNA and miRNA exhibits robust and steady expression levels within the oocyte (Watanabe et al. 2008). Immediately following fertilization, however, Dicer1 levels are significantly reduced (Cui et al. 2007; Murchison et al. 2007). Coincident with decreased Dicer1 expression, global miRNA levels were also reduced in the two-cell embryo subsequent to increased Dicer1 expression and a return of increased miRNA expression (Tang et al. 2007). Consistent with these changes in Dicer1 expression, examination of small RNA populations in oocytes by Watanabe et al. (2006) indicated that approximately 10 % and 11 % of the total non-coding RNAs are miRNA and endo-siRNA, respectively. Moreover, no observable differences between miRNA and siRNA expression patterns were seen between fully grown and metaphase II (MII) oocytes (Watanabe et al. 2006). Complete loss of Dicer1 activity by targeted deletion of the gene causes embryonic lethality (Bernstein et al. 2003), therefore to investigate miRNA/endo-siRNA activity multiple labs “floxed” Dicer1 (Harfe et al. 2005; Andl et al. 2006; Muljo et al. 2005) in order to selectively knock out Dicer1 using cell-specific promoters to drive Cre recombinase.
Two studies using an oocyte-specific zona pellucida 3 (Zp3) Cre recombinase to knock out Dicer1, set out to determine if this biogenic enzyme functions within the oocyte (Murchison et al. 2007; Tang et al. 2007). In both studies there was near complete ablation of miRNA in the oocyte (Murchison et al. 2007; Tang et al. 2007). Murchison et al. (2007) observed disorganized spindles and chromosomal abnormalities, while Tang et al. (2007) reported that fertilized oocytes were unable to proceed through the first round of cell division and that half of the mutant oocytes showed cell fragmentation. Neither of these studies indicated that Dicer1 and by inference miRNA (and endo-siRNA upon latter discovery) were critical in early oocyte development from the primary follicular stage (when Zp3 expression begins) through the late maturation processes just prior to the ovulatory surge of luteinizing hormone (LH) (Murchison et al. 2007; Tang et al. 2007). Because these Dicer1 knockout studies were inadequate to determine if Dicer1 had any function at the earliest stages of folliculogenesis and oocyte growth, Mattiske et al. knocked out Dicer1 using a Cre recombinase driven by the alkaline phosphatase liver/bone/kidney (Alpl) promoter, which is expressed in primordial germ cells (Mattiske et al. 2009). Similar to the results of the Zp3 Cre models, this study identified chromosomal and meiotic abnormalities following the LH surge, while early folliculogenesis and oocyte growth were indistinguishable from wild type littermates (Mattiske et al. 2009). A particularly interesting observation from these oocyte-specific Dicer1 deletions is that the well-established oocyte – somatic (cumulus) cell interactions, which are deemed vital for normal folliculogenesis (Gilchrist et al. 2004; Buccione et al. 1990; Eppig 2001), are not reliant of oocyte miRNA or endo-siRNA post-transcriptional mechanisms.
What remains unanswered from these studies is the extent to which miRNA or endo-siRNA are contributing to the observed phenotypes. In an effort to address this question, Suh et al. (2010) using the Zp3-Cre conditionally to knock out DiGeorge Syndrome critical region gene 8 (Dgcr8). This RNA-binding protein, in conjunction with Drosha, processes pri-miRNA to pre-miRNA within the nucleus. This protein is only present in the miRNA biosynthetic pathway (Suh et al. 2010). Similar to the previous Dicer1 knockout experiments, miRNA expression was almost entirely ablated in the oocyte (Suh et al. 2010). However, in contrast to Dicer1 knockout models, oocyte maturation following the LH surge and embryonic preimplantation development were normal (Suh et al. 2010). These results implicated endo-siRNA and not miRNA as the important players in late oocyte development. Additional confirmation of this hypothesis came from a study by Ma et al. (2010) who found that the 3′untranslated regions (3′UTRs) of mRNA upregulated after Dicer1 deletion were not enriched in miRNA binding sites. The authors concluded from this finding that miRNA would therefore have a marginal impact on the maternal transcriptome in oocyte development (Ma et al. 2010). Further research investigated the ability of siRNA to regulate expression of retrotransposons (a class of genes known to be regulated in oocytes and embryos) during development (Watanabe et al. 2006). Co-transfection of EGFP linked to retrotransposons and EGFP linked to siRNA libraries into fully grown oocytes caused more degradation of the EGFP-retrotransposons when compared to the EGFP controls (Watanabe et al. 2006). Findings from this study suggest that siRNAs can post-transcriptionally regulated retrotransposons in the oocyte. However, whether siRNA regulates retrotransposons in vivo remains unknown, since the authors used an exogenous expression system (Watanabe et al. 2008).
5.3.2 MicroRNA and Endo-siRNA in Ovarian Somatic Cells
Folliculogenesis is characterized by rapid proliferation and dramatic morphological changes in granulosa cells as they transition from squamous to cuboidal cells during the transition from primordial follicles to primary follicles and beyond. These somatic changes are also coincident with alterations in oocyte growth, maturation, and differentiation, as well as the differentiation and identification of the thecal cell layer immediately adjacent to the basement membrane, separating the granulosa cells and oocyte from the vascular and stromal tissue of the ovary. In addition to the recruitment, growth, and selection processes commonly associated with folliculogenesis, the majority of these follicles that reach the preantral and antral stages will undergo atresia, a hormonally controlled apoptotic process that limits the number of follicles/oocytes that can be released to a number consistent with that species birth rate (monoovulatory or multiovulatory/litter bearing) (Tilly et al. 1991; Richards 1980).
Ovarian folliculogenesis is tightly regulated by the pituitary hormones, follicle stimulating hormone (FSH) and LH (Richards et al. 2002a, b). These hormones function as survival factors for follicles and increase the steroidogenic capacity of the follicles. At completion of folliculogenesis, the increasing oestradiol levels produced by the dominant follicle feeds back on the hypothalamic/pituitary to generate a surge of LH, which in turn initiates ovulation (release) of the cumulus oocyte complex from the dominant follicle, and subsequent uptake by the oviduct where fertilization can occur. Following the LH surge, the follicular granulosa and theca cells undergo a rapid and dramatic transformation into luteal cells, forming the parenchymal cells of the corpus luteum. The corpus luteum in turn secretes progesterone, a hormone critical for the establishment and maintenance of the pregnancy, if fertilization occurs, and for menstrual (human and primates) and oestrous (domestic animals, rodents) cyclicity in its absence. To accomplish these dramatic transformative events, beginning at the earliest stages of ovarian development all the way through to the process of luteolysis, the cellular mechanisms must be tightly regulated. MiRNA and endo-siRNA abilities to fine-tune gene regulation post-transcriptionally may be one of the mechanisms through which this regulation takes place.
