Abstract
Methylation of several lysine residues of histones is a crucial mechanism for relatively long-term regulation of genomic activity. Recent molecular biological studies have demonstrated that the function of histone methylation is more diverse and complex than previously thought. Moreover, studies using newly available genomics techniques, such as exome sequencing, have identified an increasing number of histone lysine methylation-related genes as intellectual disability-associated genes, which highlights the importance of accurate control of histone methylation during neurogenesis. However, given the functional diversity and complexity of histone methylation within the cell, the study of the molecular basis of histone methylation-related neurodevelopmental disorders is currently still in its infancy. Here, we review the latest studies that revealed the pathological implications of alterations in histone methylation status in the context of various neurodevelopmental disorders and propose possible therapeutic application of epigenetic compounds regulating histone methylation status for the treatment of these diseases.
Keywords: epigenetic changes, histone lysine methylation, lysine methyltransferase, lysine demethylase, neurodevelopmental disorder
1. Introduction
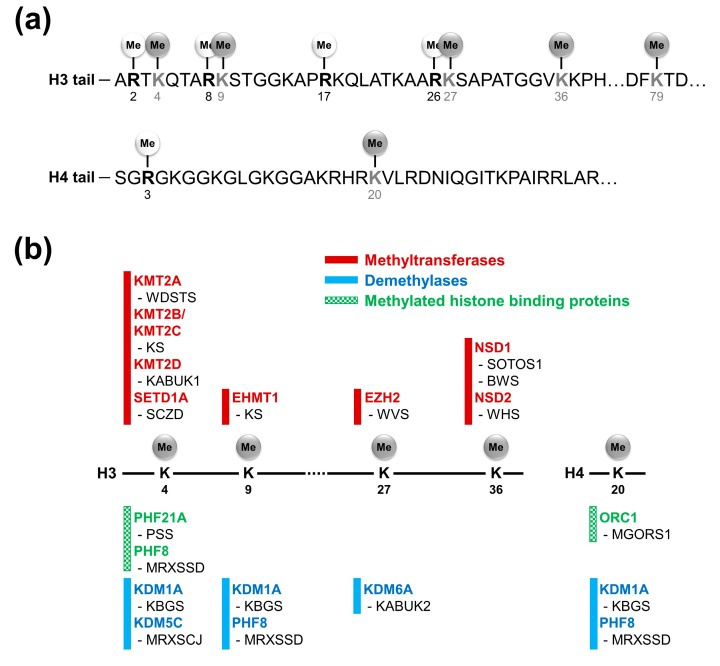
Post-translational modifications of histone proteins in eukaryotic cells serve as crucial regulatory mechanisms of gene expression and are important for maintaining genomic integrity [1,2]. The histone modifications, such as its acetylation, methylation, phosphorylation, and ubiquitination, influence genomic activity by altering the binding force of DNA to histones or by acting as marks that recruit specific histone binding proteins [2]. Among these histone modifications, methylation has been implicated in heterochromatin formation and the regulation of promoter activity [3,4]. The histone residues, on which methylation occurs, include the following lysine and arginine residues: H3 (K4, 9, 27, 36, and 79), H4K20, H3 (R2, 8, 17, and 26), and H4R3 [5,6] (Figure 1a). These methylation sites are evolutionarily well conserved [7]. A variety of histone methyltransferases (writers), histone demethylases (erasers), and methylated histone binding proteins (readers) have been identified in various eukaryotic genomes [8]. Their site-specific molecular functions have been defined by biochemical and genetic studies [2,8] (Table 1).
Figure 1.
Histone methylation and neurodevelopmental disorders: (a) histone methylation sites in the tails of histone H3 and H4; and (b) histone methyltransferases, demethylases, and methylated histone binding proteins linked with neurodevelopmental disorders. Five methylation sites were associated with several neurodevelopmental disorders. BWS, Beckwith-Wiedemann syndrome; KABUK1/2, Kabuki syndrome 1/2; KBGS, KBG syndrome; KS, Kleefstra syndrome; MGORS1, Meier-Gorlin syndrome 1; MRXSCJ, Mental retardation, X-linked, syndromic, Claes-Jensen type; MRXSSD, Siderius X-linked mental retardation syndrome; PSS, Potocki-Shaffer syndrome; SCZD, Schizophrenia; SOTOS1, Sotos syndrome 1; WDSTS, Wiedemann-Steiner syndrome; WHS, Wolf-Hirshhorn syndrome; WVS, Weaver syndrome.
