Abstract
Preeclampsia is a common pregnancy-specific vascular disorder characterized by new-onset hypertension and proteinuria during the second half of pregnancy. The etiology of the disease is unknown but predisposition to preeclampsia is in part heritable. Preeclampsia is associated with an increased risk of cardiovascular disease later in life. We have sequenced 124 candidate genes implicated in preeclampsia to pinpoint genetic variants contributing to predisposition to or protection from preeclampsia. First, targeted exomic sequencing was carried out in 500 preeclamptic women and 190 controls from the Finnish Genetics of Preeclampsia Consortium (FINNPEC) cohort. Then 122 women with a history of preeclampsia and 1,905 parous women with no such history from the National FINRISK Study were included in the analyses. We tested 146 rare and low-frequency variants and found an excess (observed 13 versus expected 7.3) nominally associated with preeclampsia (p< 0.05). The most significantly associated sequence variants were protective variants rs35832528 (E982A; p=2.49E-4, OR=0.387) and rs141440705 (R54S; p=0.003, OR=0.442) in Fms Related Tyrosine Kinase 1 (FLT1). These variants are enriched in the Finnish population with minor allele frequencies 0.026 and 0.017, respectively. They may also be associated with a lower risk of heart failure in 11,257 FINRISK women. This study provides the first evidence of maternal protective genetic variants in preeclampsia.
Keywords: Targeted exomic sequencing, candidate gene, preeclampsia, FLT1, angiogenesis
Introduction
Preeclampsia (PE) is a common vascular disorder that affects 3% of pregnant women 1. Worldwide, it annually accounts for approximately 50,000 maternal and 900,000 perinatal deaths 2, 3. The clinical characteristics are diverse and the course of the disease is unpredictable. Both a PE mother and a child born from a PE pregnancy are at higher risk for later-life cardiovascular diseases and type 2 diabetes 4.
Angiogenesis is tightly involved in the pathophysiology of PE 5. A high ratio of soluble fms related tyrosine kinase 1 (sFLT1) to placental growth factor (PlGF) is among the most promising biomarkers for predicting the onset of the disease 6. Several susceptibility loci for PE have been identified in genome-wide linkage studies 7, 8. However, linkage and candidate gene studies have been plagued with poor reproducibility.
The population of Finland is genetically unique in Europe 9. The repeated bottleneck events that caused strong founder effects, and geographic isolation over centuries have resulted in the enrichment of variants that are rare or absent in other populations 10. These features provide an opportunity to overcome many of the obstacles in studying rare enriched variants that contribute to the risk of complex diseases 11.
We designed a targeted gene sequencing protocol to screen the coding and splicing areas of genes of interest within angiogenic and vascular pathways and other putative candidate genes in a sizeable cohort of PE cases and controls from Finland. Here we explore the potential causal role of variation in candidate genes in PE.
Methods
Diagnostic criteria and patient cohorts
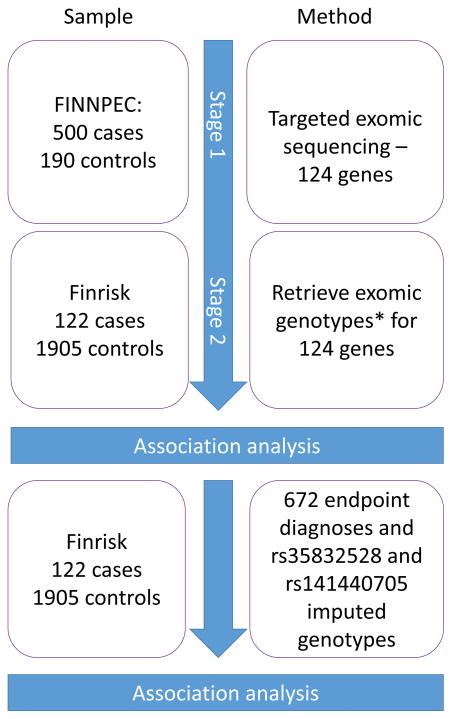
The study design is outlined in Figure 1. At Stage 1 we studied 500 non-obese (body mass index [BMI]<30 kg/m2) women with PE pregnancies and 190 non-PE controls that were matched geographically, in age, and in BMI from The Finnish Genetics of PE Consortium (FINNPEC) cohort, a case-control cohort recruited from the five Finnish University Hospitals 12. Nulliparous or multiparous women with a singleton pregnancy were eligible for the study. PE was defined as hypertension and proteinuria occurring after 20 week’s gestation. Hypertension was defined as systolic blood pressure ≥140 mm Hg or diastolic blood pressure ≥90 mm Hg after 20 weeks of gestation. Proteinuria was defined as the urinary excretion of ≥0.3 g protein in a 24-h specimen, or 0.3 g/l, or two ≥1+ readings on dipstick in a random urine determination with no evidence of urinary tract infection. All diagnoses were independently verified by a research nurse and a physician. Seven PE and one control were excluded due to a failure in genotyping and further nine cases were excluded due to non-Finnish ethnicity and ovum donation pregnancy. The characteristics of the study participants are presented in S1. All women provided a written informed consent and the FINNPEC study protocol was approved by the coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa.
