Abstract
Changes in metabolism can be initiated in response to signals received from other cells. An example of this is provided by macrophages that have been stimulated by IL-4 to become alternatively/M2 activated. In these cells, fatty acid oxidation is increased and this is critical for M2 activation. Compared to resting macrophages, M2 macrophages also exhibit changes in glucose metabolism that we have found are essential for activation. In other cell types, mTORC2 has been linked to enhanced glycolysis. We have found that mTORC2 operates in parallel with the IL-4Rα/Stat6 pathway to facilitate increased glycolysis during M2 activation. Our data strongly implicate PI3K and AKT signaling initiated by M-CSF as components in this pathway, and indicate that downstream induction of IRF4 expression plays a role in metabolic reprograming to support M2 activation. We show that loss of mTORC2 in macrophages suppresses tumor growth and decreases immunity to a parasitic nematode.
Introduction
Macrophages are tissue-resident cells that play critical roles in in a broad range of immunologic and homeostatic processes (Ginhoux et al., 2015; Wynn et al., 2013). The ability of these cells to serve multiple functions reflects their ability to express different genes in response to distinct extracellular signals including pathogen and damage associated molecular patterns and cytokines (Glass and Natoli, 2015; Wynn et al., 2013). IL-4, which can be made by a variety of innate and adaptive immune cells (Pulendran and Artis, 2012), induces a Stat6-dependent macrophage activation state referred to as M(IL-4), or M2 or “alternative” activation (Murray et al., 2014). M2 macrophages are important in immunity to parasitic helminths, tissue remodeling and wound repair, adipose tissue homeostasis, and tumor growth and metastasis.
Recent work has revealed that macrophage activation status is intrinsically linked to metabolic remodeling (O'Neill and Pearce, 2016). Initial studies established that fatty acid oxidation (FAO) and oxidative phosphorylation (OXPHOS) are enhanced in M2 macrophages, and critical for M2 activation (Huang et al., 2014; Odegaard and Chawla, 2011; Vats et al., 2006). Integrated metabolomic and transcriptomic studies revealed that the metabolic reprogramming that occurs during activation is more complex than originally envisaged, and uncovered enhanced use of glucose for UDP-GlcNAc synthesis as a metabolic signature of M2 macrophages (Jha et al., 2015). Moreover, a recent report showed that inhibition of glycolysis prevents the expression of a subset of genes that comprise the M2 activation module (Covarrubias et al., 2016).
It is now clear that manipulation of metabolic reprogramming in immune cells has therapeutic potential. Depending on context, being able promote or inhibit M2 activation could have therapeutic benefit and understanding how glucose metabolism is reprogrammed downstream of stimulation with IL-4, and how this is integrated with changes in FAO, would be an important step towards this goal. Recent work has implicated IL-4-induced signaling through AKT and mTORC1 in the regulation of glucose metabolism for M2 activation (Covarrubias et al., 2016), raising the possibility that the mTORC1 pathway may be good target for manipulating alternative activation. However, loss of Tsc1, a negative regulator of mTORC1, allows enhanced M1 and diminished M2 activation (Byles et al., 2013), indicating that the role of mTORC1 in M2 activation is context dependent (Covarrubias et al., 2016). Moreover, in brown adipose tissue mTORC2 has been shown to be responsible for AKT-induced increases in glycolysis (Albert et al., 2016), and mTORC2 has been implicated in glycolytic remodeling in tumors (Masui et al., 2015). Questions remain therefore about the role of mTOR in M2 activation and the potential for the contribution of mTORC1 and mTORC2 complexes to this process.
In light of these accumulated findings, we decided to address the roles of mTORC1 and mTORC2 in the metabolic reprograming that allows M2 activation. Our findings point to an mTORC2-mediated pathway, involving PI3K and AKT, as being essential for accentuated glucose metabolism to promote M2 activation, and implicate M-CSF as an upstream activator of this pathway. Our data indicate that this pathway profoundly influences FAO, and that its effects are mediated by interferon regulatory factor 4 (IRF4), a transcription factor that is important for M2 activation (Satoh et al., 2010), and which previously had been shown to play a critical role in metabolic reprogramming towards glycolysis during CD8+ T cell activation (Man et al., 2013; Yao et al., 2013). Our data show that IRF4 expression requires both mTORC2 and Stat6 pathways and provide an underlying mechanism to explain how glucose utilization is increased to support M2 activation.
Results
Glucose is crucial for M2 activation
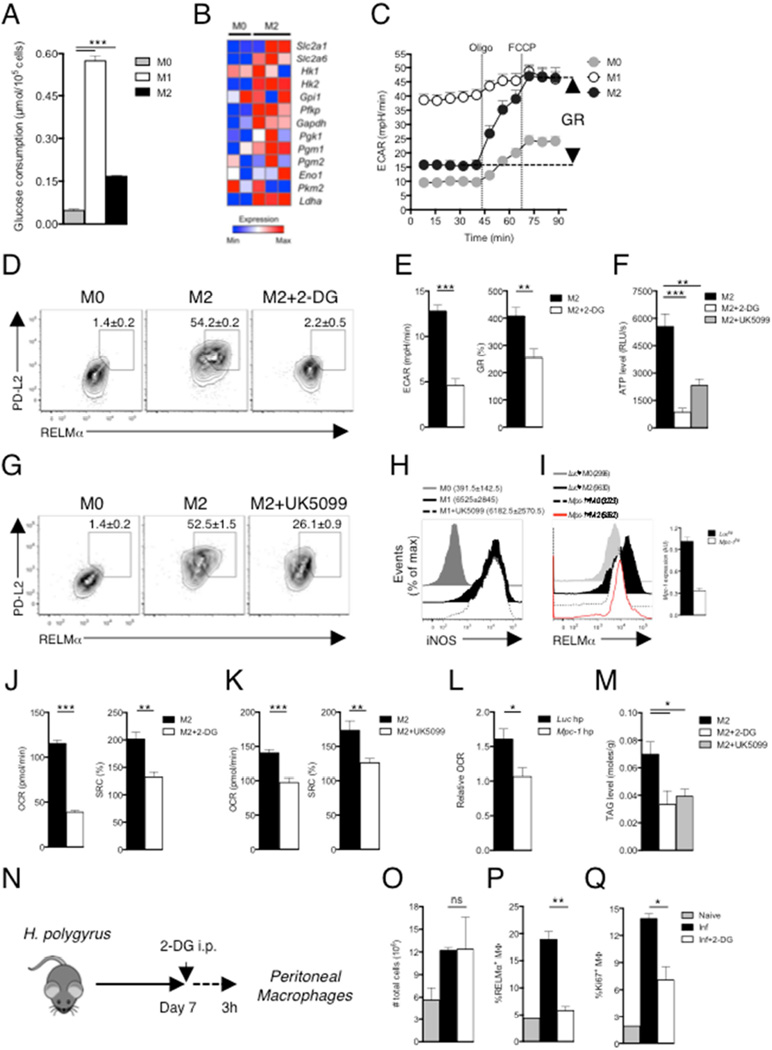
Macrophages grown from bone marrow were unstimulated (M0) or activated with IL-4 (M2) and glucose consumption and changes in extracellular acidification rates (ECAR, a measure of the production of lactic acid, an end product of cytoplasmic glucose metabolism) were assessed. Glucose consumption, expression of genes encoding enzymes in the glycolysis pathway, and basal ECAR were increased in M2 compared to M0 macrophages (Fig. 1A, B, C), and M2 macrophages exhibited increased glycolytic reserve (GR, defined as the ability to upregulate aerobic glycolysis, measureable as increased ECAR, following inhibition of mitochondrial ATP synthesis by oligomycin and inner membrane depolarization by FCCP, Fig. 1C). High GR is a measure of the increased ability of M2 cells to route pyruvate to lactate in order to meet their ATP needs through aerobic glycolysis. We asked whether increased glucose utilization is important for alternative activation by stimulating cells with IL-4 in the presence of 2-deoxyglucose (2-DG). Expression of the M2 activation markers RELMα and PD-L2, was markedly inhibited by 2-DG (Fig. 1D). Similar results were obtained when medium that lacked glucose was used (Fig. S1). As expected, 2-DG caused a reduction in basal ECAR and GR (Fig. 1E). These data confirm and expand recent findings on the importance of glucose for M2 activation (Covarrubias et al., 2016).
Figure 1. Glucose metabolism is essential for M2 macrophage activation.