Studies examining the role of miRNA and endo-siRNA in ovarian somatic cells have been conducted by three independent laboratories, who knocked out Dicer1 in granulosa cells using the anti-Müllerian hormone receptor 2-Cre (Amhr2-Cre) (Hong et al. 2008; Gonzalez and Behringer 2009; Nagaraja et al. 2008; Pastorelli et al. 2009), which is expressed in early developing granulosa cells as well as throughout the female reproductive tract (Arango et al. 2008). Conditional deletion of Dicer1 resulted in complete female sterility (Hong et al. 2008; Gonzalez and Behringer 2009; Nagaraja et al. 2008), predominantly caused by an inhospitable environment within the oviduct that killed early embryos. However, in addition to the marked morphologic and functional changes seen in the oviduct and uterus, Hong et al. (2008) observed markedly impaired ovulation rates both following natural cycles and following stimulated (PMSG+HCG) cycles. Confirmation of loss of ovarian function was also seen in the other studies (Gonzalez and Behringer 2009; Nagaraja et al. 2008). The partial loss of fertility is consistent with the lack of penetrance of the Amhr2-Cre within ovarian granulosa cells (Gifford et al. 2009).
In a follow-up study of miRNA/endo-siRNAs’ role in folliculogenesis, Lei et al. (2010) examined mouse ovarian tissues from 8-day, 8-week to 8-month old mice. Increased numbers of primordial follicles, accelerated follicular recruitment and more follicles that were degenerate were observed in the knockout animals versus control animals (Lei et al. 2010). In addition, several genes important in ovarian development, including 17alpha-hydroxylase (Cyp17a1), aromatase (Cyp19a1), growth/differentiation factor 9 (Gdf9), and bone morphogenetic protein 15 (Bmp15), amongst others, were differentially expressed in wild-type and conditional knockout ovaries (Lei et al. 2010). This study also evaluated expression of miR-503 an abundant ovarian miRNA (Takada et al. 2006), as expected loss of Dicer decreased miR-503 expression and temporal expression analysis indicated gonadotropin stimulation of follicle growth caused miR-503 levels to decrease (Lei et al. 2010). A specific role for miR-503 in ovarian function however awaits further study. In an alternative approach, Otsuka et al. (2008) used a hypomorphic Dicer1 allele to globally deplete Dicer1 to ~20 % of normal levels in mice. Female infertility was the major phenotype observed in this Dicer1 depletion model and subsequent characterization identified that the corpus luteum had reduced vascularization and that progesterone production was reduced (Otsuka et al. 2008). In conclusion, these partial and complete knockout studies indicate that maintenance of the miRNA/endo-siRNA biogenic pathway within granulosa and luteal cells is important for folliculogenesis, ovulation and luteal function. Currently, no direct studies have implicated a role for miRNA/endo-siRNA in theca cells, the other major cell type within the follicle. However, it is expected that important regulatory roles for microRNA/siRNA occur in that cell type too. Additionally, it is unclear if the phenotypes derived from the Dicer knockouts described above are due to the loss of miRNAs and/or endo-siRNAs. However, both profiling and specific functional studies have shown that miRNA are important to ovarian function (described in detail below). Nonetheless, further research using selective depletion of Dgcr8 in granulosa cells and theca-specific Cre lines should help address these open questions.
Additional evidence for miRNA-mediated post-transcriptional gene regulation as an important regulator of ovarian function can be obtained from a number of studies that have examined tissue (follicular and luteal) or cellular (granulosa) specific expression of miRNA over stages of development or from tissues at different stages during the oestrous/menstrual cycle or of pregnancy. Profiling studies have revealed that miRNA can vary their expression patterns depending on the stage of follicular development (Yao et al. 2009, 2010a; Ma et al. 2011; Fiedler 2008; Li et al. 2011; Yang et al. 2012a; McBride et al. 2012). Moreover, several of these gene expression studies have shown that certain miRNA can be under hormonal regulation in specific cell sub-types within the ovary.
Yao et al. (2009) investigated a small subset (6) of miRNA that were known to have low expression in primordial follicles, but are robustly expressed in primary follicles. These included let-7a, miR-125b and miR-143. To determine if miRNA expression is FSH regulated, an ovarian cell line, KK1, was subjected to FSH treatment for 6, 12, 24 and 48 h, after which miRNA expression was evaluated. MiR-143 was significantly reduced following FSH when compared to the untreated cells, while let-7a and miR-125b trended down (Yao et al. 2009). This study suggests that miRNA can be hormonally regulated. Subsequent work by the same laboratory using cultured rat granulosa cells identified 17 FSH-up-regulated and 14 FSH-down-regulated miRNA (Yao et al. 2010b). However, because only a few select miRNA were chosen for examination or the method utilized was an in vitro approach, the overall relevance to ovarian function is limited. Using an unbiased in vivo approach to identify miRNA that are regulated by the LH surge, our laboratory treated mice with a standard follicular stimulation protocol (PMSG +HCG) and collected granulosa cells 4 h after the surge of LH (hCG) and compared them to PMSG treatment alone (Fiedler et al. 2008). We identified ten microRNA with decreased expression (miR-483, miR-491, miR-484, miR-329, miR-433-3p, miR-532, miR-431, miR-672, miR-99b and miR-351) and three dramatically up-regulated microRNA (miR-132, miR-212 and miR-21) in granulosa cells (Fiedler et al. 2008). These findings were the first to reveal that hormones can have in vivo effects on miRNA expression patterns in specific ovarian cells. Through extensive studies from our lab (discussed below) we have begun to elucidate miR-21’s functional role and mechanism of action in granulosa cells and within the ovary.