Figure 1.
Methods and samples of study design.
*Genotyping performed by Agilent 1.1 refseq (60 cases and 4 controls), Illumina coding v1 (162 cases and 10 controls) and Nimblegen SeqCap EZ VCRome (1682 cases and 109 controls) platforms.
At Stage 2 we included in the analyses whole exome sequencing from an additional 122 women with a history of PE and 1905 parous women with no such history from the national FINRISK study cohort (FINRISK license 8/2016)13. For identifying PE and eclampsia cases in the FINRISK study cohort, we used following Finnish ICD codes in the comprehensive National Hospital Discharge Register covering years 1992 to 2007: ICD-10 (in use since 1996): O14.0, O14.1, O14.9, O15.0, O15.1, O15.2, O15.9, ICD-9 (in use from 1987–1996): 6424–6426, 6427A and ICD-8 (in use from 1968–1986) :637.03, 637.04, 637.09, 637.10, 637.99). Controls from FINRISK were all women who had given birth at least once and did not have any of these diagnoses recorded. In the final association analyses, we included genotypes of FINNPEC and FINRISK which combined totaled 609 cases and 2,092 controls. All polymorphic sites in the Stage 1 targeted sequencing were queried from the population cohort exomes. All of the National FINRISK Study methodology and ethical approvals are available online: https://www.thl.fi/en/web/thlfi-en/research-and-expertwork/population-studies/the-national-finrisk-study.
Preparation of Genomic DNA
Genomic DNA was extracted from whole blood using the NucleoSpin Blood XL DNA extraction kit (Macherey-Nagel GmbH & Co.) or the Chemagic Magnetic Separation Module I –machine (Chemagen) and subsequently stored at −20°C.
Targeted Sequencing and Capture Enrichment
Genes implicated in human biomarker studies were chosen as targeted candidate genes, as were their signalling partners and proteases that alter their function and levels. Specific single nucleotide polymorphisms (SNPs) and any neighbouring or associated gene that have been enriched in PE women based on meta-analysis, genomewide association studies (GWAS) or functional studies were included. This list of existing candidate genes was based on review of literature in January 2013 and it included genes involved in angiogenesis, such as FLT1, and pregnancy, such as pregnancy-zone protein PZP, among other relevant functional pathways. The studied genes are listed in S2.
Libraries from genomic DNA were prepared in-house (Washington University School of Medicine) 14. Enzymes were purchased from Enzymatics (Beverly, MA). Briefly, the ends of sheared genomic DNA fragments were repaired by treatment with T4 DNA Polymerase and T4 DNA Polynucleotide Kinase, which phosphorylates the 5′ hydroxyl group. An adenosine was then added to the 3′ position at each end of the DNA fragment with Taq Polymerase. Illumina adapters with an overhanging “T” were ligated onto the DNA fragment followed by bead-based size selection procedure to remove adapter-dimers and fragments below the desired size. A unique index sequence was added by PCR by targeting the two ligated universal adapters on each fragment end.
Sequence capture by hybridization was performed according to the manufacturer’s recommendations with modifications (Roche SeqCap Hybridization and Wash Kit No. 05634261001). We utilized longer blocking oligos containing an additional 7 bp inosine segment for promiscuous pairing with different index sequences. Following hybridization, captured DNA was washed and eluted according to the manufacturer’s instructions.
Sequencing and Analysis
Sequencing was performed on an Illumina HiSeq 2000 at the Washington University Genome Technology Access Center using 2×101 bp, 2×135 bp and 2×150 bp reads. We aligned sequencing to GRCh37 using bwa aln (v0.6.1-r104) and genotyped the samples using the Genome Analysis ToolKit (v2.5.2-gf57256b) Unified Genotyper 15–17.
Statistical Analyses
Quality control before meta-analysis included removal of singleton and monomorphic variants, removal of sites with >10% missing data in the targeted sequencing or a significant departure from Hardy-Weinberg equilibrium in controls (p<0.001). The combined dataset was analyzed using Fisher’s exact test which served as our primary association test. The analysis was divided into two classes: low frequency and rare variants (minor allele frequency (MAF) <10%) or common variants (MAF>10%). After quality control, 622 variants (443 variants with MAF<10% and 179 variants with MAF>10%) were advanced to the combined primary analysis. Of these, 201 were in putatively functional categories (missense, nonsense, or splice region variants) and 421 were in likely benign categories (synonymous, intronic, intergenic).