(A) Glucose uptake by bone marrow macrophages cultured for 24 hr in medium alone (M0) or with IFNγ plus LPS (M1), or with IL-4 (M2). (B) Relative expression of genes encoding glycolysis pathway enzymes. (C) ECAR at baseline and after sequential treatment with oligomycin (Oligo) and FCCP. GR, glycolytic reserve. (D) PD-L2 and RELMα expression in macrophages cultured for 24 hr in IL-4 with or without 2-DG, and in M0 macrophages. (E) Effect of 2-DG on Basal ECAR and GR in M2 macrophages. (F) Levels of ATP in macrophages cultured for 24 hr without (M0) or with (M2) IL-4 with or without 2-DG or UK5099. (G) PD-L2 and RELMα expression in macrophages cultured for 24 hr in IL-4 with or without UK5099, and in M0 macrophages. (H) iNOS expression by macrophages cultured for 24 hr in M0 or M1-polarizing conditions with or without UK5099. (I) RELMα expression in M2 macrophages transduced with Luc hpRNA or Mpc-1 hpRNA retroviruses. (J–K) Basal OCR and SRC of M2 macrophages with or without 2-DG or UK5099. (L) Relative OCR of Mpc-1 hpRNA transduced M2 macrophages, relative to untransduced M0 macrophages. (M) Levels of TAGs in macrophages cultured for 24 hr with IL-4 with or without 2-DG or UK5099. (N) Plan to assess the role of glucose in M2 macrophage activation during helminth infection. (O) The total number of PEC; (P) the frequency of RELMα+ pMacs, and (Q) the percentage of pMacs expressing the proliferation marker Ki67 (Ki67+MΦ), in mice from experiment shown in (N). PEC from naïve mice (2 per group) were controls. *P < 0.05, **P < 0.005 and ***P < 0.0001 (Student’s t-test). In D–I data are from flow cytometry. In D, G, H and I data from pooled replicates from one experiment are shown. In D, G, numbers are percentages of macrophages that are positive for both markers and in H, I, numbers are mean fluorescence intensity (MFI). Numbers in these panels represent mean ± s.e.m of data from three or more independent experiments. In A, E, F, J–M and O–Q, data are mean ± s.e.m. of technical replicates from one experiment, representative of 3 or more independent experiments. In B data from macrophages harvested from two MO cultures and three M2 cultures are shown.
For comparison, macrophages classically activated with IFN-γ plus LPS (M1 macrophages), were also examined in these experiments. As expected (O'Neill and Pearce, 2016), M1 macrophages consumed significantly more glucose than M0 cells (Fig. 1A), exhibited elevated basal ECAR, and had very little GR since they effectively run maximal glycolysis at baseline (Fig. 1C).
The fact that M2 macrophages have large GR (Fig. 1C) indicated that glucose-derived pyruvate is entering mitochondria and being used to fuel the TCA cycle and support mitochondrial ATP synthesis in these cells. Consistent with this, we found that 2-DG and UK5099 (which inhibits the mitochondrial pyruvate carrier, MPC-1; (Halestrap, 1975)) caused declines in ATP levels in M2 macrophages (Fig. 1F). In addition, like 2-DG, UK5099 caused a reduction in the IL-4-induced expression of PD-L2 and RELMα (Fig. 1G). However, it had no effect on iNOS expression in M1 macrophages, in which pyruvate is largely converted to lactate due to the inhibition of the electron transport chain by NO (Fig. 1H) (Everts et al., 2012). Targeting MPC-1 with a short hairpin RNA (shRNA) also resulted in decreased commitment to the M2 phenotype, as measured by RELMα expression (Fig. 1I).
Previous work showed a requirement for OXPHOS for M2 activation (Vats et al., 2006). We found that 2-DG, UK5099 and Mpc-1-shRNA all inhibited IL-4-induced elevations in OXPHOS as measured by oxygen consumption rates (OCR) and/or SRC (Fig. 1J,K,L). We postulated previously that de novo fatty acid synthesis (FAS) could be contributing to the fueling of FAO in M2 macrophages (Huang et al., 2014). In this scenario, we assumed that, since lipolysis is necessary for FAO and M2 activation, de novo synthesized fatty acids (FA) would first need to be incorporated into triacylglycerols (TAGs) prior to use for FAO. Indeed, expression of both Fasn and Acaca, which encode FAS enzymes, were increased in M2 macrophages (Fig. S1B). We tested whether glycolysis and mitochondrial pyruvate import could be contributing to this pathway by measuring the effects of 2-DG and UK5099 on TAGs in M2 macrophages, and found that TAG levels were diminished when glucose use was inhibited (Fig. 1M). Moreover, the FAS inhibitor C75 prevented IL-4 induced increases in RELMα expression, but had no effect on iNOS expression in M1 cells (Fig. S2C,D), and Acaca-shRNA inhibited IL-4 induced increases in PD-L2 and RELMα expression and OCR (Fig. S2E). Together, these data indicate that glycolysis and mitochondrial pyruvate import are essential for M2 activation, possibly because they are being used to fuel FAS for increased FAO and OXPHOS.
Infection of mice with the gastrointestinal helminth parasite Heligmosomoides polygyrus bakeri (H. polygyrus) evokes a Th2 response in the mesenteric LN and M2 activation of peritoneal macrophages (pMacs) (Huang et al., 2014; Reynolds et al., 2012). To determine whether glucose metabolism is critical for M2 development in vivo, we injected H. polygyrus-infected mice with 2-DG i.p. and examined pMac activation status 3 h later (Fig. 1N). Although we found that the total number of peritoneal cells, which increased as a result of infection, was not affected by 2-DG, (Fig. 1O), the percentage of pMacs (defined as CD11b+F4/80+ cells) that expressed RELMα was significantly suppressed (Fig. 1P). Further, 2-DG suppressed pMac proliferation in infected mice (Fig. 1Q). Taken together, our data suggest that enhanced glucose metabolism is essential for M2 macrophage activation.
Metabolic reprogramming necessary for M2 activation requires mTORC2 signaling
mTOR is a component of two functionally distinct protein complexes (mTORC1 and 2) which are key regulatory molecules in the control of immune cell function and energy homeostasis (Weichhart et al., 2015). mTORC1 is implicated in the regulation of glucose metabolism, but while mTORC1 has been implicated in the expression of a subset of M2 genes, constitutive activation of mTORC1 has been shown to negatively regulate alternative activation (Byles et al., 2013; Covarrubias et al., 2016), raising the possibility that mTORC2 could also be playing a role in metabolic reprogramming for M2 activation. We found that NDRG1, and AKTS473, downstream targets in the mTORC2-dependent signaling pathway (Garcia-Martinez and Alessi, 2008), and S6KT389 (S6K), a downstream target in the mTORC1 pathway, were phosphorylated in M2 macrophages, indicating that both mTORC1 and mTORC2 pathways are active in these cells (Fig. 2A). The mTOR inhibitor, Torin 1 (Liu et al., 2010), which inhibits both mTORC1 and mTORC2 as indicated by decreased phosphorylation of their respective targets, S6K and AKTs473, in macrophages responding to IL-4 (Fig. S2A) effectively suppressed M2 activation (Fig. S2B). In contrast, rapamycin (at 20 nM, at which it selectively inhibits mTORC1 (Fig. S2A)) did not inhibit M2 activation (Fig. S2B). Moreover, Torin, but not rapamycin, inhibited increased uptake of glucose by M2 compared to M0 macrophages, as measured using flow cytometry to detect uptake of the fluorescent glucose analog 2-NBDG (Fig. S2D).
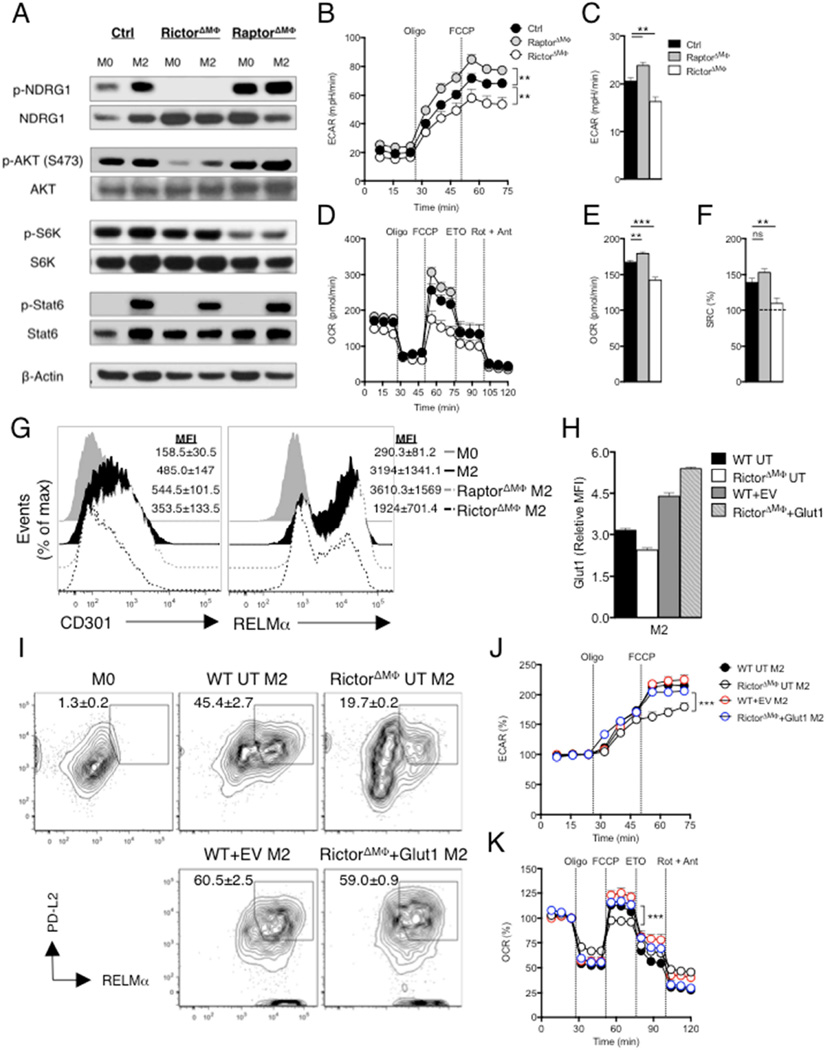
Figure 2. mTORC2 is required to promote metabolic reprogramming and cellular activation in IL-4 stimulated macrophages.