In addition to hormonal regulation, studies have shown that intra-ovarian factors can regulate miRNA expression within ovarian cells. Yao et al. (2010a) investigated the effect of transforming growth factor β1 (TGFβ1), a factor known to play a role in follicular development and female fertility, on miRNA expression in pre-antral granulosa cells. After 6 h of TGFβ1 treatment of cultured pre-antral granulosa cells, 16 miRNAs were differentially expressed compared to control treated cells (Yao et al. 2010a). Of these 16, 13 were down-regulated and 3 were up-regulated including miR-712, miR-224 and miR-764-3p. Further investigation (described below in detail) implicated miR-224 in the regulation of granulosa cell proliferation (Yao et al. 2010a). In an alternative approach, granulosa cells were transfected with miRNA mimics and steroid synthesis was evaluated (Sirotkin et al. 2009). Of the 80 miRNAs tested, progesterone, testosterone and oestrogen synthesis decreased following transfection of 36, 57, and 51 of the individual miRNAs into cultured human granulosa cells respectively. Conversely, 10, 1 and no miRNAs were able to stimulate progesterone, testosterone and oestrogen synthesis, respectively. These experiments however are difficult to interpret, for several reasons: First, it is unclear if the changes in steroid output are a result of changes in production or if it is related to cellular viability, i.e. proliferation and apoptosis were not investigated, but were shown to be changed in a subsequent paper by the same group (Sirotkin et al. 2009) and secondly, the level of over-expression achieved was not reported. It is very likely that these cells were exposed to supra-physiologic levels of miRNA, which could lead to dramatic off-target effects.
Lastly, using microarray analysis, Ma et al. (2011) investigated miRNA expression patterns in regressed and non-regressed bovine corpora lutea (CL). They identified 13 differentially expressed miRNAs, 7 of which had preferential expression in non-regressed CL, while 6 miRNAs were elevated in regressed CL. MiR-378, a miRNA implicated in regulation of apoptosis, was one of the miRNAs that was up-regulated in the healthy non-regressing CL (Ma et al. 2011). Examination of miR-378 across the luteal phase indicated that its expression was greatest in mid and late stages and it dropped appreciably in regressing CL. Conversely a putative target gene, interferon gamma receptor 1, showed no change in mRNA levels, but its protein levels increased in regressing CL, prompting the conclusion that miR-378 might regulate this gene’s expression (Ma et al. 2011). The failure to demonstrate an actual cause and effect relationship, through further experimentation, however should temper this conclusion. It remains to be shown if miR-378 is a regulator of apoptosis and interferon gamma receptor 1 expression or merely if miR-378 expression is correlated with regression. Additional studies from another lab (discussed below) implicated this miRNA in having a functional role in folliculogenesis.
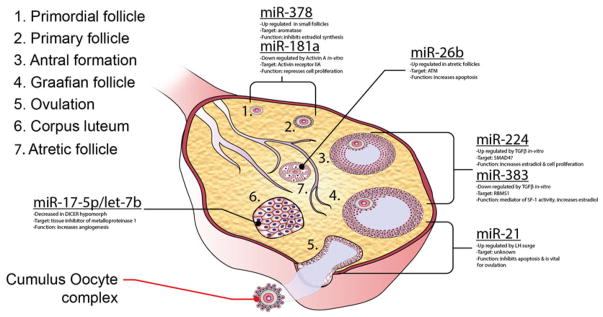
Together the profiling studies implicate a number of miRNAs in functional roles throughout the ovarian cycle. The following paragraphs describe the miRNA that have been individual evaluated with respect to ovarian somatic cell function (Fig. 5.1).
Fig. 5.1.
Schematic showing the different stages of follicles, ovulation and the corpus luteum where individual miRNA are expressed or where loss of miRNA action (by targeted or pharmacologic depletion) has been implicated in ovarian somatic tissue function
MicroRNA-378 (miR-378)
Bioinformatics analysis indicated the presence of several miRNA recognition sites within the 3′UTR of the porcine aromatase gene and expression analysis within small (1–3 mm) and large (3–6 mm) porcine follicles showed that miR-378 expression was approximately two-times greater in small follicles (Xu et al. 2011). To determine if miR-378 was able to directly regulate aromatase expression, miR-378 was overexpressed in granulosa cells isolated from both small and large follicles using a lentiviral expression system (Xu et al. 2011). Expression of miR-378 decreased aromatase levels with a concomitant decrease in oestradiol release (Xu et al. 2011). Similarly, inhibition of endogenous miR-378 levels increased aromatase levels and oestradiol production. Lastly, using the 3′UTR of aromatase linked to a luciferase reporter Xu et al. (2011) revealed that miR-378 regulates aromatase through direct binding to its 3′UTR. These observations therefore support a role for miR-378 in the regulation of aromatase, and point to the question of what regulates miR-378 expression in granulosa cells. Interestingly, recent studies of miR-378 and miR-378*, which are both are produced from the same pre-miRNA, but contain different seed sequences and thus target different genes, indicate that these miRNAs, which are located within the intron of the peroxisome proliferator-activated receptor γ coactivator 1β (Pgc1β) are equally co-expressed with this gene and counterbalance the metabolic actions of PGC1β, which includes metabolism and mitochondrial biogenesis (Carrer et al. 2012). Thus, this miRNA’s ability to regulate ovarian aromatase linked with its direct connection to expression of PGC1β, a key metabolic regulatory protein, may provide a potential mechanism of integration of metabolism and ovarian function. It is well known that energy balance is essential for optimal ovarian function and that metabolic pathologies such as obesity, diabetes and other metabolic syndromes can detrimentally affect ovarian function (Brewer and Balen 2010).