Table 1.
The names of the histone methylation-related factors mentioned in this paper and their synonyms.
| Symbol | Previous Symbol | Synonym(s) | Residue | Function |
|---|---|---|---|---|
| ASH1L | ASH1L | ASH1, ASH1L1, huASH1, KMT2H | H3K36 | Methyltransferase |
| DOT1L | DOT1L | DOT1, KIAA1814, KMT4 | H3K79 | Methyltransferase |
| EHMT1 | EHMT1 | bA188C12.1, Eu-HMTase1, FLJ12879, KIAA1876, KMT1D | H3K9 | Methyltransferase |
| EHMT2 | BAT8, C6orf30 | Em:AF134726.3, G9A, KMT1C, NG36/G9a | H3K9 | Methyltransferase |
| EZH1 | EZH1 | KIAA0388, KMT6B | H3K27 | Methyltransferase |
| EZH2 | EZH2 | ENX-1, EZH1, KMT6, KMT6A | H3K27 | Methyltransferase |
| KDM1A | AOF2, KDM1 | BHC110, KIAA0601, LSD1 | H3K4, H3K9, H4K20 | Demethylase |
| KDM2A | FBXL11, KDM2A | CXXC8, DKFZP434M1735, FBL11, FBL7, FLJ00115, JHDM1A, KIAA1004, LILINA | H3K36 | Demethylase |
| KDM2B | FBXL10, KDM2B | CXXC2, Fbl10, JHDM1B, PCCX2 | H3K36 | Demethylase |
| KDM3A | JMJD1, JMJD1A, KDM3A | JHMD2A, KIAA0742, TSGA | H3K9 | Demethylase |
| KDM3B | C5orf7, JMJD1B, KDM3B | KIAA1082, NET22 | H3K9 | Demethylase |
| KDM4A | JMJD2, JMJD2A, KDM4A | JHDM3A, KIAA0677, TDRD14A | H3K9, H3K36 | Demethylase |
| KDM4B | JMJD2B, KDM4B | KIAA0876, TDRD14B | H3K9, H3K36 | Demethylase |
| KDM4C | JMJD2C, KDM4C | GASC1, KIAA0780, TDRD14C | H3K9, H3K36 | Demethylase |
| KDM5A | JARID1A, KDM5A, RBBP2 | - | H3K4 | Demethylase |
| KDM5C | JARID1C, KDM5C, MRX13, SMCX | DXS1272E, XE169 | H3K4 | Demethylase |
| KDM6A | KDM6A, UTX | - | H3K27 | Demethylase |
| KDM6B | JMJD3, KDM6B | KIAA0346 | H3K27 | Demethylase |
| KMT2A | KMT2A, MLL | ALL-1, CXXC7, HRX, HTRX1, MLL1A, TRX1 | H3K4 | Methyltransferase |
| KMT2B | KMT2B | CXXC10, HRX2, KIAA0304, MLL1B, MLL2, MLL4, TRX2, WBP7 | H3K4 | Methyltransferase |
| KMT2C | KMT2C, MLL3 | HALR, KIAA1506 | H3K4 | Methyltransferase |
| KMT2D | KMT2D, MLL2, TNRC21 | ALR, CAGL114, MLL4 | H3K4 | Methyltransferase |
| KMT5A | KMT5A, SETD8 | PR-Set7, SET07, SET8 | H4K20 | Methyltransferase |
| KMT5B | KMT5B, SUV420H1 | CGI-85 | H4K20 | Methyltransferase |
| KMT5C | KMT5C, SUV420H2 | MGC2705 | H4K20 | Methyltransferase |
| NSD1 | STO | ARA267, FLJ22263, KMT3B | H3K36 | Methyltransferase |