Bone marrow macrophages were from WT (Ctrl), RictorΔMΦ and RaptorΔMΦ mice. (A) Activity of mTORC1 and mTORC2 was assessed by immunoblot analysis for the indicated phosphoproteins from M0 macrophages, or macrophages stimulated for 3 hr with IL-4 (M2). (B) ECAR of M2 macrophages at baseline and following sequential treatment with oligomycin and FCCP. (C) Basal ECAR of M2 macrophages. (D) OCR of M2 macrophages at baseline and following sequential treatment with oligomycin, FCCP, etomoxir (ETO) and rotenone + antimycin. (E–F) Basal OCR and SRC of M2 macrophages. (G) Expression of CD301 and RELMα in M2 macrophages, assessed by flow cytometry. MFI: mean fluorescence intensity. (H–K) WT or RictorΔMΦ macrophages were either untransduced (UT) or transduced with retrovirus encoding a control reporter gene (EV-Ctrl)) or a reporter gene plus the Slc2a1 sequence (encodes Glut1), and stimulated with IL-4. Glut1 expression (H), and expression of PD-L2 and RELMα (I) were measured by flow cytometry. M0 WT macrophages were included in as a control in (I). ECAR (J) and OCR (K) were measured sequentially before and following the addition of inhibitors as indicated. In B-F, H, J and K, data are mean ± s.e.m. of technical replicates from one experiment representative of two or more independent experiments. Data in A, G and I are from pooled replicates from one experiment representative of two or more independent experiments. Numbers in G and I are mean MFI ± s.e.m (G) or mean % ± s.e.m, of date from two or more experiments. **P < 0.005 and ***P < 0.0001.
We next examined the response to IL-4 of macrophages in which key components of the mTORC2 and mTORC1 complexes, namely Rictor and Raptor, were deleted. Phosphorylation of NDRG1(Garcia-Martinez and Alessi, 2008) was lost in macrophages from Rictorfl/flLysMcre (RictorΔMΦ) mice, but increased in IL-4 stimulated macrophages from Raptorfl/flLysMcre (RaptorΔMΦ) mice (Fig. 2A), suggesting that mTORC1 might restrain mTORC2 activation in M2 macrophages. We also found that phosphorylation of AKTs473 was greatly diminished in RictorΔMΦ macrophages, but not in RaptorΔMΦ macrophages (Fig. 2A). As anticipated, S6K phosphorylation was diminished in RaptorΔMΦ macrophages but not RictorΔMΦ macrophages (Fig. 2A). Deletion of neither Raptor nor Rictor impaired IL-4 induced Stat 6 phosphorylation (Fig. 2A), indicating that Stat6 activation in M2 macrophages is mTOR-independent. We found that RictorΔMΦ M2 macrophages had lower GR and basal ECAR and RaptorΔMΦ M2 macrophages had higher GR and basal ECAR than control M2 macrophages (Fig. 2B,C). These changes in metabolism in RictorΔMΦ cells were linked to reduced IL-4-induced expression of genes encoding glycolysis pathway enzymes (Fig. S2E).
We showed that glycolysis and mitochondrial pyruvate import are essential for increased OXPHOS in M2 cells (Fig. 1). We reasoned that if mTORC2 is controlling glucose usage, RictorΔMΦ macrophages should exhibit diminished changes in OXPHOS following stimulation with IL-4. Consistent with this control and RaptorΔMΦ M2 macrophages behaved similarly to each other in a mitochondrial fitness test, but RictorΔMΦ M2 macrophages had significantly diminished baseline OCR and SRC (Fig. 2D,E,F). Moreover, the sensitivity of SRC to etomoxir (ETO), which inhibits mitochondrial Carnitine palmitoyl transferase-1 (Cpt1), and was apparent in control and RaptorΔMΦ M2 macrophages, was largely lost in RictorΔMΦ M2 macrophages (Fig. 2D), suggesting that FAO was diminished in the absence of Rictor. As expected, ETO inhibited M2 activation, and in our hands this result was recapitulated when Cpt1a expression was suppressed using a Cpt1a-hpRNA (Fig S2,G)
Finally, we asked whether deletion of Rictor affects the expression of M2 genes. We found that IL-4 induced expression of CD301, RELMα, Arg1, Ym1, Il10, Lipa, Cd36, Fabp4, Pparg and Ppargc1b was diminished when Rictor was deleted (Fig.2G; S2H). In contrast, expression of CD301 and RELMα was increased over control M2 levels when RaptorΔMΦ macrophages were stimulated with IL-4 (Fig. 2G). Raptor deletion had no effect on Ym1, Il10, Lipa, Cd36, Pparg or Ppargc1b expression, but did diminish expression of Arg1 and Fabp4 (Fig. S2H).
Our data indicate that mTORC2 controls M2 activation by regulating glucose metabolism and this in turn has effects on FAO. To further assess this, we asked whether enforcing expression of Glut1 (Slc2a1) would rescue the ability of RictorΔMΦ macrophages to become alternatively activated. We found that enforced expression of Glut1 (Fig. 2H) reversed the phenotype of RictorΔMΦ macrophages, allowing them to express levels of PD-L2 and RELMα in response to IL-4 that were equivalent to those expressed by IL-4 stimulated WT macrophages (Fig. 2I). Overexpression of Glut1 resulted in increased GR and SRC (Fig. 2J,K).
The PI3K, mTORC2, AKT pathway in M2 macrophages
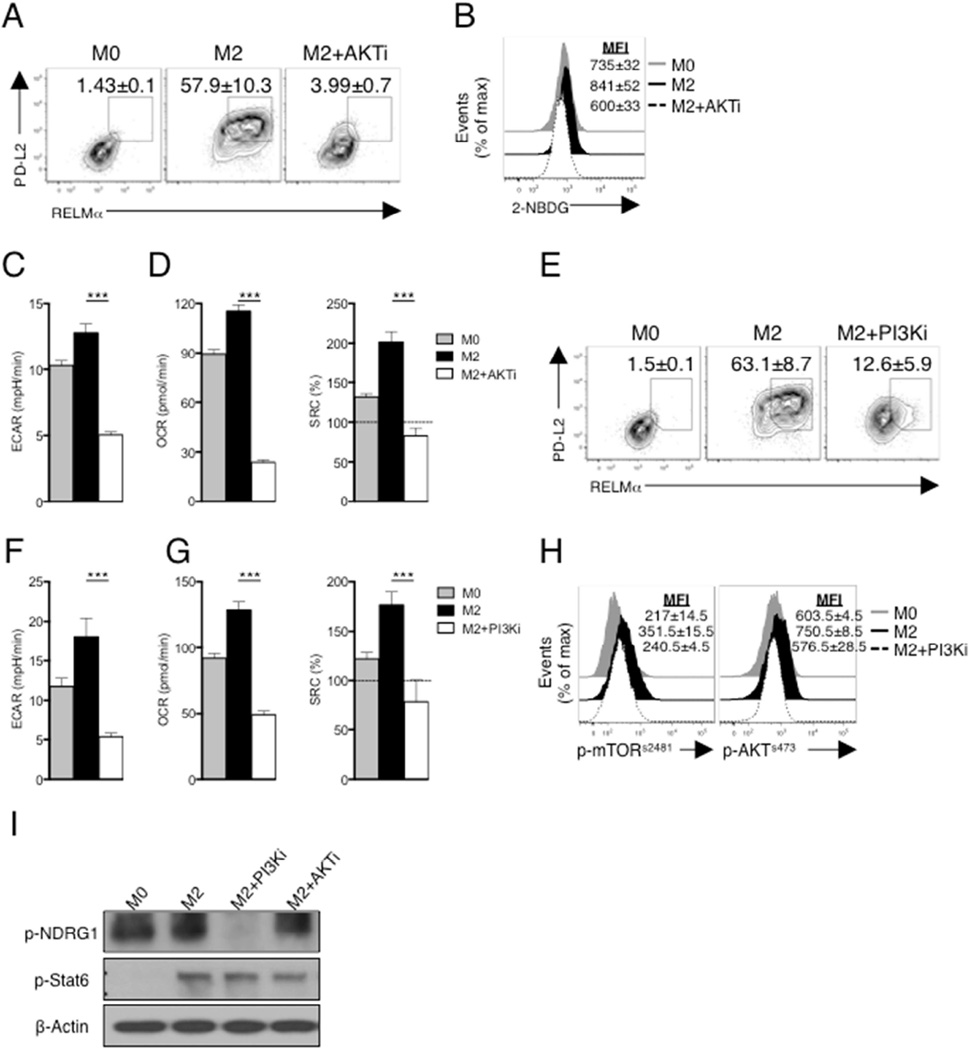
In recent work, we found that AKT is essential for regulating glycolytic metabolism in dendritic cells (Everts et al., 2014), and previous work has shown that AKT is important for M2 activation (Byles et al., 2013; Ruckerl et al., 2012), and increased glycolysis in these cells (Covarrubias et al., 2016). In keeping with these results, the AKT inhibitor triciribine suppressed M2 activation as assessed by PD-L2 and RELMα expression (Fig. 3A) and simultaneously blocked increases in glucose uptake and ECAR (Fig. 3B,C). Consistent with the functional link between glucose usage and OXPHOS in M2 activation, triciribine inhibited IL-4 induced increases in basal OCR and SRC (Fig. 3D). Our data collectively point to a pathway in which mTORC2-mediated phosphorylation of AKT is critical for M2 activation.