The two SNPs with strongest associations were further compared to 672 predefined epidemiological diagnoses derived from national health care registries using imputed genotypes of 11,257 FINRISK women. Genotypes were imputed using a combined panel of Finnish whole genome sequences and 1000 Genomes phase I reference with a high imputation confidence (info metric > 0.97) 18, 19. Associations were calculated using logistic regression (SNPTEST v2.5.2, EM algorithm) and then further confirmed with Fisher’s exact test given the rarity of variants and several diagnoses. Due to the explorative nature of this part of the analysis, no multiple testing corrections to estimates were performed. Heart failure was identified using codes I50, 4289X, and 42700 in Finnish ICD-10, ICD-9, and ICD-8, respectively.
Data were analysed using PlinkSeq, Plink 20 and R. Kaviar21 and VEP Build 37 was used in additional annotations 22. Loss of function (LoF) analyses were conducted for each gene with associating variants in silico by the Loss of Function – tool (https://github.com/ensembl-variation/VEP_plugins/blob/master/LoFtool.pm). The following annotations were calculated: LoF score < 0.2 is probably damaging, LoF score 0.2–0.7 is possibly damaging, LoF score < 0.7 is benign. In addition to appropriate statistical probability tests, odds ratios (OR) with 95% confidence intervals (CI95) were calculated for all variants.
Results
Results of the association analyses are listed in Table 1. For these studies, genotypes from Stage 1 (FINNPEC) were combined with a population cohort in Stage 2 (FINRISK) for joint analysis. Forty-one variants demonstrated nominal association (p<0.05; 34 uncommon and 7 common). The tail of the p-value distribution of benign variants was as expected (0 results with p<0.001; expected ≤ 1 and 2 results with p<0.01; expected ≤ 4.2), suggesting that the overall study design and quality control were successful. Among the 201 putatively functional variants, one had p<0.001 (alpha=0.05) compared to the 0/421 observed among benign variants. The lack of inflated p-values points out that confounders such as stratification are not causing false positives.
Table 1.
The observed variants with significant associations to preeclampsia. Counts of minor alleles are given for the combined data; 615 cases and 2,094 controls, and per FINNPEC data set (in parentheses); 487 of 500 women with PE and 187 of 190 control women passed quality control.
| RSID number | Gene name | P-value* | OR (95% confidence interval) | MAF in total sample | Count with minor allele ncases/ncontrols FINNPEC minor allele count in parentheses (cases/controls) |
HWE | Consequence (distance from exon, base pairs) | LoFtool† |
|---|---|---|---|---|---|---|---|---|
| rs35832528 | FLT1 | 2.49E-4 | 0.387 (0.205 – 0.678) | 0.010 | 14/122 (7/6) | 1 | missense variant, E982A | 0.463 |
| rs141440705 | FLT1 | 0.003 | 0.442 (0.233 – 0.779) | 0.010 | 14/106 (8/5) | 1 | missense variant, R54S | 0.463 |
| rs61758484 | CORIN | 0.003 | 2.658 (1.320 – 5.261) | 0.013 | 17/22 (16/1) | 1 | non coding transcript exon variant, E10K | 0.201 |
| rs34106916 | ANGPTL1 | 0.006 | 3.186 (1.325 – 7.597) | 0.008 | 12/13 (8/3) | 1 | synonymous variant, Q103Q | 0.658 |
| rs61759670 | CORIN | 0.010 | 2.082 (1.147 – 3.693) | 0.016 | 21/35 (18/4) | 1 | missense variant, Y907T | 0.201 |
| rs80338240 | JAG1 | 0.010 | 0.256 (0.051 – 0.806) | 0.004 | 3/40 (2/4) | 1 | intron variant (−11) | 0.006 |
| rs147998709 | GPR98 | 0.011 | 13.757 (1.360 – 675.490) | 0.003 | 4/1 (3/0) | 0.004 | intron variant (+24) | 0.977 |
| rs2290843 | ADAM12 | 0.011 | 0.726 (0.561 – 0.932) | 0.071 | 83/383 (63/33) | 1 | synonymous variant, T326T | 0.320 |
| rs61760500 | CORIN | 0.011 | 2.550 (1.179 – 5.382) | 0.010 | 14/19 (11/3) | 1.001 | intron variant (+13) | 0.201 |