| NSD2 | WHSC1 | KMT3G, MMSET | H3K36 | Methyltransferase |
| NSD3 | WHSC1L1 | FLJ20353, KMT3F, WHISTLE | H3K36 | Methyltransferase |
| ORC1 | ORC1L | HSORC1, PARC1 | H4K20 | Recognition |
| PHF2 | - | CENP-35, JHDM1E, KDM7C, KIAA0662 | H3K9, H4K20 | Demethylase |
| PHF8 | - | JHDM1F, KDM7B, KIAA1111, ZNF422 | H3K9, H4K20/H3K4 | Demethylase/Recognition |
| PHF21A | - | BHC80, BM-006, KIAA1696 | H3K4 | Recognition |
| RIOX1 | C14orf169 | FLJ21802, JMJD9, MAPJD, NO66 | H3K4, H3K36 | Demethylase |
| SETD1A | - | KIAA0339, KMT2F, Set1 | H3K4 | Methyltransferase |
| SETD1B | - | KIAA1076, KMT2G, Set1B | H3K4 | Methyltransferase |
| SETD2 | - | FLJ23184, HIF-1, HYPB, KIAA1732, KMT3A | H3K36 | Methyltransferase |
| SETD3 | C14orf154 | FLJ23027 | H3K4, H3K36 | Methyltransferase |
| SETDB1 | SETDB1 | ESET, KG1T, KIAA0067, KMT1E, TDRD21 | H3K9 | Methyltransferase |
| SETMAR | - | Mentase | H3K4, H3K36 | Methyltransferase |
| SMYD2 | - | HSKM-B, KMT3C, ZMYND14 | H3K4, H3K36 | Methyltransferase |
| SUV39H1 | SUV39H | KMT1A | H3K9 | Methyltransferase |
| SUV39H2 | SUV39H2 | KMT1B FLJ23414 | H3K9 | Methyltransferase |
| UTY | UTY | KDM6AL, KDM6C | H3K27 | Demethylase |
| The names of the proteins are followed by HUGO Gene Nomenclature Committee (http://www.genenames.org/) | ||||
Dysregulation of epigenetic modifications are associated with various human diseases, including neurodevelopmental disorders [9,10]. In particular, an increasing number of mutations in histone lysine methylation-related genes have been identified as intellectual disability-associated genes by exome sequencing with patients’ samples [11,12,13,14] (Figure 1b and Table 2). This highlights the importance of proper control of histone methylation during neurogenesis. In the current article, we provide an overview of the latest updates on the pathological implication of alterations in histone lysine methylation status in terms of neurodevelopmental disorders. Through this, we try to predict the future direction of research on this emerging field.
Table 2.
Neurodevelopmental disorders caused by mutations in histone methylation-related genes.