MicroRNA-224 (miR-224)
As mentioned above, the effect of the TGFβ superfamily and specifically TGFβ1 on cultured ovarian granulosa cell miRNA expression identified a number of differentially expressed miRNAs. The functional consequence of one of the three highly induced microRNAs, miR-224, was further evaluated in granulosa cells and the authors implicated miR-224 in the regulation of SMAD4 expression via a series of miR-224 inhibitor and over-expression experiments (Yao et al. 2010a). The authors also found that loss or gain of miR-224 activity through inhibitors/mimics decreased and increased cell proliferation and oestradiol synthesis, respectively (Yao et al. 2010a). These cellular changes were associated with increased (miR-224 inhibitor) and decreased (miR-224 mimic) SMAD4 levels, respectively. Reconciling these results with the observations of Wang et al. (2011) is however difficult, as they show that direct loss of SMAD4 decreases cell proliferation and oestrogen synthesis, the opposite effect shown when miR-224 is used to modulate SMAD4 levels. It is possible that miR-224 may be mediating its effects on cell proliferation and oestradiol synthesis through the regulation of other genes. Interestingly, in a follow-up study examining what regulates miR-224 expression, Liang et al. (2013) clearly showed involvement of p53 and p65 in miR-224 expression. Surprisingly though, in this follow-up paper they only showed a minimal effect of miR-224 inhibition on SMAD4 levels. Ultimately, these in vitro experiments await confirmation in an in vivo context.
MicroRNA-383 (miR-383)
Yin et al. initially confirmed that TGFβ signalling suppresses expression of miR-383 along with the sarcoglycan zeta (Sgcz) gene. Mir-383 is embedded within one of the Sgcz introns, thus implicating the Sgcz gene promoter in miR-383 expression (Yin et al. 2012a). Transfection of miR-383 mimics into granulosa cells led to a dose-dependent increase in oestradiol (Yin et al. 2012a). In their comprehensive study, Yin et al. (2012a) further elucidated a possible mechanism of action for miR-383 within the follicle, identifying a direct target, RNA binding motif, single stranded interacting protein 1 (RBMS1), that is decreased at both the mRNA and protein levels following overexpression of miR-383 (Yin et al. 2012a). Additionally, cMyc, a downstream target of RBMS1, decreased following miR-383 overexpression and cMyc inhibition and over-expression in granulosa cells increased and decreased estradiol production, respectively (Yin et al. 2012a). Furthermore over-expression of cMyc was able to mitigate the effects of over-expression of miR-383 or silencing of RBMS1 on oestradiol synthesis. Lastly, the impact of steroidogenic factor 1 (SF1), a transcription factor known to regulate steroidogenesis in gonadal tissue, on Sgcz/miR-383 expression was examined (Yin et al. 2012a). Knockdown of Sf1 decreased the primary and mature forms of miR-383 as well as Sgcz mRNA expression, follow-up luciferase and ChIP experiments confirmed that SF1 regulates Sgcz and mir-383 gene expression via direct binding to its promoter (Yin et al. 2012a). This investigation reveals a unique role for miR-383 in mediating SF1 actions in granulosa cell oestradiol synthesis via a direct target, Rbms1, and a target of cMYC.
MicroRNA-21 (miR-21)
miR-21 is up-regulated in virtually every known cancer and as a result has been the subject of many studies that have investigated both its function and its direct targets in tumours and cancer cell lines (Kumarswamy et al. 2011). Reports from our lab have revealed that, in addition to its pathological functions in cancer, miR-21 has a physiological role in the ovary (Fiedler et al. 2008; Carletti et al. 2010). Originally identified as an LH/hCG-regulated gene within murine ovarian granulosa cells (Fiedler et al. 2008), subsequent work has shown it to be up-regulated in ovine granulosa cells during the periovulatory period (McBride et al. 2012). Expression analysis throughout the periovulatory period (the time period between onset of LH/hCG stimulation and ovulation) in mice indicated that LH transcriptionally regulated pri-mir-21 expression (Carletti et al. 2010). Furthermore, miR-21’s potential regulatory role in cultured granulosa cells was examined and it was observed that complementary locked nucleic acid oligonucleotide inhibitors of miR-21 were able to induce apoptosis as evidenced by increased cleaved caspase 3 and annexin 5 staining (Carletti et al. 2010). These observations are consistent with miR-21’s known anti-apoptotic role in cancer cells (Kumarswamy et al. 2011). Interestingly, analysis of a number of bonafide miR-21 targets identified in other cells and tissues were not found to change in granulosa cells (Carletti et al. 2010). Lastly, we demonstrated that loss of miR-21 activity in vivo in mice undergoing ovarian stimulation (PMSG/hCG) caused a pronounced reduction in ovulation rates when compared to control treated animals, and the presence of trapped oocytes within corpora lutea (Carletti et al. 2010). Current work in the laboratory has identified additional pathways and target genes that are modulated by miR-21 expression (unpublished Carletti, Fitzgerald and Christenson). Linking these genes to ovarian function in vivo remains to be tested.
A study by Mase et al. (2012) also suggests that miR-21 is present in human granulosa cell lines, that it is highly expressed, and that over 80 % of the miRNA bound to EIF2C2 (a major component of the RNA induced silencing complex) was miR-21 (Mase et al. 2012). This latter finding suggests that a vast majority of miRNA-mediated post-transcriptional gene regulation may be occurring through miR-21 in these cell lines (Mase et al. 2012). Lastly, in a recent examination of GV and MII pig oocytes, miR-21 was shown to be substantially expressed in the MII oocytes (Yang et al. 2012a).
MicroRNA-181a (miR-181a)
Activins, which are produced by the ovary, are important for follicular development and maintaining female fertility (Pangas et al. 2007). Zhang et al. (2013) recently showed that activin A treatment of cultured mural granulosa cells caused a reduction in miR-181a expression in a dose dependent manner. Over-expression of miR-181a in cultured granulosa cells resulted in decreased granulosa cell proliferation through direct interaction with the 3′UTR of the activin receptor IIA (Acvr2a) mRNA (Zhang et al. 2013). Additionally, target genes downstream of ACVR2A were shown to be regulated by miR-181a in the expected direction. These studies further showed that replacement of ACVR2A was able to compensate for miR-181a-mediated suppression. The authors also examined miR-181a expression in ovarian follicles, demonstrating that miR-181a expression decreased in murine follicles as they progressed from primary, preantral to antral stages. Lastly, these investigators observed in a very small sample of patients with premature ovarian failure (POF) that miR-181a was elevated 4.5-fold in the blood of the POF patients (Zhang et al. 2013). This study provides a wealth of information and points to miR-181a as an important regulator of ovarian function. It will be interesting to determine whether these effects are seen in other species and whether miR-181a mediates a similar effect in vivo.