| rs147942437 | ADAM28 | 0.012 | 5.181 (1.226 – 24.998) | 0.004 | 6/4 (5/0) | 1.002 | missense variant, L449P | 0.994 |
| rs13406336 | ACVR1 | 0.013 | 2.423 (1.129 – 5.062) | 0.011 | 14/20 (13/2) | 1.003 | missense variant, A15G | 0.138 |
| rs142436579 | ADAM28 | 0.014 | 0.366 (0.129 – 0.852) | 0.005 | 6/56 (4/3) | 1.004 | missense variant, R219S | 0.994 |
| rs140437272 | INHBE | 0.014 | 0.477 (0.236 – 0.881) | 0.013 | 12/85 (11/6) | 1.005 | missense variant, P27L | 0.786 |
| rs201756397 | FLT4 | 0.014 | Inf (1.283 – Inf) | 0.001 | 3/0 (2/0) | 1.006 | synonymous variant, E926E | 0.023 |
| rs80069610 | GPR98 | 0.016 | 2.499 (1.122 – 5.411) | 0.010 | 13/18 (11/2) | 1.007 | synonymous variant, V1101V | 0.977 |
| rs139608664 | INHA | 0.018 | 5.716 (1.110 – 36.859) | 0.003 | 5/3 (4/0) | 1.008 | synonymous variant, S225R | 0.046 |
| rs4556933 | ACVR1C | 0.019 | 0.847 (0.735 – 0.976) | 0.323 | 1157/381 (314/120) | 0.113 | synonymous variant, F38F | 0.076 |
| rs41302834 | GPR98 | 0.022 | 3.451 (1.031 – 11.557) | 0.007 | 7/7 (7/2) | 1.009 | missense variant, D1944N | 0.977 |
| rs3736061 | FLT4 | 0.023 | 0.803 (0.665 – 0.973) | 0.135 | 485/175 (143/39) | 0.868 | synonymous variant, L252L | 0.023 |
| rs3741849 | PZP | 0.039 | 1.309 (1.004 – 1.696) | 0.068 | 88/235 (71/21) | 1 | synonymous variant, K563K, splice region variant | 0.988 |
| rs34307240 | LCT | 0.026 | 2.201 (1.038 – 4.515) | 0.010 | 14/22 (12/2) | 1.011 | missense variant, D106E | 0.571 |
| rs36032184 | INHA | 0.027 | 1.906 (1.058 – 3.346) | 0.017 | 21/38 (20/3) | 1.012 | synonymous variant, G109G | 0.046 |
| rs2228048 | TGFBR2 | 0.029 | 0.672 (0.459 – 0.963) | 0.030 | 38/191 (27/13) | 0.450 | synonymous variant, N354N | 0.060 |
| rs138819536 | INHBA-AS1 | 0.032 | 4.311 (0.926 – 21.758) | 0.005 | 5/4 (4/1) | 1 | missense variant, R229Q | 0.043 |
| rs1466360 | ADAM12 | 0.033 | 1.152 (1.010 – 1.315) | 0.414 | 1852/504 (404/152) | 0.691 | intron variant (+30) | 0.320 |
| rs56133834 | TEK | 0.034 | 3.450 (0.921 – 12.934) | 0.004 | 6/6 (6/0) | 1 | synonymous variant, E986E | 0.046 |
| rs140593977 | TREX1 | 0.034 | 3.448 (0.920 – 12.928) | 0.004 | 6/6 (6/0) | 1 | downstream gene variant (−44) | 0.824 |
| rs1466361 | ADAM12 | 0.034 | 1.152 (1.010 – 1.316) | 0.414 | 1810/501 (404/152) | 0.691 | intron variant (−46) | 0.320 |
| rs148671842 | EHD3 | 0.035 | 2.372 (0.992 – 5.459) | 0.007 | 11/16 (8/2) | 1 | synonymous variant, E188E | 0.139 |
| rs115734907 | KDR | 0.035 | 0.695 (0.483 – 0.979) | 0.029 | 42/205 (28/11) | 1 | intron variant (+12) | 0.196 |
| rs1554286 | IL10 | 0.037 | 1.203 (1.009 – 1.438) | 0.153 | 761/191 (145/60) | 0.231 | intron variant (+18) | 0.538 |
| rs2453040 | NOTCH2 | 0.037 | 0.818 (0.676 – 0.992) | 0.141 | 482/172 (144/45) | 0.423 | intron variant (−45) | 0.016 |
| rs116951780 | LCT | 0.038 | 10.333 (0.829 – 541.218) | 0.001 | 3/1 (2/0) | 1 | synonymous variant, NMD transcript variant, A921A | 0.571 |
| rs138894008 | TEK | 0.038 | 10.333 (0.829 – 541.218) | 0.002 | 3/1 (3/0) | 1 | missense variant, R479H | 0.046 |
| rs148588802 | FLT1 | 0.038 | 10.328 (0.829 – 540.960) | 0.002 | 3/1 (3/0 | 1 | intron variant (+40) | 0.463 |
| rs200071734 | FLT4 | 0.040 | 10.114 (0.811 – 529.805) | 0.002 | 3/1 (3/0) | 1 | missense variant, V157M | 0.023 |
| rs150123876 | ANGPT4 | 0.042 | 0.348 (0.090 – 0.968) | 0.007 | 4/39 (3/7) | 1 | missense variant, R25H | 0.773 |
| rs368518386 | FLT4 | 0.046 | 5.581 (0.799 – 61.819) | 0.003 | 4/2 (4/0) | 1 | intron variant (−43) | 0.023 |
| rs3736062 | FLT4 | 0.046 | 1.547 (0.988 – 2.374) | 0.024 | 33/74 (26/6) | 1 | synonymous variant, Y531Y | 0.023 |
| rs7830 | NOS3 | 0.049 | 0.877 (0.769 – 1.002) | 0.434 | 1635/525 (411/172) | 0.346 | intron variant (+11) | 0.817 |
| rs61763183 | FLT1 | 0.050 | 1.653 (0.967 – 2.754) | 0.020 | 24/50 (20/7) | 1 | downstream gene variant (+73) | 0.463 |
Non-adjusted p-values.