| Disorder | OMIM | Symptom | Gene | Residue | Function |
|---|---|---|---|---|---|
| Beckwith-Wiedemann syndrome (BWS) | 130650 | Pediatric overgrowth disorder involving a predisposition to tumor development | NSD1 | H3K36 | Methyltransferase |
| Kabuki syndrome 1 | 147920 | Congenital mental retardation, postnatal dwarfism, peculiar faces, broad and depressed nasal tip, large prominent earlobes, cleft or high-arched palate, scoliosis, short fifth finger, and persistence of finger pads | KMT2D KDM6A | H3K4 H3K27 | Methyltransferase Demethylase |
| Kabuki syndrome 2 (KABUK1/2) | 300867 | ||||
| KBG syndrome (KBGS) | 148050 | Macrodontia of the upper central incisors, distinctive craniofacial findings, short stature, skeletal anomalies, neurologic involvement that includes global developmental delay, seizures, and intellectual disability | KDM1A | H3K4 H3K9 H4K20 | Demethylase Demethylase Demethylase |
| Kleefstra syndrome (KS) | 610253 | Severe mental retardation, hypotonia, epileptic seizures, flat face with hypertelorism, synophrys, anteverted nares, everted lower lip, carp mouth with macroglossia, and heart defects | KMT2B, KMT2C EHMT1 | H3K4 H3K4 H3K9 | Methyltransferase Methyltransferase Methyltransferase |
| Meier-Gorlin syndrome 1 (MGORS1) | 224690 | Severe intrauterine and postnatal growth retardation, microcephaly, bilateral microtia, and aplasia or hypoplasia of the patellae | ORC1 | H4K20 | Recognition |
| Mental retardation, X-linked, syndromic, Claes-Jensen type (MRXSCJ) | 300534 | Severe mental retardation, slowly progressive spastic paraplegia, facial hypotonia, and maxillary hypoplasia | KDM5C | H3K4 | Demethylase |
| Potocki-Shaffer syndrome (PSS) | 601224 | Craniofacial abnormalities, developmental delay, intellectual disability, multiple exostoses, and biparietal foramina | PHF21A | H3K4 | Recognition |
| Schizophrenia (SCZD) | 181500 | Hallucinations and delusions, severely inappropriate emotional responses, disordered thinking and concentration, erratic behavior, as well as social and occupational deterioration | SETD1A | H3K4 | Methyltransferase |
| Siderius X-linked mental retardation syndrome (MRXSSD) | 300263 | Mental retardation, a repaired cleft lip, a long face with broad nasal tip, long hands with long thin fingers, and flat feet with long thin toes | PHF8 | H3K4 H3K9 H4K20 | Recognition Demethylase Demethylase |
| Sotos syndrome 1 (SOTOS1) | 117550 | Excessively rapid growth, acromegalic features, and non-progressive cerebral disorder with mental retardation | NSD1 | H3K36 | Methyltransferase |
| Weaver syndrome (WVS) | 277590 | Pre- and postnatal overgrowth, accelerated osseous maturation, characteristic craniofacial appearance, and developmental delay, broad forehead and face, ocular hypertelorism, prominent wide philtrum, micrognathia, deep horizontal chin groove, and deep-set nails | EZH2 | H3K27 | Methyltransferase |
| Wiedemann-Steiner syndrome (WDSTS) | 605130 | Hypertrichosis cubiti associated with short stature, consistent facial features, including long eyelashes, thick or arched eyebrows with a lateral flare, down slanting and vertically narrow palpebral fissures, mild to moderate intellectual disability, behavioral difficulties, and hypertrichosis on the back | KMT2A | H3K4 | Methyltransferase |
| Wolf-Hirschhorn syndrome (WHS) | 194190 | Pre- and postnatal growth deficiency, developmental disability of variable degree, characteristic craniofacial features, and a seizure disorder | NSD2 | H3K36 | Methyltransferase |
2. Histone Lysine Methylations and Related Factors
In most cases, methylation of histone H3 lysine 4 (H3K4me) is primarily found at enhancers and promoters of actively transcribed genes, and the methylation status of genes (i.e., mono-, di-, tri-methylation) correlates with its transcriptional activity [15,16]. Members of the lysine methyl transferase 2 (KMT2) family catalyze the addition of methyl groups to H3K4 at the post-translational level, while lysine demethylases (KDMs) remove the methyl groups. This dynamically modulates chromatin structures [17,18]. The KMT2 family, which is highly conserved throughout eukaryotes, can be evolutionarily divided into three subgroups (i.e., KMT2A and KMT2B, KMT2C and KMT2D, and SETD1A and SETD1B) [4,19]. In addition, SMYD2 and SETD3 also have been identified as H3K4 methyltransferases, and eight KDMs are reported to target the H3K4me [4,20].
Methylation of histone H3 lysine 9 (H3K9me) is associated with both heterochromatin formation and gene silencing in euchromatin [2]. H3K9me acts as a binding sit Counseling, e for HP1 [21,22] which forms a complex with chromatin-modifying factors crucial for heterochromatin formation when recruited to H3K9me [23,24]. In the euchromatic region, H3K9me contributes to HP1-mediated gene silencing [25]. H3K9me is catalyzed by several methyltransferases, such as EHMT1, EHMT2, SUV39H1, SUV39H2, SETDB1, dimeric EHMT1-EHMT2, and the PRDM family, and erased by the following lysine demethylases: KDM1, KDM3, KDM4, PHF2, and PHF8 [8,26,27].