MicroRNA-26b (miR-26b)
Lin et al. (2012) compared miRNA expression in healthy, early and late stage atretic preovulatory porcine follicles. Using stringent conditions, 20 nonredundant miRNA were shown to be differentially expressed in healthy as well as both groups of atretric follicles with equal numbers being up-and down-regulated. Further characterization of miR-26b indicated that cultured porcine granulosa cells transfected with a miR-26b mimic were apoptotic as shown by anti-annexin V staining and evidence of dead cells in medium that did not occur in the controls treated cells. Lastly, the authors determined that ataxia telangiectasia mutated (Atm), a gene involved in repairing double-strand DNA breaks, was a direct target of miR-26b and that the number of DNA breaks increased in granulosa cells following treatment with miR-26b mimics (Lin et al. 2012). As noted by the authors, these experiments yielded a number of other interesting miRNA candidates that might be involved in atresia. Studies aimed at determining whether any of these identified miRNAs are involved in atresia and whether the atretic process is consistent for follicles of different stages remain to be performed.
Let-7b and miR-17-5p
In their study using the Dicer1 hypomorph, Otsuka et al. (2008) demonstrated that ovarian bursal injection of let-7b and miR-17-5p was able to partially restore the vascular defect caused by the overall loss of miRNAs. Additionally, the authors showed that the expression of a putative target, tissue inhibitor of metalloproteinase 1 (Timp1), changed in response to over-expression of either miRNA. Luciferase reporter assays showed that this is a direct effect on the 3′UTR of the Timp1 gene (Otsuka et al. 2008). The role of these particular miRNAs in ovarian function has not been further pursued, however, they would be interesting candidates for misregulation in luteal phase insufficiencies that result from neovascular defects (Boutzios et al. 2013).
Other microRNAs Implicated in Granulosa Cell Function
The following two miRNAs, miR-23a and miR-145, have been demonstrated to have effects on cultured human granulosa cells, but as yet have not been shown to change in ovarian tissues, thus limiting the interpretation of whether these effects are confined to a cell culture system or whether these miRNA play important in vivo roles in ovarian function. Transfection of human granulosa cells with miR-23a caused a decrease in XIAP (X-linked inhibitor of apoptosis protein) and caspase-3 protein levels, but an increase in cleaved caspase-3.miR-23a expression in granulosa cells may be causing apoptosis through inhibition of XIAP (Yang et al. 2012b). Interestingly, the rationale to examine miR-23a, originated from a biomarker study examining serum miRNA in POF patients (Zhou et al. 2011). Yan et al. (2012) showed that miR-145 directly regulates the expression of the activin receptor 1b. Additionally, miR-145 was shown to regulate cultured granulosa cell proliferation mediated partially through the ACVR1B/SMAD2/Activin A pathway.
5.4 Long Non-coding RNA in Ovarian Somatic Tissues
Currently, no systematic analyses of lncRNA have been reported in ovarian tissues. However, with the advent of emerging technologies such as stranded sequencing and the identification of numerous lncRNA in other tissues and the subsequent development of PCR and hybridization arrays, this will be shortly rectified. Currently, the only specific reported evidence for an ovarian lncRNA is the steroidogenic acute regulatory protein natural antisense transcript Star-NAT (Castillo et al. 2011). This RNA was found to bear full sequence complementarity to the spliced Star sense 3.5-kb transcript and is expressed in steroidogenic tissues such as testis, adrenal gland, brain and ovary (Castillo et al. 2011). Over expression of Star-NAT followed by cyclic AMP stimulation led to a decrease in StAR protein expression and concurrent decrease in progesterone production, likely through a posttranscriptional mechanism (Castillo et al. 2011).
5.5 Non-coding RNAs Involved in Ovarian Diseases
5.5.1 Ovarian Cancer
A role for small RNAs in the aetiology of ovarian cancer has been hypothesized by numerous laboratories and the literature in this area is extensive (see review Di Leva and Croce 2013). In the following two paragraphs we discuss two comprehensive studies that provide strong evidence that non-coding RNAs play critical components to the aetiology of ovarian cancer. Zhang et al. (2006) analysed 283 miRNA loci in 93 primary ovarian cells and 16 cell lines derived from ovarian cancer. Of these loci, 105 (37.1 %) were shown to have significant copy number alterations. Significantly, the mir-17–92 miRNA cluster located on chromosome 13q31, which has been described as oncogenic and is frequently amplified in multiple cancer types, was also increased in ovarian cancer (Knuutila et al. 1998). The mir-15a and mir-16 locus located on human chromosome 13q14, was lost in 23.9 % of the ovarian cancer cell types tested (Zhang et al. 2006). Zhang et al. also reported that in 24.8 % of ovarian cancer cell types, Dicer1 and Argonaute 2 (Ago2), genes that are critical for miRNA biogenesis and function, had increased copy numbers (Zhang et al. 2006). In contrast to the study performed by Zhang et al. (2006), which showed increased copy numbers of Dicer1 in ovarian cancer cell types, Merritt et al. (2008) show that there was variability in Dicer1 and Drosha expression in ovarian cancer specimens and that low expression of these genes was highly correlative with advanced tumour stage and low survival rate. In support of these findings, previous studies where Dicer1 was knocked down increased tumour formation in vivo was observed (Kumar et al. 2007).
Numerous investigations (far beyond what can be covered in this chapter) have gone beyond profiling and identified specific miRNA that might participate in cancer or are implicated in chemotherapeutic roles in ovarian cancer (Li et al. 2010). As an example, Yang et al. (2008) identified that patients who were resistant to chemotherapy, exhibited significantly reduced expression of let-7i and expression of this miRNA was correlated with low survival rate in women undergoing chemotherapy. Overexpression of let-7i in cultured ovarian cancer cells increased resistance to the chemotherapy drug cisplatin when compared to control treated cells (Yang et al. 2008). Studies of miR-200a in ovarian cancer cells showed that restoration of this miRNA led to decreased binding of laminin and increased sensitivity to paclitaxel, a chemotherapeutic drug (Cochrane et al. 2010). In support of these findings, previous research has shown that miRNA can have pharmacologic/chemotherapeutic functions in tumours (Blower et al. 2008) and that chemoresistance in ovarian cancer and ovarian cancer cells is associated with a distinct miRNA profile (Sorrentino et al. 2008; Boren et al. 2009). Lastly, a number of lncRNA have identified by in situ hybridization and RT-PCR analysis to be mis-regulated in ovarian cancer cells, these include the lncRNA XIST, H19 RNA, steroid receptor activator (SRA), BC200 RNA, and, more recently, a sno-lncRNA (Ariel et al. 1995; Hussein-Fikret and Fuller 2005; Yin et al. 2012b).