LoF tool - loss of function tool
MAF-minor allele frequency
HWE-Hardy-Weinberg Equilibrium
Focusing on 146 putatively functional (missense or truncating) rare and low-frequency variants, we observed an excess (observed 13, expected 7.3) of nominally associated variants (p<0.05). The strongest associations observed were in FLT1, with two missense variants associated to PE: NM_002019.4:p.E982A (rs35832528; p-value=2.5E-4, odds ratio (OR)=0.387, withstands correction for multiple testing for the 146 rare and low-frequency coding variants), and NM_001159920.1:p.R54S (rs141440705; p-value=0.0027, OR=0.44). These variants are enriched in the Finnish population, with minor allele frequencies being 0.026 (1000 Genomes MAF=0.0014) and 0.017 (1000 Genomes MAF=0.0008), respectively (www.sisuproject.fi).
E982A and R54S were examined across registry-derived disease endpoints. Both of the assessed SNPs protected from heart failure (Fisher’s exact p-value=0.007 for both, E982A: OR=0.368 95%CI=0.164–0.830, cases/controls=301/10956, minor allele (G) count in cases/controls=6/593; R54S: OR=0.340 95%CI=0.140–0.826) cases/controls=297/10960, minor allele (G) count in cases/controls=5/543).
Discussion
We discovered low frequency protective genetic variants in FLT1 that contributed to lower PE risk. We also found associating genetic variants in known candidate genes using a targeted sequencing approach.. These FLT1 variants may also be associated with lower risk of heart failure.
FLT1 codes for vascular endothelial growth factor receptor 1 (VEGFR1). It consists of seven immunoglobulin (Ig)-like domains in an extracellular ligand-binding region, a transmembrane segment and a cytoplasmic region containing a tyrosine kinase (TK) domain 23. VEGFR1 is essential for survival by negatively regulating the levels of endogenous vascular endothelial growth factor (VEGF). Internalization and signaling of functional VEGF receptors will enhance angiogenic growth of blood vessels 24. Sustaining angiogenesis is necessary for circulation and anti-angiogenic treatment causes cardiovascular morbidity 25. There are multiple isoforms of VEGFR1 of which the soluble forms have been implicated in PE. The soluble forms of VEGFR1 only contain the extracellular parts of the protein encoded by the first 13 of 30 exons. In preeclampsia, an excess of VEGFR1 of placental origin has been recorded 26. Furthermore, after healthy endothelium is restored, elevated levels of soluble VEGFR1 are observed in women with a history of PE 27.
Increased levels of sFLT1/sVEGFR1 have also been indicated in peripartum cardiomyopathy where high levels of sFLT1 correlate with the symptoms’ severity 28. Similarly, heart failure after myocardial infarction independent of pregnancy is reflected in extreme levels of sFLT1 29. Rhee et al. report extreme sFLT1 levels in at least the 95th percentile in a cases of heart failure during pregnancy 30. Although further research is required to assess the effect of underlying risk factor profile in preeclamptic women’s increased risk of heart failure in later life, it was recently shown in a meta-analysis of results from seven studies that 3.6-fold increase in risk of heart failure is associated with preeclampsia, particularly during the time period of 1–10 years after preeclampsia 30. Preeclampsia increases the risk of peripartum cardiomyopathy and it has been suggested that sFLT1 may be toxic to the heart. Also the driver of the cardiac dysfunction in susceptible preeclamptics is likely mediated by antiangiogenic factors 31. Our results indicate that FLT1 variants that protect from preeclampsia may also protect from heart failure thereby adding to the growing body of evidence that imbalance of angiogenic factors may be the link between preeclampsia and cardiovascular morbidity in later life in susceptible individuals. Novel therapeutic options for these individuals may include blocking sFlt1 production, or neutralizing antibodies against angiogenic proteins 32, 33.