Figure 3. PI3K/AKT signaling is essential for M2 activation.
(A) PD-L2 and RELMα expression by macrophages cultured for 24 hr in the absence (M0) or presence of IL-4 (M2) plus or minus triciribine (AKTi). (B) 2-NBDG staining of macrophages treated as in panel A. (C,D) Basal ECAR, basal OCR and SRC of macrophages treated as in A. (E) Expression of PD-L2 and RELMα by M0 macrophages, or by M2 macrophages with or without LY294002 (PI3Ki) for 24 hr. (F,G) Basal ECAR, basal OCR and SRC in macrophages treated as in panel E. (H) Phosphorylation of mTORs2481 (p-mTORs2481) and AKTs473 (p- AKTs473) in macrophages treated as in panel E. (I) Phosphorylation of NDRG1 and Stat6 from unstimulated macrophages (M0) or macrophages stimulated with IL-4 (M2) for 3 hr in the presence or absence of PI3Ki and AKTi, assessed by immunoblot analysis. Data in A,B, E and H are from flow cytometry, and are from individual experiments, but numbers represent mean % or mean MFI, ± s.e.m, of data from three or more independent experiments. In C, D, F and G, data are mean ± s.e.m. of technical replicates from one experiment representative of three or more independent experiments. Data in I are from one experiment representative of three independent experiments. ***P < 0.0001.
PI3K is implicated in M2 activation, and PI3K has been shown to directly activate mTORC2 (Weichhart et al., 2015; Zinzalla et al., 2011). We therefore asked whether PI3K plays a role in activating mTORC2 in M2 activation. We found that the PI3K inhibitor LY294002 strongly suppressed IL-4 induced expression of PD-L2 and RELMα (Fig. 3E) and associated elevations in basal ECAR, and basal OCR and SRC (Fig. 3F,G). mTORs2481 phosphorylation, a marker of mTORC2 activation (Copp et al., 2009), was increased in M2 compared to M0 macrophages, and this effect was diminished by inhibition of PI3K (Fig. 3H). Moreover, induced phosphorylation of AKTs473 in M2 macrophages was prevented by inhibition of PI3K (Fig. 3H). We also found that phosphorylation of the mTORC2 target NDRG1 was inhibited by LY294002 (Fig. 3H). AKT inhibition had no effect on NRDG1 phosphorylation (Fig. 3I), confirming that AKT activation is occurring downstream of mTORC2 in M2 macrophages. Consistent with previous reports, inhibition of neither PI3K nor AKT had any measurable effect on Stat6 phosphorylation (Fig. 3I) (Covarrubias et al., 2016; Munugalavadla et al., 2005), indicating that the PI3K/mTORC2/AKT pathway is occurring in parallel to the Stat6 pathway following IL-4 stimulation.
In all, our findings suggest a pathway in which, following stimulation with IL-4, mTORC2 is activated by PI3K, and then itself activates AKT, and that this pathway is important for the changes in metabolism that are essential for M2 activation.
M-CSF synergizes with IL-4 to drive mTORC2 activation
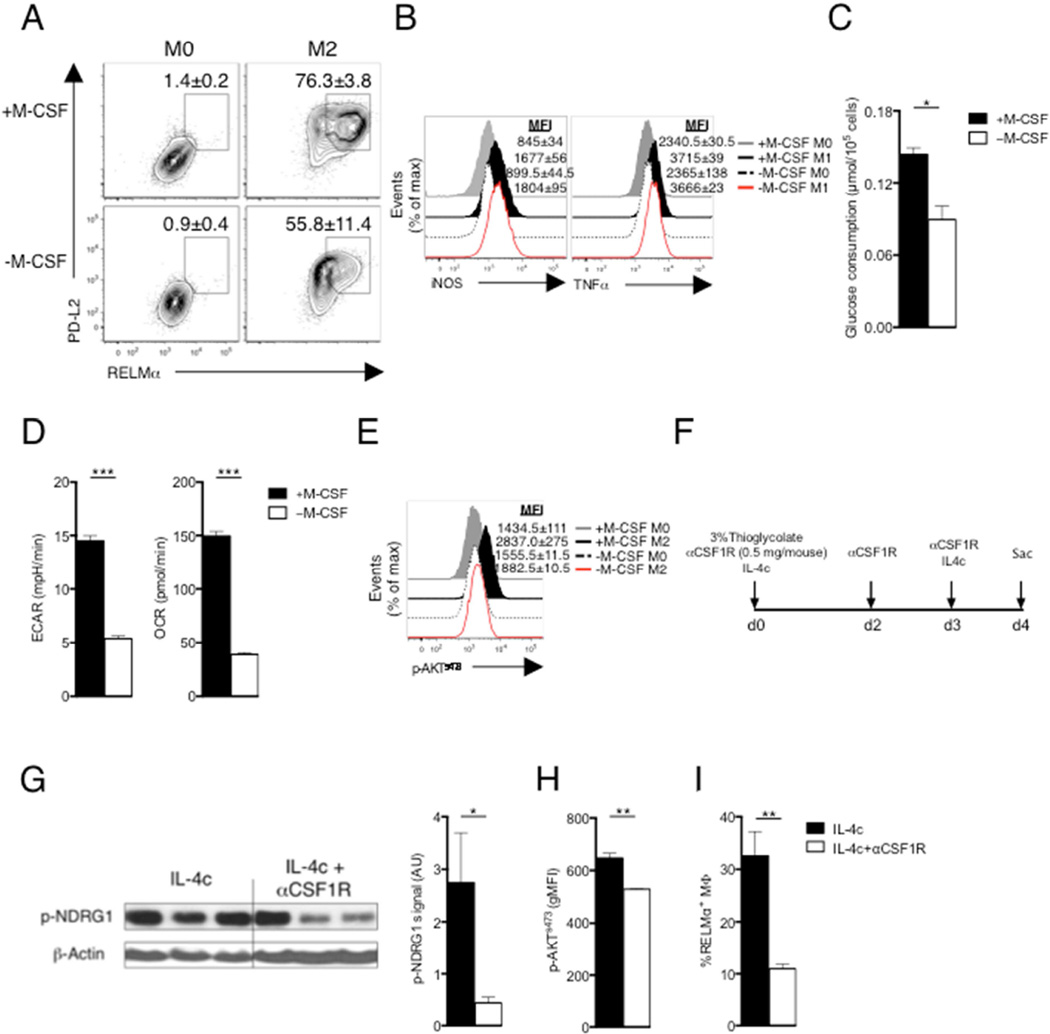
Given the recognized role of PI3K in growth factor initiated signaling (Zinzalla et al., 2011), and our finding that it is critical for mTORC2 activation in M2 macrophages, we hypothesized that the growth factor, macrophage colony-stimulating factor (M-CSF), which plays an important role for proliferation, differentiation and survival of macrophages, and which is used to grow the bone marrow-derived macrophages used herein, could be synergizing with IL-4 to activate mTORC2. This is plausible since previous work indicated that M-CSF can induce AKTs473 phosphorylation (Heller et al., 2008), and control glucose uptake by macrophages (Chang et al., 2009). We found that commitment to expression of RELMα and PD-L2 in response to IL-4 was significantly reduced when M-CSF was withdrawn (Fig, 4A). This treatment had no effect on M1 activation as measured by expression of iNOS and TNF-α (Fig. 4B). Reduced M2 activation in the absence of M-CSF was associated with reduced glucose consumption, ECAR, and OCR (Fig. 4C,D). That this was due to a failure of mTORC2 activation was indicated by diminished AKTs473 phosphorylation when M-CSF was withdrawn prior to IL-4 stimulation (Fig. 4E).
Figure 4. M-CSF is essential to regulate mTORC2 signaling in M2 macrophages.
(A) Expression of PD-L2 and RELMα in macrophages cultured for 24 hr with (M2) or without (M0) IL-4 or M-CSF. (B) Expression of iNOS and TNF-α in macrophages cultured for 24 hr with (M1) or without (M0) IFN-γ + LPS or M-CSF. (C) Glucose consumption, and (D) basal ECAR and basal OCR of M2 macrophages treated as in (A). (E) Level of phosphorylated AKTs473 in macrophages as in A, assessed by flow cytometry. MFI values are shown. (F) Scheme to examine the effect of blocking M-CSF/CSF1R interaction on M2 activation in vivo. (G) Phosphorylated NDRG1 (p-NDRG1) in pMacs was measured by immunoblot; band density was normalized to loading controls and is presented in arbitrary units (AU). (H) Level of AKTs473, phosphorylation, and (I) frequency of RELMα+ cells, in pMacs. Data in A,B, E, H and I are from, flow cytometry and in A, B and E are from one experiment representative, but numbers represent mean % (A) or MFI (B,E) values, ± s.e.m, from three independent experiments. In C,D and G–I, data are mean ± s.e.m. of technical replicates from one experiment representative of three or more independent experiments. *P < 0.05, **P < 0.005 and ***P < 0.0001.
Next we assessed the contribution of M-CSF to M2 activation in vivo. We elicited pMacs with thioglycolate in the presence or absence of IL-4/anti-IL-4 complexes (which initiate M2 activation in vivo), and/or a neutralizing anti-CSF1R antibody (Fig. 4F). Compared to pMacs recovered from mice injected with IL-4c alone, pMacs from mice injected with IL-4c plus anti-CSF1R antibody had diminished levels of phosphorylation of NDRG1 (Fig. 4G) and AKTs473 (Fig. 4H) and significantly diminished expression of RELMα (Fig. 4I)
Together, our data indicate that M-CSF synergizes with IL-4 to induce M2 activation by promoting mTORC2 signaling and downstream metabolic reprogramming.