5.5.2 MicroRNA in Ovarian Fertility Disorders
Polycystic ovary syndrome is the most prevalent female infertility disease with ~5–10 % of reproductive aged women being affected. This disease is characterized as an oligo-or anovulatory disease with elevated systemic androgen levels with or without presence of multiple cystic follicles on the ovary (Ehrmann 2005; Franks 1995). In a recent study examining miRNA within follicular fluid of patients undergoing assisted reproductive technology, Sang et al. (2013) identified ~120 miRNAs, including several highly upregulated (miR-132 and miR-21), which were previously identified as LH-regulated in murine granulosa cells (Fiedler et al. 2008). Characterization of seven of the more highly expressed miRNAs in control patients and in PCOS patients, indicated that miRNA-132 and miR-320 were significantly lower expressed in PCOS patients (Sang et al. 2013). The putative targets and any possible roles for these miRNAs remain to be determined, as does a more comprehensive analysis of the cellular source of these small RNAs.
In another study, miR-93, -133 and -223, which all target GLUT4, the insulin-sensitive glucose transporter, were evaluated in adipose tissue of PCOS patients (Chen et al. 2013). The authors go on to demonstrate a direct effect of miR-93 on GLUT4 and that this miRNAs is over-expressed in PCOS patients and women with insulin resistance and hypothesized that this miRNA may partially explain the overlap of these two conditions (Chen et al. 2013). In a rodent PCOS model, rats were given dihydrotestosterone (DHT) to induce a PCOS-like phenotype and ~25 highly expressed miRNAs were found in both control and DHT ovaries (Hossain et al. 2013). Interestingly, of the differentially expressed RNAs several were shown to be selectively expressed in theca cells (Hossain et al. 2013). This is an important observation as theca cells are the predominant source of elevated androgens in PCOS women and these studies provide evidence that dysregulation of miRNA expression should be further evaluated in human ovarian theca cells.
Currently, other diseases of ovarian origin such as POF and ovarian hyperstimulation, which occurs as a consequence of assisted reproductive technology, have not been carefully examined with respect to changes in small or long non-coding RNA as a causative agent or as reactive genes, i.e. potential biomarkers, with the exception of the serum miRNA biomarker study of POF previously described (Zhou et al. 2011).
In conclusion the current literature indicates that a number of regulatory small RNAs are present within ovarian tissues. A select group of these small RNAs have been shown to be modulated by growth factors and steroids in cell culture systems, with only a few miRNAs having been shown to change in vivo and/or be hormonally (gonadotropin) regulated in vivo. Individual target genes for a select few miRNA have elucidated, yet it remains very likely that this represents a very small proportion of the target mRNAs these miRNA regulate. Lastly, the presence and study of long non-coding RNAs and the recently identified circular RNAs has not yet systematically studied in ovarian tissues and this area is sure to be an interesting topic in the coming years.
Acknowledgments
Grant support: Supported by a grant from the National Institutes of Health (HD061580; LKC).
References
- Ahn HW, et al. MicroRNA transcriptome in the newborn mouse ovaries determined by massive parallel sequencing. Mol Hum Reprod. 2010;16(7):463–471. doi: 10.1093/molehr/gaq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andl T, et al. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16(10):1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango NA, et al. A mesenchymal perspective of müllerian duct differentiation and regression in Amhr2-lacZ mice. Mol Reprod Dev. 2008;75(7):1154–1162. doi: 10.1002/mrd.20858. [DOI] [PubMed] [Google Scholar]
- Ariel I, et al. The imprinted H19 gene as a tumor marker in bladder carcinoma. Urology. 1995;45(2):335–338. doi: 10.1016/0090-4295(95)80030-1. [DOI] [PubMed] [Google Scholar]
- Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Blower PE, et al. MicroRNAs modulate the chemo-sensitivity of tumor cells. Mol Cancer Ther. 2008;7(1):1–9. doi: 10.1158/1535-7163.MCT-07-0573. [DOI] [PubMed] [Google Scholar]
- Boren T, et al. MicroRNAs and their target messenger RNAs associated with ovarian cancer response to chemotherapy. Gynecol Oncol. 2009;113(2):249–255. doi: 10.1016/j.ygyno.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Boutzios G, Karalaki M, Zapanti E. Common pathophysiological mechanisms involved in luteal phase deficiency and polycystic ovary syndrome. Impact on fertility. Endocrine. 2013;43(2):314–317. doi: 10.1007/s12020-012-9778-9. [DOI] [PubMed] [Google Scholar]
- Brewer CJ, Balen AH. The adverse effects of obesity on conception and implantation. Reproduction. 2010;140(3):347–364. doi: 10.1530/REP-09-0568. [DOI] [PubMed] [Google Scholar]
- Buccione R, Schroeder AC, Eppig JJ. Interactions between somatic cells and germ cells throughout mammalian oogenesis. Biol Reprod. 1990;43(4):543–547. doi: 10.1095/biolreprod43.4.543. [DOI] [PubMed] [Google Scholar]
- Carletti M, Christenson LK. MicroRNA in the ovary and female reproductive tract. J Anim Sci. 2009;87(14 suppl):E29–E38. doi: 10.2527/jas.2008-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carletti MZ, Fiedler SD, Christenson LK. MicroRNA 21 blocks apoptosis in mouse periovulatory granulosa cells. Biol Reprod. 2010;83(2):286. doi: 10.1095/biolreprod.109.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer M, et al. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci U S A. 2012;109(38):15330–15335. doi: 10.1073/pnas.1207605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo AF, et al. Hormone-dependent expression of a steroidogenic acute regulatory protein natural antisense transcript in MA-10 mouse tumor Leydig cells. PLoS One. 2011;6(8):e22822. doi: 10.1371/journal.pone.0022822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-H, et al. MiRNA-93 inhibits GLUT4 and is overexpressed in adipose tissue of polycystic ovary syndrome patients and women with insulin resistance. Diabetes. 2013;62:2278–2286. doi: 10.2337/db12-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, et al. Microarray analyses of newborn mouse ovaries lacking Nobox. Biol Reprod. 2007;77(2):312–319. doi: 10.1095/biolreprod.107.060459. [DOI] [PubMed] [Google Scholar]