Histone H3 lysine 27 methylation (H3K27me) is a repressive chromatin mark that is involved in gene silencing during development and X-chromosome inactivation [28,29]. H3K27me is associated with the repression of developmental regulator genes in human and murine embryonic stem cells (ESCs) [30,31]. Intriguingly, a variety of promoters characteristically contain both H3K4me3 (an activating mark) and H3K27me (a repressive mark) in pluripotent ESCs, which is referred to as “bivalency.” The change in the bivalent situation is associated with differentiation [32]. H3K27me, catalyzed by EZH1 or EZH2 containing Polycomb Repressive Complex (PRC) 2, is a binding site for PRC1 to compact chromosomes [33]. KDM6A, KDM6B, and UTY have been identified as erasers of H3K27me [8].
A role of methylation on histone H3 lysine 36 (H3K36me) has initially been reported in the activation of genes in various systems [34]. However, H3K36me also functions in various processes, including alternative splicing [35], dosage compensation [36], DNA damage response [37], and transcriptional repression [38], depending on the chromatin context. H3K36me is tightly regulated by multiple KMTs and KDMs [20]. In vitro and in vivo studies, to date, have demonstrated that there are the following eight types of KMTs regulating H3K36 methylation levels in humans: SETD2, SETD3, NSD1, NSD2, NSD3, ASH1L, SMYD2, and SETMAR [20]. Although all H3K36-specific methyltransferases contain highly conserved SET domains, the patterns of H3K36 methylation vary. Most H3K36 KMTs preferentially mono- and di-methylate the residue, whereas SETD2 is the only enzyme that catalyzes H3K36me3 and requires mono- or di-methylated H3K36 for its function [39]. Conversely, methylated H3K36 can be demethylated by six KDMs. The H3K36 KDMs, which all belong to the Jumonji protein family, contain the conserved JmjC domain consisting of the following three groups: JHDM1 (KDM2A, KDM2B), JHDM3 (KDM4A, KDM4B, KDM4C), and RIOX1 [40]. JHDM1 is specific for H3K36me1/me2 demethylation, whereas JHDM3 uses H3K36 and H3K9 residues as substrates for the me2/me3-specific demethylation [41]. Similarly, in addition to H3K36me2/me3-specific activity, RIOX1 preferentially demethylates H3K4me1/me3 residues [42].
Histone H3 lysine 79 methylation (H3K79me) is associated with a diverse range of cellular processes including telomeric silencing, cellular development, cell-cycle checkpoint, DNA repair, and transcription regulation [43]. However, only one H3K79-specific KMT is known, with no KDM for H3K79 demethylation reported to date. DOT1L is the sole enzyme that is responsible for all three forms of H3K79 methylation in humans [44]. In addition, DOT1L is unique because it is the only non-SET domain containing methyltransferase, which has been identified to date [18].
Methylation on Histone H4 lysine 20 (H4K20me) displays various biological processes depending on its methylated levels. H4K20me1 is associated with transcriptional activation, appearing in the most highly transcribed group of genes with other core modifications at active promoters [45]. H4K20me2 has distinct roles, such as marking points of replication origin and damage response in the DNA [46,47]. Conversely, H4K20me3 is associated with transcriptional repression at promoters and silencing of repetitive DNA and transposons [45,48]. H4K20me is catalyzed by three enzymes, with activities restricted to specific methylation states. KMT5A, the first identified H4K20 methyltransferase, is the only H4K20me1 enzyme [49]. H4K20me1 can be further di- and tri-methylated by KMT5B and KMT5C [50]. Similarly, several distinct demethylases are involved in the removal of specific H4K20me. PHF8 acts as a demethylase for H4K20me1 [51]. Intriguingly, as previously described, PHF8 is the KDM that has additional activities towards H3K9me1 and H3K9me2 [8]. In addition, LSD1n, an alternatively spliced form of KDM1A, demethylates H4K20me1 and H4K20me2 [52], while PHF2 displays demethylase activity on H4K20me3 [53].