The mTORC2 pathway enhances the expression of Irf4, which is crucial for metabolic reprogramming in M2 macrophages
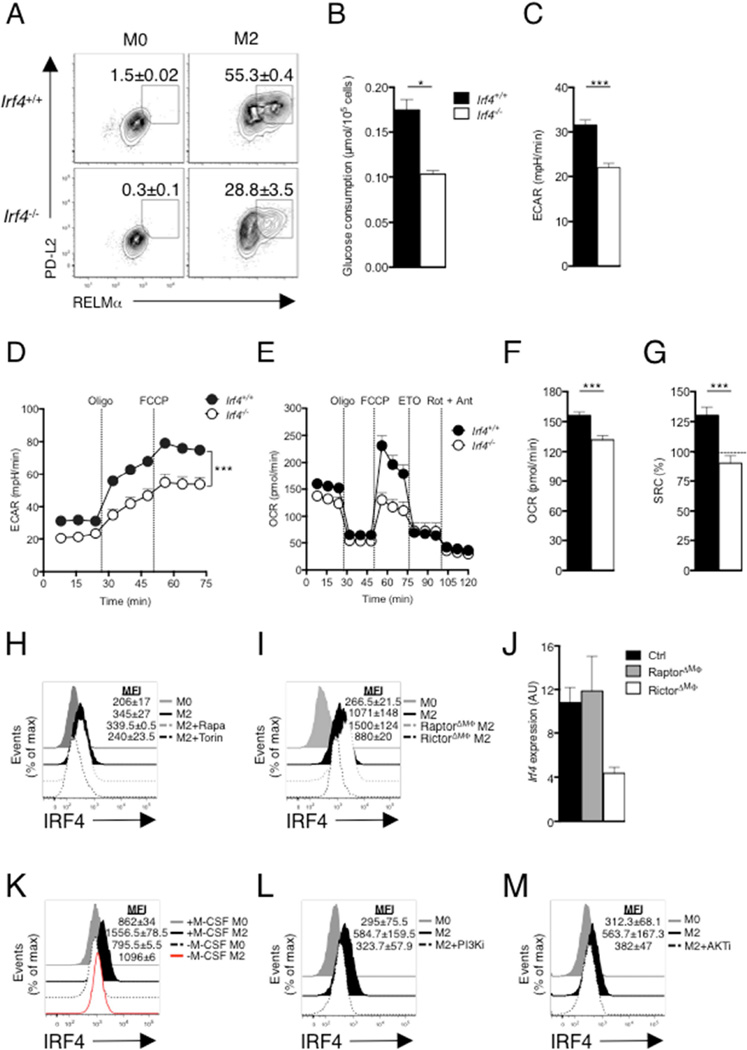
IRF4 has been reported to play roles in alternative macrophage activation (Satoh et al., 2010) and in mTOR-mediated regulation of glycolysis in CD8+ T cells (Man et al., 2013; Yao et al., 2013). We hypothesized that IRF4 could be playing a role in M2 activation through its ability to regulate metabolism. As expected, M2 activation, as measured by increased expression of PD-L2, RELMα (Fig. 5A), Arg1, Ym1, Il10, Lipa, Cd36, Fabp4, Pparg and Ppargc1b (Fig. S3A) was diminished in Irf4−/− compared to WT macrophages. IRF4 deficiency had no effect on iNOS expression, indicating that this transcription factor is not important for M1 activation (Fig. S3B). Irf4−/− macrophages exhibited defects in IL-4-induced increases in glucose consumption, baseline ECAR, and GR (Fig. 5B–D), and failed to upregulate expression of genes encoding key regulators of glycolysis, including Slc2a1, Hk1, Hk2, Gpi1, Gapdh, Pfkp and Ldha (Fig. S3C). Moreover, IL-4-stimulated Irf4−/− macrophages lacked etomoxir-sensitive SRC (Fig. 5E), and had low basal OCR and SRC, indicating that they had not committed to FAO (Fig. 5E–G). Thus IRF4 plays a role in the metabolic reprogramming that supports M2 activation
Figure 5. IRF4 mediates glucose metabolism to promote M2 activation.
Bone marrow macrophages were from WT (Irf4+/+) and Irf4−/− mice. (A) Expression of PD-L2 and RELMα in macrophages cultured for 24 hr without (M0) or with (M2) IL-4. (B) Glucose uptake and (C) basal ECAR after macrophage culture in IL-4 for 24 hr. (D) ECAR of macrophages cultured for 24 hr in IL-4, followed by sequential treatment with oligomycin and FCCP. (E) OCR of macrophages cultured for 24 hr in IL-4, followed by sequential treatment with oligomycin, FCCP, etomoxir and rotenone + antimycin. (F) OCR and (G) SRC of macrophages after culture in IL-4 for 24 hr. (H) IRF4 expression by macrophages after culture without or with IL-4 for 24 hr plus or minus rapamycin (Rapa; 20 nM) or Torin 1 (Torin; 100 nM). (I) IRF4 expression by Raptor or Rictor deficient macrophages after culture without or with IL-4 for 24 hr. (J) Expression of Irf4 in IL-4 stimulated WT macrophages, or macrophages lacking Raptor or Rictor, as measured by qRT-PCR (expression normalized to WT M0 macrophages and presented in arbitrary units, AU). (K–M) IRF4 expression by macrophages after culture without (M0) or with (M2) IL-4 or M-CSF (K), PI3Ki (L) or AKTi (M) for 24 hr. Data in A, H, I, K–M are from flow cytometry and are from individual experiments, but numbers represent mean % (A) or MFI, ± s.e.m., of data from 3 or more independent experiments. In B–G, data are mean ± s.e.m. values from technical replicates from one experiment, representative of three or more independent experiments). *P < 0.05, **P < 0.005 and ***P < 0.0001.
We asked whether mTOR was involved in IRF4 expression in M2 macrophages. We found that Torin but not rapamycin strongly inhibited IRF4 expression (Fig. 5H), and that levels of IRF4 were reduced in IL-4 stimulated Rictor-deficient, but increased in IL-4 stimulated Raptor-deficient M2 macrophages (Fig. 5I). This reflected diminished transcription of Irf4 in the absence of Rictor (Fig. 5J). Our data therefore support a role for mTORC2 in the expression of IRF4 in macrophages stimulated with IL-4.
We reasoned that if IRF4 expression is induced via the mTORC2 pathway, it should also be sensitive to inhibition of other key components of this pathway that are important for M2 activation, including M-CSF, PI3K and AKT. Consistent with this, we found that removal of M-CSF from IL-4-stimulated macrophages, or inhibition of PI3K or AKT, all inhibited IRF4 expression (Fig. 5K–M). Conversely, IL-4-stimulated phosphorylation of AKTs473 and Stat6 were similar in WT cells and macrophages from Irf4fl/fl-LysMcre mice (Fig. S3D,E), indicating that major IL-4 initiated upstream signaling events are occurring normally in the absence of IRF4.
While our data support a role for mTORC2 in M2 macrophages, it is accepted that signaling via Stat6 is critical for this activation process. We found that Stat6−/− macrophages were unable to express RELMα and PD-L2 in response to IL-4 (Fig. S3F), but also failed to increase expression of IRF4 (Fig. S3G). This did not reflect an effect of the absence of Stat6 on mTORC2-dependent signaling, since NDRG1 and AKTs473 were equivalently phosphorylated in IL-4 stimulated WT and Stat6−/− macrophages (Fig. S3H). Our findings indicate that IRF4 expression relies on both the Stat6 and mTORC2 pathways, and is critical for metabolic remodeling and M2 activation in IL-4 stimulated macrophages. While our data point towards a role for CSFR1 in initiating mTORC2 activation, activation of PI3K downstream of the common γ chain of the IL-4 specific type I IL-4R (Heller et al., 2008) could also be contributing to this activation pathway. In this context, it is intriguing that IL-13 has been reported to be less effective than IL-4 at promoting M2 activation despite the fact that it induces equivalent signaling through Stat6, since the type II IL-4R which recognizes IL-13 (in addition to IL-4) is not coupled to PI3K/AKT (Heller et al., 2008). We found that in pMacs isolated from mice that had been injected with µg-equivalent amounts of IL-4c or IL-13c, RELMα and IRF4 expression was lower in the IL-13 stimulated vs. IL-4 stimulated cells (Fig. S4A,B). Similar results were observed in a comparison of IL-4 vs. IL-13 stimulated bone marrow derived macrophages (Fig. S4C and data not shown). This was despite near-equivalent Stat6 phosphorylation in IL-13 vs. IL-4 stimulated cells (Fig. S4D), but reflected a relative lack of activation of the mTORC2 pathway, measured by mTORS2481 and AKTS473 phosphorylation, in the IL-13 stimulated cells (Fig. S4E,F).