- Cochrane DR, et al. Loss of miR-200c: a marker of aggressiveness and chemoresistance in female reproductive cancers. J Oncol. 2010;2009 doi: 10.1155/2010/821717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X-S, Shen X-H, Kim N-H. Dicer1 expression in preimplantation mouse embryos: involvement of Oct3/4 transcription at the blastocyst stage. Biochem Biophys Res Commun. 2007;352(1):231–236. doi: 10.1016/j.bbrc.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Di Leva G, Croce CM. The role of microRNAs in the tumorigenesis of ovarian cancer. Front Oncol. 2013;3:153. doi: 10.3389/fonc.2013.00153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ehrmann DA. Polycystic ovary syndrome. N Engl J Med. 2005;352(12):1223–1236. doi: 10.1056/NEJMra041536. [DOI] [PubMed] [Google Scholar]
- Eppig JJ. Oocyte control of ovarian follicular development and function in mammals. Reproduction. 2001;122(6):829–838. doi: 10.1530/rep.0.1220829. [DOI] [PubMed] [Google Scholar]
- Fiedler SD, et al. MicroRNA expression within periovulatory mural granulosa cells. Biology Reproduction. 2008;79:1030–1037. doi: 10.1095/biolreprod.108.069690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks S. Polycystic ovary syndrome. N Engl J Med. 1995;333(13):853–861. doi: 10.1056/NEJM199509283331307. [DOI] [PubMed] [Google Scholar]
- Gifford JAH, Hunzicker-Dunn ME, Nilson JH. Conditional deletion of beta-catenin mediated by Amhr2cre in mice causes female infertility. Biol Reprod. 2009;80(6):1282–1292. doi: 10.1095/biolreprod.108.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist RB, Ritter LJ, Armstrong DT. Oocyte–somatic cell interactions during follicle development in mammals. Anim Reprod Sci. 2004;82:431–446. doi: 10.1016/j.anireprosci.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Gonzalez G, Behringer RR. Dicer is required for female reproductive tract development and fertility in the mouse. Mol Reprod Dev. 2009;76(7):678–688. doi: 10.1002/mrd.21010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, et al. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102(31):10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X, et al. Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology. 2008;149(12):6207–6212. doi: 10.1210/en.2008-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MM, et al. Altered expression of miRNAs in a dihydrotestosterone-induced rat PCOS model. J Ovarian Res. 2013;6(1):36. doi: 10.1186/1757-2215-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein-Fikret S, Fuller PJ. Expression of nuclear receptor coregulators in ovarian stromal and epithelial tumours. Mol Cell Endocrinol. 2005;229(1–2):149–160. doi: 10.1016/j.mce.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Knuutila S, et al. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol. 1998;152(5):1107. [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39(5):673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011;8(5):706–713. doi: 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, et al. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009;19(10):1776–1785. doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, et al. The regulatory role of Dicer in folliculogenesis in mice. Mol Cell Endocrinol. 2010;315(1):63–73. doi: 10.1016/j.mce.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SD, et al. The role of microRNAs in ovarian cancer initiation and progression. J Cell Mol Med. 2010;14(9):2240–2249. doi: 10.1111/j.1582-4934.2010.01058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, et al. Repertoire of porcine microRNAs in adult ovary and testis by deep sequencing. Int J Biol Sci. 2011;7(7):1045. doi: 10.7150/ijbs.7.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, et al. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8(1):166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, et al. Transcriptional cooperation between p53 and NF-κB p65 regulates microRNA-224 transcription in mouse ovarian granulosa cells. Mol Cell Endocrinol. 2013;370:119–129. doi: 10.1016/j.mce.2013.02.014. [DOI] [PubMed] [Google Scholar]
- Lin F, et al. miR-26b promotes granulosa cell apoptosis by targeting ATM during follicular atresia in porcine ovary. PLoS ONE. 2012;7(6):e38640. doi: 10.1371/journal.pone.0038640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luense LJ, et al. Developmental programming: gestational testosterone treatment alters fetal ovarian gene expression. Endocrinology. 2011;152(12):4974–4983. doi: 10.1210/en.2011-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, et al. MicroRNA activity is suppressed in mouse oocytes. Curr Biol. 2010;20(3):265–270. doi: 10.1016/j.cub.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, et al. Microarray analysis of differentially expressed microRNAs in non-regressed and regressed bovine corpus luteum tissue; microRNA-378 may suppress luteal cell apoptosis by targeting the interferon gamma receptor 1 gene. J Appl Genet. 2011;52(4):481–486. doi: 10.1007/s13353-011-0055-z. [DOI] [PubMed] [Google Scholar]
- Mase Y, et al. MiR-21 is enriched in the RNA-induced silencing complex and targets COL4A1 in human granulosa cell lines. Reprod Sci. 2012;19:1030–1040. doi: 10.1177/1933719112442245. [DOI] [PubMed] [Google Scholar]
- Mattiske DM, Han L, Mann JR. Meiotic maturation failure induced by DICER1 deficiency is derived from primary oocyte ooplasm. Reproduction. 2009;137(4):625–632. doi: 10.1530/REP-08-0475. [DOI] [PubMed] [Google Scholar]
- McBride D, et al. Identification of miRNAs associated with the follicular-luteal transition in the ruminant ovary. Reproduction. 2012;144(2):221–233. doi: 10.1530/REP-12-0025. [DOI] [PubMed] [Google Scholar]
- Merritt WM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359(25):2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo SA, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202(2):261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, et al. Critical roles for Dicer in the female germline. Genes Dev. 2007;21(6):682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja AK, et al. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol Endocrinol. 2008;22(10):2336–2352. doi: 10.1210/me.2008-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M, et al. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J Clin Invest. 2008;118(5):1944. doi: 10.1172/JCI33680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangas SA, et al. Intraovarian activins are required for female fertility. Mol Endocrinol. 2007;21(10):2458–2471. doi: 10.1210/me.2007-0146. [DOI] [PubMed] [Google Scholar]