3. Neurodevelopmental Disorders Related with Histone Lysine Methylations
3.1. H3K4 Methylation
3.1.1. KMT2A and Wiedemann-Steiner Syndrome
Mutations in KMT2A were reported to be associated with Wiedemann-Steiner syndrome (WDSTS; OMIM 605130), an extremely rare neurodevelopmental condition accompanied by microcephaly, short stature, autism-like phenotype, and aggression [54]. Interestingly, these abnormal brain functions were recapitulated in KMT2A heterozygous mutant mice, which displayed profound deficits in long-term contextual fear memory [55,56]. In particular, neuronal ablation of KMT2A in the postnatal forebrain and adult prefrontal cortex exhibited increased anxiety and robust cognitive deficits in mice. In the same study, the analyzing H3K4me3 level and the gene expression profiles in KMT2A-deficient cortical neurons revealed that the homeodomain transcription factor, MEIS2, was repressed in these mice. Moreover, MEIS2 knockdown in prefrontal cortex phenocopied memory defects elicited by the deletion of KMT2A [57], thus proposing a critical role of MEIS2 in the pathogenesis of WDSTS.
3.1.2. KMT2D and Kabuki Syndrome 1
The most well-studied neurodevelopmental disorder associated with dysregulated H3K4me is Kabuki syndrome 1 (KABUK1; OMIM 147920), which is a rare congenital syndrome characterized by a distinctive face (a reminiscent of the make-up of actors Kabuki, traditional Japanese music-drama) and mental retardation with additional features including autism, seizure, and microcephaly [58]. Heterozygous mutations in KMT2D were found in more than 50% of patients with KABUK1, with the majority of mutations resulting in the premature termination of the protein product. In addition, mutations in KDM6A, an H3K27me demethylase gene, were also reported to contribute to less than 10% of this syndrome, and this type is referred as Kabuki syndrome 2 (KABUK2; OMIM 300867) [59,60,61,62]. Recently, Bögershausen et al. identified two mutations in RAP1A/B, which encode the Ras family small GTPases, in patients with KABUK1 by whole exome sequencing [61]. The authors also demonstrated that mutant RAP1 morphant phenocopied KDM6A and KMT2D mutants in zebrafish, and that the MEK/ERK pathway signaling was perturbed in RAP1- and KMT2D-defective cells. Interestingly, these phenotypes were rescued by treatment with an MEK inhibitor. On the other hands, the reduction in neurogenesis and hippocampal memory defects exhibited in a KABUK1 mouse model were ameliorated by the treatment with a histone deacetylase (HDAC) inhibitor, AR-42 [63]. Furthermore, a ketogenic diet rescued hippocampal memory defects through the elevation of beta-hydroxybutyrate, an endogenous HDAC inhibitor, in the same mice model [64]. Taken together, these results potentially provide diverse therapeutic directions to treat, or at least mitigate, the symptoms of KABUK1.
3.1.3. SETD1A and Schizophrenia
Extensive exome sequencing from over 200 patients with schizophrenia (SCZD; OMIM 181500) revealed two de novo mutations in SETD1A, which likely cause malfunction of SETD1A activity [65]. Furthermore, a strong association between the loss-of-function mutation of SETD1A and SCZD was confirmed by analyzing the whole exome sequencing of over 4000 patients with SCZD [66]. Interestingly, a recent bioinformatic analysis demonstrated that in addition to mutations in the protein coding region, mutations in the regulatory elements of SETD1A also contributed to the etiology of SCZD. De novo synonymous mutations within frontal cortex-derived DNase I-hypersensitive sites were enriched in SCZD, and SETD1A was identified as the highest statistical significant gene [67].