Loss of mTORC2 signaling in macrophages suppresses tumor growth and decreases immunity to a parasitic nematode
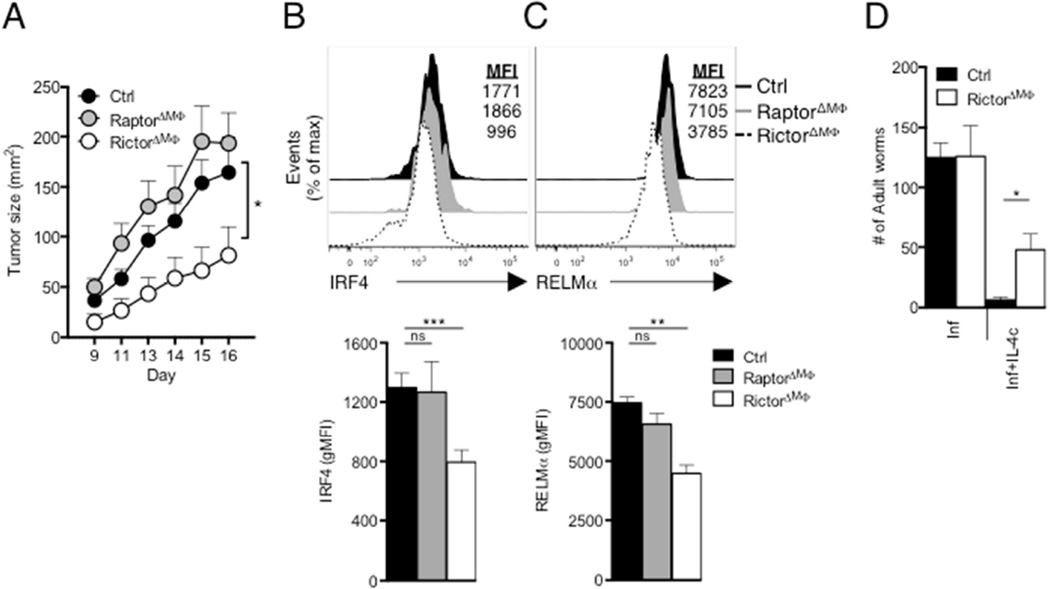
Tumor growth is supported by tumor-associated macrophages (TAMs) that exhibit M2 like properties (Noy and Pollard, 2014). To examine the role of mTOR in TAMS, we implanted B16 melanoma cells into RictorΔMΦ, RaptorΔMΦ and control (LysMCre) mice, and tracked tumor growth. Tumors grew more slowly in RictorΔMΦ mice than in WT and RaptorΔMΦ mice (Fig. 6A) and TAM (defined as CD45+CD11b+CD64+F4/80+ cells) M2 activation, as measured by RELMα and IRF4 expression, was significantly diminished in RictorΔMΦ mice (Figs. 6B,C). We did not see a difference in the number of TAMs in tumors isolated from RictorΔMΦ or RaptorΔMΦ mice compared to WT mice, but did measure increased expression of genes encoding IFN-γ, TNFα and iNOS, which are linked to tumor control, and reduced expression of Il10, which inhibits anti-tumor immunity (Fig. S5A,B) (Noy and Pollard, 2014). We found no difference in the infiltration of tumors by myeloid-derived suppressor cells (MDSCs; defined as CD45+CD11b+Gr1+ cells; Fig. S5C), which suggested the slower growth rate of tumors in RictorΔMΦ mice was not due to diminished infiltration of MDSCs.
Figure 6. Loss of mTORC2 activity in macrophages suppresses tumorigenesis and inhibits protective immunity against H. polygyrus.
(A) Growth profile of tumors following inoculation of 2×105 B16-OVA melanoma cells into WT (Ctrl), RaptorΔMΦ or RictorΔMΦ mice. (B) Top, IRF4 expression by TAMS from day 16 tumors from from Ctrl, RaptorΔMΦ and RictorΔMΦ. Bottom, geometric MFI of IRF4 staining shown in top panel. (C) Top, RELMα expression by TAMs, as in (B). Bottom, gMFI of RELMα. staining shown in top panel. (D) Adult H. polygyrus counts from infected WT (Ctrl) and RictorΔMΦ mice which on days 9, 11, 13 and 15 after infection were injected with PBS (Inf) or IL-4c (Inf+IL-4c), followed by analysis on day 16. Data are mean ± s.e.m. of five to six individually analyzed mice/group in one experiment, and representative of two independent experiments (A–C), or from one experiment representative of two independent experiments (mean ± s.e.m. of three to five mice per group) (D). *P < 0.05, **P < 0.005 and ***P < 0.0001.
Finally, we asked whether loss of mTORC2 signaling in macrophages could affect resistance to H. polygyrus. Primary infection with H. polygyrus in B6 mice induces a Th2 response, but nevertheless is chronic (Reynolds et al., 2012). However, injection of IL-4c into infected mice enhances type 2 immunity and M2 activation resulting in elimination of the parasites (Huang et al., 2014). Consistent with this, the ability of IL-4c to eliminate worms from infected RictorΔMΦ mice, was diminished compared to in control mice (Fig. 6D). Increased basal OCR in pMacs from infected mice following IL-4c injection (Huang et al., 2014) was diminished in infected RictorΔMΦ mice (Fig. S6A). Moreover, glycolysis in pMacs, measured by ECAR, increased as a result of infection in control but not infected RictorΔMΦ mice (Fig. S6B) (Injection of IL-4c had no effect on ECAR in these experiments). Failure of metabolic reprograming in Rictor-deficient macrophages in infected/IL-4c-injected mice correlated with diminished expression of RELMα and IRF4 (Fig. S6C,D).
Together, our findings indicate that diminished mTORC2 signaling negatively affects macrophage M2 activation, with an associated gain in resistance to tumor progression, and loss of resistance to helminth parasites.
Discussion
Changes in key metabolic regulatory events in immune cells can be initiated not only by changes in nutrient and oxygen conditions, but also in response to the presence of danger signals or antigen, or instructional signals received from other cells. For example in macrophages stimulated by IL-4 to become alternatively activated, FAO increases to support OXPHOS and these processes are critical for full M2 activation (Huang et al., 2014; Vats et al., 2006). Compared to M0 macrophages, M2 macrophages also exhibit changes in glucose metabolism. mTOR plays a central role in integrating nutrient availability, and growth factor and immune factor initiated signaling with metabolic demand (Sengupta et al., 2010; Weichhart et al., 2015; Yang and Chi, 2012), processes that are important as immune cells move from quiescent to activated states. Recent reports have implicated mTOR signaling in macrophage polarization, indicating that mTORC1 (Byles et al., 2013; Covarrubias et al., 2015; Festuccia et al., 2014) can play both positive and negative roles in M2 cell activation depending on context (Byles et al., 2013; Covarrubias et al., 2016), and revealing a role for mTORC1 in an AKT-dependent pathway that regulates glucose metabolism in these cells (Covarrubias et al., 2016). However, the role of mTORC2, which has been implicated in regulating glycolysis in other cell types (e.g. brown adipocytes (Albert et al., 2016)), has not been examined in detail in macrophages. Here we report that mTORC2 operates in parallel with the canonical IL-4Rα/Stat6 pathway to facilitate increased glycolysis during M2 activation. Our data implicate PI3K and AKT signaling initiated by M-CSF as components in this pathway, and indicate that downstream induction of IRF4 expression plays a role in facilitating metabolic reprograming to support M2 activation.
We found that increased glucose uptake is critical for M2 activation since 2-DG prevented IL-4 from inducing/sustaining M2 activation even in the presence of TAGs and FA. We previously reported that M2 activation can occur in the absence of external FA sources as long as glucose is present, and suggested that glucose can be used to synthesize FA for FAO (Huang et al., 2014). Our findings here support this contention since inhibition of glycolysis, or of pyruvate entry into mitochondria, or of FAS, led variously to reductions in OCR, SRC and etomoxir-sensitive oxygen consumption, and diminished cellular ATP levels supporting the view that glucose is bioenergetically important for M2 macrophages. This interpretation is consistent with renewed interest in the ability of increased flux through FAS to concomitantly drive high levels of FAO and glycolysis for ATP regeneration (Cader et al., 2016). It is important to note that a similar pathway of FAO supported by FAS operates in T cells (O'Sullivan et al., 2014). It is also of note that a key intermediate in the synthesis of FA from glucose is acetyl-CoA, which is made from citrate by the enzyme Acly. Recent reports showed that Acly is important for M2 activation and postulated that this was due to the importance of acetyl-CoA in histone acetylation (Covarrubias et al., 2016), but our data suggest that it may also be important because of its role in FAS.
Our data indicate that mTOR is critical for M2 activation. However, while mTORC1 is important for enhanced glycolysis in activated T cells (Delgoffe et al., 2009; Pollizzi et al., 2015), its role in M2 macrophages is less clear, since enhanced mTORC1 activity caused by deletion of Tsc1 enhances M1 activation and inhibits M2 activation (Byles et al., 2013; Covarrubias et al., 2015) whereas in the presence of Tsc1, mTORC1 has been reported to be important for M2 activation (Covarrubias et al., 2016). We found that mTORC1 is active in M2 macrophages, since S6K is phosphorylated in these cells. Moreover, results from experiments in which rapamycin was used to inhibit mTORC1, and in which mTORC1 was inactivated by Raptor deletion, support the view that mTORC1 antagonizes M2 activation to some extent even in the presence of Tsc1, since in IL-4 stimulated cells glycolysis and OXPHOS, along with the expression of CD301 and RELMα were increased in the absence of Raptor. Nevertheless, in our studies expression of Arg1 was decreased in the Raptor-deficient IL-4 stimulated cells, which is consistent with previous reports, but so was expression of Fabp4, which previously was shown to be induced by IL-4 in a Raptor-independent manner (Covarrubias et al., 2016).