- Pastorelli LM, et al. Genetic analyses reveal a requirement for Dicer1 in the mouse urogenital tract. Mamm Genome. 2009;20(3):140–151. doi: 10.1007/s00335-008-9169-y. [DOI] [PubMed] [Google Scholar]
- Richards JS. Maturation of ovarian follicles: actions and interactions of pituitary and ovarian hormones on follicular cell differentiation. Physiol Rev. 1980;60(1):51–89. doi: 10.1152/physrev.1980.60.1.51. [DOI] [PubMed] [Google Scholar]
- Richards JAS, et al. Expression of FKHR, FKHRL1, and AFX genes in the rodent ovary: evidence for regulation by IGF-I, estrogen, and the gonadotropins. Mol Endocrinol. 2002a;16(3):580. doi: 10.1210/mend.16.3.0806. [DOI] [PubMed] [Google Scholar]
- Richards JS, et al. Novel signaling pathways that control ovarian follicular development, ovulation, and luteinization. Recent Prog Horm Res. 2002b;57(1):195–220. doi: 10.1210/rp.57.1.195. [DOI] [PubMed] [Google Scholar]
- Ro S, et al. Cloning and expression profiling of small RNAs expressed in the mouse ovary. RNA. 2007;13(12):2366–2380. doi: 10.1261/rna.754207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Q, et al. Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J Clin Endocrinol Metab. 2013;98(7):3068–3079. doi: 10.1210/jc.2013-1715. [DOI] [PubMed] [Google Scholar]
- Sirotkin AV, et al. Identification of MicroRNAs controlling human ovarian cell steroidogenesis via a genome-scale screen. J Cell Physiol. 2009;219(2):415–420. doi: 10.1002/jcp.21689. [DOI] [PubMed] [Google Scholar]
- Sorrentino A, et al. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol Oncol. 2008;111(3):478. doi: 10.1016/j.ygyno.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Suh N, et al. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol. 2010;20(3):271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, et al. Mouse microRNA profiles determined with a new and sensitive cloning method. Nucleic Acids Res. 2006;34(17):e115. doi: 10.1093/nar/gkl653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21(6):644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly JL, et al. Involvement of apoptosis in ovarian follicular atresia and postovulatory regression. Endocrinology. 1991;129(5):2799–2801. doi: 10.1210/endo-129-5-2799. [DOI] [PubMed] [Google Scholar]
- Torley KJ, et al. Expression of miRNAs in ovine fetal gonads: potential role in gonadal differentiation. Reprod Biol Endocrinol. 2011;9:2. doi: 10.1186/1477-7827-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripurani SK, et al. Cloning and analysis of fetal ovary microRNAs in cattle. Anim Reprod Sci. 2010;120(1):16–22. doi: 10.1016/j.anireprosci.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Veiga-Lopez A, et al. Developmental programming: gestational bisphenol-A treatment alters trajectory of fetal ovarian gene expression. Endocrinology. 2013;154(5):1873–1884. doi: 10.1210/en.2012-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, et al. Interference RNA-based silencing of endogenous SMAD4 in porcine granulosa cells resulted in decreased FSH-mediated granulosa cells proliferation and steroidogenesis. Reproduction. 2011;141(5):643–651. doi: 10.1530/REP-10-0098. [DOI] [PubMed] [Google Scholar]
- Watanabe T, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20(13):1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453(7194):539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Xu S, et al. Micro-RNA378 (miR-378) regulates ovarian estradiol production by targeting aromatase. Endocrinology. 2011;152(10):3941–3951. doi: 10.1210/en.2011-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, et al. MicroRNA-145 suppresses mouse granulosa cell proliferation by targeting activin receptor IB. FEBS Lett. 2012;586(19):3263–3270. doi: 10.1016/j.febslet.2012.06.048. [DOI] [PubMed] [Google Scholar]
- Yang N, et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68(24):10307–10314. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CX, et al. Small RNA profile of the cumulusoocyte complex and early embryos in the pig. Biol Reprod. 2012a;87(5):117. doi: 10.1095/biolreprod.111.096669. [DOI] [PubMed] [Google Scholar]
- Yang X, et al. Differentially expressed plasma microRNAs in premature ovarian failure patients and the potential regulatory function of mir-23a in granulosa cell apoptosis. Reproduction. 2012b;144(2):235–244. doi: 10.1530/REP-11-0371. [DOI] [PubMed] [Google Scholar]
- Yao N, et al. A network of miRNAs expressed in the ovary are regulated by FSH. Front Biosci. 2009;14:3239–3245. doi: 10.2741/3447. [DOI] [PubMed] [Google Scholar]
- Yao G, et al. MicroRNA-224 is involved in transforming growth factor-β-mediated mouse granulosa cell proliferation and granulosa cell function by targeting Smad4. Mol Endocrinol. 2010a;24(3):540–551. doi: 10.1210/me.2009-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N, et al. Follicle-stimulating hormone regulation of microRNA expression on progesterone production in cultured rat granulosa cells. Endocrine. 2010b;38(2):158–166. doi: 10.1007/s12020-010-9345-1. [DOI] [PubMed] [Google Scholar]
- Yin M, et al. Transactivation of microRNA-383 by steroidogenic factor-1 promotes estradiol release from mouse ovarian granulosa cells by targeting RBMS1. Mol Endocrinol. 2012a;26(7):1129–1143. doi: 10.1210/me.2011-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin QF, et al. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012b;48(2):219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Zhang L, et al. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci. 2006;103(24):9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, et al. MicroRNA-181a suppresses mouse granulosa cell proliferation by targeting activin receptor IIA. PLoS ONE. 2013;8(3):e59667. doi: 10.1371/journal.pone.0059667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Rajkovic A. MicroRNAs and mammalian ovarian development. In: Chegini N, editor. Seminars in reproductive medicine. © Thieme Medical Publishers; Stuttgart: 2008. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhu Y, Zhang SH, Wang HM, Wang SY, Yang XK. MicroRNA expression profiles in premature ovarian failure patients and its potential regulate functions. Chin J Birth Health Hered. 2011;19:20–22. [Google Scholar]