3.1.4. H3K4me Demethylases and Neurodevelopmental Disorders
Given the intimate association between H3K4 methylation and neurodevelopment disorders, it is rational to assume that KDMs that are responsible for demethylation of H3K4me can be also mutated in neurodevelopmental disorders. Indeed, homozygous missense mutation in KDM5A has been reported in an individual with intellectual disability [68]. Furthermore, KDM5C, another H3K4 demethylase coding gene, has been recurrently mutated in patients with mental retardation, X-linked, syndromic, Claes-Jensen type (MRXSCJ; OMIM 300534) [68,69,70]. Intriguingly, KDM5C has been shown to be transcriptionally regulated by ARX, a homeobox transcription factor, which is frequently mutated in X-linked mental retardation and epilepsy [71,72,73,74]. Additionally, a missense mutation in amine oxidase domain of KDM1A has been reported in patients with mixed features of KABUK1 and KBG syndrome (KBGS; OMIM 148050), which are characterized by macrodontia, distinctive craniofacial findings, and intellectual disability [75]. It is noteworthy that KDM1A catalyzes the demethylation of mono- and di-methylated H3K4, while other KDMs can demethylate H3K4me1/2/3 [76].
3.1.5. PHF21A and Potocki-Shaffer Syndrome
Besides H3K4me writers and erasers, PHF21A, an unmethylated H3K4 reader, was associated with a neurodevelopmental disorder. PHF21A was translocated in patients with Potocki-Shaffer syndrome (PSS; OMIM 601224), characterized by multiple exostoses, parietal foramina, intellectual disability, and craniofacial anomalies [77,78,79]. This translocation commonly results in deletion of the PHD domain coding region of PHF21A, suggesting that dictation of unmethylated H3K4 is crucial for its functions. Accordingly, the deficiency of head development was observed in PHF21A morpholino-injected zebrafish, and this defect was rescued by injection of human PHF21A mRNA [78]. In addition, PHF21A, in combination with KDM1A, is a key component of the BHC complex, which is involved in the repression of neuron-specific genes [80]. Furthermore, SCN3A, a KDM1A target gene, was derepressed, and LSD1 occupancy at the SCN3A promoter was reduced in PHF21A-translocated lymphoblastoid cell lines [78], hence proposing the idea that interplay between KDM1A and PHF21A is indispensable for normal brain development.
3.2. H3K9 Methylation
3.2.1. EHMT1 and Kleefstra Syndrome
Mutations in EHMT1, a gene encoding H3K9 methyltransferase, have been associated with Kleefstra syndrome (KS; OMIM 610253) which is characterized by intellectual disability, childhood hypotonia, and distinctive facial features [81,82]. Previously, this syndrome was known as the 9q Subtelomeric Deletion syndrome, in which minimal critical deleted region comprises EHMT1 [83]. In agreement with the role of EHMT1 on neurodevelopment in human, both Drosophila EHMT mutants and EHMT1 heterozygous knockout mice showed deficits in dendrite branching, learning, and memory [84,85]. Recent studies revealed the functions of EHMT1 in neurons, which may explain the phenotypes of patients and animal models of KS. A study measuring network and single cell activity in cortical cultures showed that EHMT1 is important for cortical neuronal network development [86]. Additionally, EHMT1 mediates homeostatic synaptic scaling, which stabilizes the activity of neural networks by balancing excitation and inhibition [87]. Interestingly, recent studies using exome sequencing revealed that the KS phenotypic spectrum was also linked to mutations in KMT2B and KMT2C [88,89], and these suggest that complicated epigenetic modules might underlie the pathogenesis of KS.
3.2.2. PHF8 and Siderius X-Linked Mental Retardation Syndrome
Siderius X-linked mental retardation syndrome (MRXSSD; OMIM 300263) is an X-linked intellectual disability condition; patients display mental retardation, a long face and broad nasal tip, and cleft lip and palate [90,91]. MRXSSD has been associated with mutations in PHF8 [91,92,93]. Interestingly, PHF8 has a histone lysine demethylase activity towards three different methylated lysines on histones, H3K9me1/2 and H4K20me1 [94,95,96], and also functions as a trimethylated H3K4 reader [94].