Like mTORC1, mTORC2 has been linked to enhanced glycolysis in other cell types, largely due to its ability to activate AKT (Albert et al., 2016; Masui et al., 2014a, b, 2015). Our data indicate that there is tonal mTORC2 activity in resting macrophages, since both NDRG1 and AKTs473 are phosphorylated. However, AKTs473 phosphorylation is increased in M2 vs. M0 macrophages, indicating that mTORC2 activity is upregulated following exposure to IL-4. Loss of mTORC2 function through deletion of Rictor led to inhibition of M2 activation and diminished glycolysis and OXPHOS. Deletion of Rictor in hepatocytes has also been reported to result in reduced glycolysis and FAS coupled to loss of AKTs473 phosphorylation (Hagiwara et al., 2012). Our observations are consistent with the findings in T cells that mTORC2 is critical for AKTs473 phosphorylation and Th2 cell development. Th2 polarization is also driven by IL-4 through Stat6, but despite its effect on Th2 polarization, deletion of Rictor was reported to have no effect on Stat6 phosphorylation (Delgoffe et al., 2011; Lee et al., 2010). This indicates that in Th2 cells, as in M2 macrophages, the Stat6-dependent and mTORC2-dependent pathways operate in parallel to promote changes in cellular function. (Heller et al., 2008).
PI3K and/or AKT have either directly or indirectly been implicated in M2 activation previously (Byles et al., 2013; Covarrubias et al., 2016; Ruckerl et al., 2012). Our findings that PI3K activates mTORC2 and that mTORC2 phosphorylates AKT, and that all three are important for M2 activation, provide context for these earlier findings. Our data suggest that full activation of this pathway is dependent on M-CSF/CSF-1R. This is consistent with previous reports that enhancement of macrophage survival by M-CSF is dependent on PI3K and AKT, and linked to the ability to increase glucose uptake by promoting Glut1 expression (Chang et al., 2009). Since our data show that under constant M-CSF stimulation the addition of IL-4 leads to increased glycolysis, it is likely that M-CSF and IL-4 synergistically promote the activation of the mTORC2 signaling axis and the metabolic reprogramming which are essential for M2 activation. This interpretation is supported by our finding that, unlike IL-4, IL-13 fails to activate the mTORC2 pathway. This may represent the critical difference between the strength of alternative activation signal delivered by IL-4 versus IL-13.
Our data point to the induced expression of IRF4, a transcription factor controlling M2 activation (Satoh et al., 2010), as being a key function of mTORC2 to promote metabolic reprogramming during M2 activation. IRF4 is expressed in M2 macrophages and has been shown to regulate the expression of glycolytic genes (Man and Kallies, 2015; Man et al., 2013; Yao et al., 2013), suggesting a conserved link between mTOR, IRF4 and metabolic reprogramming in immune cells. Our data show that IRF4 expression in M2 macrophages is dependent on both IL-4/Stat6 and M-CSF/mTORC2 and this is compatible with the fact that in T cells IRF4 also integrates disparate extracellular signals from cytokine receptors, T cell receptor, and nutrient sensing pathways to sustain metabolic remodeling and appropriate cellular function.
Infiltration by macrophages is a common feature of many solid tumors and M2 activated TAMs are recognized as playing an important role in allowing tumors to progress and metastasize (Noy and Pollard, 2014; Wynn et al., 2013). We found that tumor growth was delayed in mice in which Rictor was deleted in macrophages, and that in these tumors TAM M2 marker expression was muted and pro-inflammatory mediator expression was increased. Others have reported that CSF-1R blockade leads to suppression of TAM M2 activation and reductions in tumor progression (Pyonteck et al., 2013), and cancer therapies based on the inhibition of M-CSF/CSF-1R interactions are in clinical trials (Ries et al., 2015). Our findings that M-CSF promotes mTORC2 activation and that mTORC2 is important for TAM M2 activation are generally consistent with these reports. The finding that mice lacking Rictor in macrophages developed fewer M2 macrophages in response to infection with H. polygyrus or injection of IL-4 support a role of mTORC2 in metabolic reprogramming and M2 macrophage activation.
We believe that our data from targeting numerous points in the mTORC2 pathway are consistent with a role for mTORC2 signaling in metabolic reprograming during M2 activation. Our data indicate that alternative macrophage activation in response to IL-4 requires not only signaling through the canonical Stat6 pathway, but also input from the mTORC2 pathway, and suggest that this signal is initiated synergistically by IL-4 and M-CSF and mediated by AKT downstream of mTORC2. We propose that induction of expression of IRF4 is ultimately the mechanism through which glucose metabolism is promoted to support FAO and OXPHOS. Fuller understanding of this process will require mechanistic insights from approaches such as ChiP-seq to identify genes targeted by IRF4 in IL-4 stimulated cells, and genome-wide assays to broadly identify mTORC2-dependent changes, including those at genes discussed herein. Our findings point to mTORC2 as a potential target for the therapeutic manipulation of M2 activation in vivo. This approach may have value in the treatment of cancers and the inhibition of detrimental tissue changes such as fibrosis that are associated with allergies and atopy.
Experimental Procedures
Animals and in vivo experiments
Mice were orally infected with H. polygyrus and parasite burden was quantitated as described (Huang et al., 2014). Infected mice were injected with 2-DG or PBS i.p. for 3 hr, and pMacs were harvested and analyzed, or with IL-4c on days 9, 11, 13 and 15 after infection. For CSF1R blockade, mice were injected i.p. with thioglycollate (Sigma) immediately prior to injection with IL-4c with anti-CSF1R mAb or control IgG. For comparisons of effects of IL-4 and IL-13,mice were injected i.p. with IL-4c or IL-13c, or PBS, on days 0 and 3, and pMacs were harvested by lavage on day 4. For tumor experiments, B16-OVA melanoma cells were injected s.c. Tumors were measured every other day and excised post mortem at day 16 post-implantation; single cell suspensions were made for analysis. Total PECs and pMacs were determined by cell counting in combination with flow cytometry. Cells were maintained on ice until use or analysis.
Preparation of macrophages from bone marrow and macrophage activation
Marrow was flushed from bones, dissociated by pipetting, and grown in mouse M-CSF in complete medium for 7 d after which macrophages were stimulated with IL-4, IL-13 or LPS plus IFN-γ with or without 2-DG, UK5099, rapamycin, Torin 1, LY294002, triciribine (AKTi) or C75 for 24 hr. For M-CSF withdrawal, bone marrow macrophages were transferred to M-CSF-free medium for 24 hr prior to IL-4 stimulation. Macrophages were harvested 24 hr later for analysis.
Retroviral transduction
Sequences for luciferase, Mpc-1, Cpt1a and Acaca short hairpin RNAs (shRNAs) were obtained from Open Biosystems and cloned into MSCV-LMP retroviral vector encoding huCD8 or green fluorescent protein (GFP) as a reporter. For overexpression, Slc2a1 sequence (Glut1) was cloned into MSCV-IRES retroviral vector, encoding GFP as a reporter (Chang et al., 2015). Day 3 bone marrow macrophage cultures were spin infected with retrovirus (Everts et al., 2014). At day 7 of culture, macrophages were harvested and transduced cells were identified by huCD8 or GFP expression.
Metabolism assays
ECAR and OCR measurements were made using an XF-96 Extracellular Flux Analyzer (Seahorse Bioscience) as described (Huang et al., 2014). Glucose, ATP and TAGs were measured using commercially available kits.
Flow Cytometry
Cells on ice were blocked with anti-CD16/32 before surface staining with Ab to CD45, CD11b, F4/80, CD64, CD301, PD-L2, and Ly-6G/Ly-6C. For intracellular staining, cells were fixed and permeabilized and stained with rabbit anti-RELMα, mouse anti-NOS2, mouse anti-TNFα, mouse ant-phospho-AKT Ser473 (p-AKTs473), mouse anti-phospho-Stat6 Tyr641, rabbit anti-phospho-mTOR Ser2481 (p-mTORs2481), anti-Glut1, mouse anti-IRF4, or mouse anti-Ki67, followed by incubation with appropriate fluorochrome-conjugated secondary antibodies. Cells were also stained with 2-NBDG. Data were acquired by flow cytometry and analyzed with FlowJo v.9.5.2 (Tree Star).
Cell fractionation and Western blotting
Cells were lysed in buffer containing 1% Triton X-100 and 0.1% SDS with protease and phosphatase inhibitors. Anti-phospho-p70 S6 kinase (p-S6K), anti-p70 S6 kinase (S6K), anti-phospho-NDRG1 Thr346 (p-NDRG1), anti-NDRG1, anti-p-AKTs473, anti-AKT, anti-p-Stat6, anti-Stat6 and anti-β-actin were used and detected with peroxidase-linked secondary antibody followed by ECL Western Blotting Detection Reagents (GE Healthcare). Images were analyzed using ImageJ (NIH).
Gene expression
The Taqman method was used for real-time PCR with primers from Applied Biosystems. The expression of mRNA was normalized to that of mRNA encoding β-actin. Analysis of reported RNAseq data set (Huang et al., 2014) was used for data in Fig. 1B and S1B.
Statistical analysis
Comparisons for two groups were calculated using One-way ANOVA and, where indicated, unpaired two-tailed Student’s t-tests. Differences were considered significant when p-values were below 0.05.