From the observed associating variants in FLT1, rs141440705 causing R54S is more likely of functional importance. This is because it results in a polar change (pos->neutral) within the Ig-like domain 1, a functional part of the protein 34. Rs141440705 is located in the last nucleotide of an exon. Therefore, the variant may affect splicing as well as the coded amino acid sequence.
Epidemiological studies with many of the protective associations from severe cardiovascular diseases are strongest for R54S, supporting the suggestion of functional significance. The other variant resulting in the amino acid substitution E982A is not included in the s-FLT1 or the VEGFR1 isoform 2, which is the dominant isoform in the placenta. Even so, since VEGFR1 isoform 1 (canonical sequence) is expressed in vascular endothelium, E982A might have an important role in pregnancy in mediating decidual blood flow and in remodeling the spiral arteries.
We also found support for the role of several other candidate genes in PE. Corin has been previously indicated in PE with population-specific variants 35. Stepanian et al. report very similar odds ratios of approximately 2.5 at two intronic SNPs, rs2271036 and rs2271037 to our finding at rs61759670 and rs61760500 35. Rs13406336 and rs4556933 in activin A receptor type 1C (ACVR1C) have been previously listed as a candidate SNPs, although no significant association to PE was established in a Norwegian population 36. Interestingly, rs7830 features in numerous studies that pinpoint the association of nitric oxide synthase 3 (NOS3) to a spectrum of multifactorial diseases. Most relevantly, a haplotype including rs7830 was found to protect women from pregnancy hypertension and PE 37. Rs1554286 in interleukin 10 (IL-10) has been shown to belong to an intronic haplotype, which strongly predisposes women of Bahraini Arab population to idiopathic recurrent miscarriages 38. Our findings support the hypothesis that common immunological etiology may be shared between recurrent miscarriages and PE 39, 40.
Perspectives
To our knowledge, this study provides the first evidence that maternal FLT1 sequence variants associate with lower PE susceptibility. Further research is required to pinpoint the mechanism of protection from PE and heart failure and the effect of FLT1 variants on gene transcription. Genetic associations may open new avenues of drug development once the functional consequences of our findings are further deciphered. Genetic protection from PE due to FLT1 variants may also protect these women from heart failure in later life.
Supplementary Material
Novelty and significance.
1. What Is New?
We have established a genetic association between PE mothers and the FLT1 gene.
The same variants may also protect Finnish women from heart failure in later life
2. What Is Relevant?
Preeclampsia is a common hypertensive disorder of the pregnancy.
Identification of protective variants in FLT1, a known candidate gene, opens an opportunity for drug development to target the women most at risk of the disease.
3. Summary of the conclusions of the study
Genetic variants within the maternal Flt1 protect Finnish women from preeclampsia.
Acknowledgments
We thank the participants of the FINNPEC study as well as those in the National FINRISK Study. The FinMetSeq-project provided most of the FINRISK genotypes for which we are grateful.
Funding
This study was supported by Jane and Aatos Erkko Foundation, The Academy of Finland (121196 and 278941), Sigrid Jusélius and Alfred Kordelin Foundations and National Institutes of Health grants U54 HL112303 (JPA), RO1 GM099111-20A1(JPA), and P30 AR048335 (EDOR). Finnish Medical Foundation, University of Helsinki Funds, Special State Subsidy for Health Research (EVO funding), Sakari and Päivikki Sohlberg Foundation, Novo Nordisk Foundation, Signe and Ane Gyllenberg Foundation, and Foundation for Pediatric Research contributed to FINNPEC sample collection. JK is a recipient of The Royal Society Wolfson Research Excellence award. MT is a recipient of the Ruth L. Kirschstein National Research Service Award
Footnotes
Disclosures
None.