Supplementary Material
Acknowledgments
We thank Dr. Ramon Klein Geltink for careful reading of the manuscript and helpful discussions, Drs. Ken Murphy, Skip Virgin, Chih-Hao Chang and Joe Urban for generously providing mice, reagents or parasites, and Erica Lantelme, and Dorjan Brinja for expert FACS assistance. The work was supported by National Institutes of Health grants to E.J.P. (AI110481 and CA164062), E.L.P. (CA18112502), J.D.S. (P30 DK020579), NWO VENI-91614087 and Marie Curie CIG-631585 to B.E., and an American Heart Association Postdoctoral Fellowship to S.C.-C.H,
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
S.C.-C.H., B.E., E.L.P., J.D.S., E.J.P. designed the research and analyzed and interpreted the data. S.C-C.H. and A.M.S. performed experiments. M.C. provided essential discussion. S.C.-C.H. and E.J.P. wrote the manuscript.
References
- Albert V, Svensson K, Shimobayashi M, Colombi M, Munoz S, Jimenez V, Handschin C, Bosch F, Hall MN. mTORC2 sustains thermogenesis via Akt-induced glucose uptake and glycolysis in brown adipose tissue. EMBO Mol Med. 2016;8:232–246. doi: 10.15252/emmm.201505610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byles V, Covarrubias AJ, Ben-Sahra I, Lamming DW, Sabatini DM, Manning BD, Horng T. The TSC-mTOR pathway regulates macrophage polarization. Nature communications. 2013;4:2834. doi: 10.1038/ncomms3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cader MZ, Boroviak K, Zhang Q, Assadi G, Kempster SL, Sewell GW, Saveljeva S, Ashcroft JW, Clare S, Mukhopadhyay S, et al. C13orf31 (FAMIN) is a central regulator of immunometabolic function. Nat Immunol. 2016;17:1046–1056. doi: 10.1038/ni.3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Qiu J, O'Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ, et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Hamilton JA, Scholz GM, Masendycz P, Macaulay SL, Elsegood CL. Phosphatidylinostitol-3 kinase and phospholipase C enhance CSF-1-dependent macrophage survival by controlling glucose uptake. Cellular signalling. 2009;21:1361–1369. doi: 10.1016/j.cellsig.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Copp J, Manning G, Hunter T. TORC-specific phosphorylation of mammalian target of rapamycin (mTOR): phospho-Ser2481 is a marker for intact mTOR signaling complex 2. Cancer research. 2009;69:1821–1827. doi: 10.1158/0008-5472.CAN-08-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias AJ, Aksoylar HI, Horng T. Control of macrophage metabolism and activation by mTOR and Akt signaling. Seminars in immunology. 2015;27:286–296. doi: 10.1016/j.smim.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias AJ, Aksoylar HI, Yu J, Snyder NW, Worth AJ, Iyer SS, Wang J, Ben-Sahra I, Byles V, Polynne-Stapornkul T, et al. Akt-mTORC1 signaling regulates Acly to integrate metabolic input to control of macrophage activation. Elife. 2016;5 doi: 10.7554/eLife.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B, Amiel E, Huang SC, Smith AM, Chang CH, Lam WY, Redmann V, Freitas TC, Blagih J, van der Windt GJ, et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKvarepsilon supports the anabolic demands of dendritic cell activation. Nat Immunol. 2014;15:323–332. doi: 10.1038/ni.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B, Amiel E, van der Windt GJ, Freitas TC, Chott R, Yarasheski KE, Pearce EL, Pearce EJ. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood. 2012;120:1422–1431. doi: 10.1182/blood-2012-03-419747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia WT, Pouliot P, Bakan I, Sabatini DM, Laplante M. Myeloid-specific Rictor deletion induces M1 macrophage polarization and potentiates in vivo pro-inflammatory response to lipopolysaccharide. PloS one. 2014;9:e95432. doi: 10.1371/journal.pone.0095432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) The Biochemical journal. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol. 2015;17:34–40. doi: 10.1038/ni.3324. [DOI] [PubMed] [Google Scholar]
- Glass CK, Natoli G. Molecular control of activation and priming in macrophages. Nat Immunol. 2015;17:26–33. doi: 10.1038/ni.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara A, Cornu M, Cybulski N, Polak P, Betz C, Trapani F, Terracciano L, Heim MH, Ruegg MA, Hall MN. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell metabolism. 2012;15:725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Halestrap AP. The mitochondrial pyruvate carrier. Kinetics and specificity for substrates and inhibitors. The Biochemical journal. 1975;148:85–96. doi: 10.1042/bj1480085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller NM, Qi X, Junttila IS, Shirey KA, Vogel SN, Paul WE, Keegan AD. Type I IL-4Rs selectively activate IRS-2 to induce target gene expression in macrophages. Science signaling. 2008;1:ra17. doi: 10.1126/scisignal.1164795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SC, Everts B, Ivanova Y, O'Sullivan D, Nascimento M, Smith AM, Beatty W, Love-Gregory L, Lam WY, O'Neill CM, et al. Cell-intrinsic lysosomal lipolysis is essential for macrophage alternative activation. Nat Immunol. 2014;15:846–855. doi: 10.1038/ni.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha AK, Huang SC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Chang JW, Wang J, Kang SA, Thoreen CC, Markhard A, Hur W, Zhang J, Sim T, Sabatini DM, Gray NS. Discovery of 1-(4-(4-propionylpiperazin-1-yl)-3-(trifluoromethyl)phenyl)-9-(quinolin-3-yl)benz o[h][1,6]naphthyridin-2(1H)-one as a highly potent, selective mammalian target of rapamycin (mTOR) inhibitor for the treatment of cancer. Journal of medicinal chemistry. 2010;53:7146–7155. doi: 10.1021/jm101144f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man K, Kallies A. Synchronizing transcriptional control of T cell metabolism and function. Nature reviews. Immunology. 2015;15:574–584. doi: 10.1038/nri3874. [DOI] [PubMed] [Google Scholar]
- Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, Pellegrini M, Belz GT, Smyth GK, Febbraio MA, et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol. 2013;14:1155–1165. doi: 10.1038/ni.2710. [DOI] [PubMed] [Google Scholar]
- Masui K, Cavenee WK, Mischel PS. mTORC2 dictates Warburg effect and drug resistance. Cell Cycle. 2014a;13:1053–1054. doi: 10.4161/cc.28377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui K, Cavenee WK, Mischel PS. mTORC2 in the center of cancer metabolic reprogramming. Trends Endocrinol Metab. 2014b;25:364–373. doi: 10.1016/j.tem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui K, Cavenee WK, Mischel PS. mTORC2 and Metabolic Reprogramming in GBM: at the Interface of Genetics and Environment. Brain Pathol. 2015;25:755–759. doi: 10.1111/bpa.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munugalavadla V, Borneo J, Ingram DA, Kapur R. p85alpha subunit of class IA PI-3 kinase is crucial for macrophage growth and migration. Blood. 2005;106:103–109. doi: 10.1182/blood-2004-10-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, Qiu J, Smith AM, Lam WY, DiPlato LM, et al. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, Chawla A. Alternative macrophage activation and metabolism. Annual review of pathology. 2011;6:275–297. doi: 10.1146/annurev-pathol-011110-130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollizzi KN, Patel CH, Sun IH, Oh MH, Waickman AT, Wen J, Delgoffe GM, Powell JD. mTORC1 and mTORC2 selectively regulate CD8(+) T cell differentiation. J Clin Invest. 2015;125:2090–2108. doi: 10.1172/JCI77746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Artis D. New paradigms in type 2 immunity. Science. 2012;337:431–435. doi: 10.1126/science.1221064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, Olson OC, Quick ML, Huse JT, Teijeiro V, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds LA, Filbey KJ, Maizels RM. Immunity to the model intestinal helminth parasite Heligmosomoides polygyrus. Seminars in immunopathology. 2012;34:829–846. doi: 10.1007/s00281-012-0347-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries CH, Hoves S, Cannarile MA, Ruttinger D. CSF-1/CSF-1R targeting agents in clinical development for cancer therapy. Current opinion in pharmacology. 2015;23:45–51. doi: 10.1016/j.coph.2015.05.008. [DOI] [PubMed] [Google Scholar]
- Ruckerl D, Jenkins SJ, Laqtom NN, Gallagher IJ, Sutherland TE, Duncan S, Buck AH, Allen JE. Induction of IL-4Ralpha-dependent microRNAs identifies PI3K/Akt signaling as essential for IL-4-driven murine macrophage proliferation in vivo. Blood. 2012;120:2307–2316. doi: 10.1182/blood-2012-02-408252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Molecular cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Wagner RA, Greaves DR, Murray PJ, Chawla A. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell metabolism. 2006;4:13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T, Hengstschlager M, Linke M. Regulation of innate immune cell function by mTOR. Nature reviews. Immunology. 2015;15:599–614. doi: 10.1038/nri3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Chi H. mTOR and metabolic pathways in T cell quiescence and functional activation. Seminars in immunology. 2012;24:421–428. doi: 10.1016/j.smim.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Buzo BF, Pham D, Jiang L, Taparowsky EJ, Kaplan MH, Sun J. Interferon regulatory factor 4 sustains CD8(+) T cell expansion and effector differentiation. Immunity. 2013;39:833–845. doi: 10.1016/j.immuni.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.