References
- 1.Lisonkova S, Joseph KS. Incidence of preeclampsia: Risk factors and outcomes associated with early-versus late-onset disease. Am J Obstet Gynecol. 2013;209:544. doi: 10.1016/j.ajog.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. World Health Report: Make every mother and child count. Geneva, Switzerland: World Health Organization; 2005. [Google Scholar]
- 3.Duley L. The global impact of pre-eclampsia and eclampsia. Semin Perinatol. 2009;33:130–137. doi: 10.1053/j.semperi.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Staff AC, Redman CW, Williams D, Leeson P, Moe K, Thilaganathan B, Magnus P, Steegers EA, Tsigas EZ, Ness RB, Myatt L, Poston L, Roberts JM Global Pregnancy Collaboration (CoLab) Pregnancy and long-term maternal cardiovascular health: Progress through harmonization of research cohorts and biobanks. Hypertension. 2016;67:251–260. doi: 10.1161/HYPERTENSIONAHA.115.06357. [DOI] [PubMed] [Google Scholar]
- 5.Agarwal I, Karumanchi SA. Preeclampsia and the anti-angiogenic state. Pregnancy Hypertens. 2011;1:17–21. doi: 10.1016/j.preghy.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeisler H, Llurba E, Chantraine F, Vatish M, Staff AC, Sennstrom M, Olovsson M, Brennecke SP, Stepan H, Allegranza D, Dilba P, Schoedl M, Hund M, Verlohren S. Predictive value of the sFlt-1:PlGF ratio in women with suspected preeclampsia. N Engl J Med. 2016;374:13–22. doi: 10.1056/NEJMoa1414838. [DOI] [PubMed] [Google Scholar]
- 7.Laivuori H, Lahermo P, Ollikainen V, Widen E, Haiva-Mallinen L, Sundstrom H, Laitinen T, Kaaja R, Ylikorkala O, Kere J. Susceptibility loci for preeclampsia on chromosomes 2p25 and 9p13 in Finnish families. Am J Hum Genet. 2003;72:168–177. doi: 10.1086/345311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chappell S, Morgan L. Searching for genetic clues to the causes of pre-eclampsia. Clin Sci (Lond) 2006;110:443–458. doi: 10.1042/CS20050323. [DOI] [PubMed] [Google Scholar]
- 9.Lundmark PE, Liljedahl U, Boomsma DI, Mannila H, Martin NG, Palotie A, Peltonen L, Perola M, Spector TD, Syvanen AC. Evaluation of HapMap data in six populations of European descent. Eur J Hum Genet. 2008;16:1142–1150. doi: 10.1038/ejhg.2008.77. [DOI] [PubMed] [Google Scholar]
- 10.Nevanlinna HR. The Finnish population structure. A genetic and genealogical study. Hereditas. 1972;71:195–236. doi: 10.1111/j.1601-5223.1972.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 11.Lim ET, Wurtz P, Havulinna AS, et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014:10. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jääskeläinen T, Heinonen S, Kajantie E, Kere J, Kivinen K, Pouta A, Laivuori H for the FINNPEC Study Group. Cohort profile: The Finnish genetics of pre-eclampsia consortium (FINNPEC) BMJ Open. 2016;6:e013148. doi: 10.1136/bmjopen-2016-013148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borodulin K, Vartiainen E, Peltonen M, Jousilahti P, Juolevi A, Laatikainen T, Mannisto S, Salomaa V, Sundvall J, Puska P. Forty-year trends in cardiovascular risk factors in Finland. Eur J Public Health. 2015;25:539–546. doi: 10.1093/eurpub/cku174. [DOI] [PubMed] [Google Scholar]
- 14.Triebwasser M. Washington University in St. Louis, Arts & Sciences Electronic Theses and Dissertations; Paper 427. 2015. Excessive complement activation due to genetic haploinsufficiency of regulators in multiple human diseases. [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chheda H, Palta P, Pirinen M, McCarthy S, Walter K, Koskinen S, Salomaa V, Daly M, Durbin R, Palotie A, Aittokallio T, Ripatti S. Whole-genome view of the consequences of a population bottleneck using 2926 genome sequences from Finland and United Kingdom. Eur J Hum Genet. 2017;25:477–484. doi: 10.1038/ejhg.2016.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glusman G, Caballero J, Mauldin DE, Hood L, Roach JC. Kaviar: An accessible system for testing SNV novelty. Bioinformatics. 2011;27:3216–3217. doi: 10.1093/bioinformatics/btr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the ensembl API and SNP effect predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibuya M. Structure and dual function of vascular endothelial growth factor receptor-1 (flt-1) Int J Biochem Cell Biol. 2001;33:409–420. doi: 10.1016/s1357-2725(01)00026-7. [DOI] [PubMed] [Google Scholar]
- 24.Boulanger CM. Endothelium. Arterioscler Thromb Vasc Biol. 2016;36:e26–31. doi: 10.1161/ATVBAHA.116.306940. [DOI] [PubMed] [Google Scholar]
- 25.Abdel-Qadir H, Ethier JL, Lee DS, Thavendiranathan P, Amir E. Cardiovascular toxicity of angiogenesis inhibitors in treatment of malignancy: A systematic review and meta-analysis. Cancer Treat Rev. 2017;53:120–127. doi: 10.1016/j.ctrv.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Maynard SE, Min JY, Merchan J, Lim KH, Li J, Mondal S, Libermann TA, Morgan JP, Sellke FW, Stillman IE, Epstein FH, Sukhatme VP, Karumanchi SA. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J Clin Invest. 2003;111:649–658. doi: 10.1172/JCI17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuzcu ZB, Asicioglu E, Sunbul M, Ozben B, Arikan H, Koc M. Circulating endothelial cell number and markers of endothelial dysfunction in previously preeclamptic women. Am J Obstet Gynecol. 2015;213:533. doi: 10.1016/j.ajog.2015.06.043. [DOI] [PubMed] [Google Scholar]
- 28.Damp J, Givertz MM, Semigran M, et al. Relaxin-2 and soluble Flt1 levels in peripartum cardiomyopathy: Results of the multicenter IPAC study. JACC Heart Fail. 2016;4:380–388. doi: 10.1016/j.jchf.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onoue K, Uemura S, Takeda Y, et al. Usefulness of soluble fms-like tyrosine kinase-1 as a biomarker of acute severe heart failure in patients with acute myocardial infarction. Am J Cardiol. 2009;104:1478–1483. doi: 10.1016/j.amjcard.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 30.Wu P, Haththotuwa R, Kwok CS, Babu A, Kotronias RA, Rushton C, Zaman A, Fryer AA, Kadam U, Chew-Graham CA, Mamas MA. Preeclampsia and future cardiovascular health: A systematic review and meta-analysis. Circ Cardiovasc Qual Outcomes. 2017:10. doi: 10.1161/CIRCOUTCOMES.116.003497. Epub 2017 e003497. [DOI] [PubMed] [Google Scholar]
- 31.Shahul S, Medvedofsky D, Wenger JB, Nizamuddin J, Brown SM, Bajracharya S, Salahuddin S, Thadhani R, Mueller A, Tung A, Lang RM, Arany Z, Talmor D, Karumanchi SA, Rana S. Circulating antiangiogenic factors and myocardial dysfunction in hypertensive disorders of pregnancy. Hypertension. 2016;67:1273–1280. doi: 10.1161/HYPERTENSIONAHA.116.07252. [DOI] [PubMed] [Google Scholar]
- 32.Thadhani R. Inching towards a targeted therapy for preeclampsia. Hypertension. 2010;55:238–240. doi: 10.1161/HYPERTENSIONAHA.109.143933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maynard SE, Karumanchi SA. Angiogenic factors and preeclampsia. Semin Nephrol. 2011;31:33–46. doi: 10.1016/j.semnephrol.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christinger HW, Fuh G, de Vos AM, Wiesmann C. The crystal structure of placental growth factor in complex with domain 2 of vascular endothelial growth factor receptor-1. J Biol Chem. 2004;279:10382–10388. doi: 10.1074/jbc.M313237200. [DOI] [PubMed] [Google Scholar]
- 35.Stepanian A, Alcais A, de Prost D, Tsatsaris V, Dreyfus M, Treluyer JM, Mandelbrot L ECLAXIR Study Group. Highly significant association between two common single nucleotide polymorphisms in CORIN gene and preeclampsia in Caucasian women. PLoS One. 2014;9:e113176. doi: 10.1371/journal.pone.0113176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roten LT, Johnson MP, Forsmo S, Fitzpatrick E, Dyer TD, Brennecke SP, Blangero J, Moses EK, Austgulen R. Association between the candidate susceptibility gene ACVR2A on chromosome 2q22 and pre-eclampsia in a large Norwegian population-based study (the HUNT study) Eur J Hum Genet. 2009;17:250–257. doi: 10.1038/ejhg.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muniz L, Luizon MR, Palei AC, Lacchini R, Duarte G, Cavalli RC, Tanus-Santos JE, Sandrim VC. eNOS tag SNP haplotypes in hypertensive disorders of pregnancy. DNA Cell Biol. 2012;31:1665–1670. doi: 10.1089/dna.2012.1768. [DOI] [PubMed] [Google Scholar]
- 38.Qaddourah RH, Magdoud K, Saldanha FL, Mahmood N, Mustafa FE, Mahjoub T, Almawi WY. IL-10 gene promoter and intron polymorphisms and changes in IL-10 secretion in women with idiopathic recurrent miscarriage. Hum Reprod. 2014;29:1025–1034. doi: 10.1093/humrep/deu043. [DOI] [PubMed] [Google Scholar]
- 39.Redman CWG, Sargent IL. Immunology of pre-eclampsia. American Journal of Reproductive Immunology. 2010;63:534–543. doi: 10.1111/j.1600-0897.2010.00831.x. [DOI] [PubMed] [Google Scholar]
- 40.Robertson SA, Bromfield JJ, Tremellen KP. Seminal ‘priming’ for protection from pre-eclampsia-a unifying hypothesis. J Reprod Immunol. 2003;59:253–265. doi: 10.1016/s0165-0378(03)00052-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.