Loss of a PHF8 homolog in Caenorhabditis elegans resulted in axon guidance defects via the alteration of Hedgehog-like signaling [97]. Furthermore, injection of zebrafish PHF8 morpholino caused brain and craniofacial development defects [96], thus suggested a critical role of histone methylation dynamics regulated by PHF8 in MRXSSD. However, surprisingly, a recent study showed that Phf8-deficient mice had no obvious developmental defects and cognitive impairment, while Phf8-deficient primary cells had reduced the proliferative potential [98]. The results in mice indicated that MRXSSD is not simply caused by a single PFH8 mutation, but rather by its combination with other genetic or environmental factors at the same time. The different phenotypes exhibited by some animal models and varying degrees of intellectual disability of human patients with MRXSSD can be attributed to the various targets and complex functions of PHF8.
3.3. H3K27 Methylation
EZH2 and Weaver Syndrome
Weaver syndrome (WVS; OMIM 277590) is an autosomal dominant disorder characterized by overgrowth and intellectual disability [99,100,101]. Exome sequencing studies identified EZH2 as a causative gene of WVS [102,103]. EZH2 interacts with EED to form PRC2, which is an H3K27me3 methyltransferase complex [104]. Interestingly, mutations in EED were found in individuals displaying symptoms similar to those of WVS [14,105], suggesting that the dysregulation of H3K27 methylation is responsible for these symptoms.
Several studies have shown that EZH2 deficiencies in animal models induced abnormal neurogenesis in the cerebral cortex [106], cerebellum [107], and spinal cord [108] during embryonic development. Moreover, EZH2 is also implicated in adult hippocampal neurogenesis [109]. The alteration of neurogenesis induced by EZH2 deficiencies has been associated with various neurogenic processes, such as the reduction of neural progenitor cell proliferation [108,109,110], cell fate change [106,107,111,112,113], and neuronal migration [114,115,116]. These results suggest that EZH2-induced H3K27 methylation plays an important role in various processes of neurodevelopment, dysfunction of which might be closely related to intellectual disability in patients with WVS.
3.4. H3K36 Methylation
3.4.1. NSD1 Defects in Sotos Syndrome 1 and Beckwith-Wiedemann Syndrome
Recent studies demonstrated that disrupted levels or patterns of H3K36 methylation can cause a range of human diseases, including neurodevelopmental disorders. Among them, Sotos syndrome 1 (SOTOS1; OMIM 117550) represents an important human model system for studying the neurodevelopmental outcome of epigenetic dysregulation, which is caused by mutations in NSD1 [117]. SOTOS1 is an autosomal dominant disorder characterized by pre- and postnatal overgrowth, facial dysmorphism, macrocephaly, and non-progressive neurological delay [118]. Interestingly, amplified genomic events of NSD1 resulted in the opposite phenotypic outcome of SOTOS1, so that duplication in NSD1 led to reversed clinical phenotypes of SOTOS1 with microcephaly, as well as delayed bone age, indicating the importance of proper NSD1 expression during brain development [119]. In addition, it was shown that neuroblastoma and glioma may occur in human in the absence of NSD1 function [120]. Although the MAPK/ERK pathway was mapped as a downstream signaling pathway of NSD1-related overgrowth of stature in SOTOS1 [121], until recently, the molecular mechanisms how dysregulated NSD1 affects the mental retardation in SOTOS1 patients remains elusive. To date, two Sotos-like overgrowth syndromes called as Sotos syndrome 2 (SOTOS2; OMIM 614753) and 3 (SOTOS3; OMIM 617169) have been reported, which are caused by mutations in the NFIX and APC2 genes, respectively [122,123]. Among the products of the two genes, APC2, a WNT signaling pathway regulator, has recently been suggested as a crucial target of NSD1, of which defects may cause the intellectual disability associated with SOTOS [123]. In the mouse model system, Apc2 deficiency caused impaired learning and memory abilities along with an abnormal head shape. In addition, Nsd1 knockdown downregulated endogenous Apc2 expression, and defective neuronal phenotypes caused by the knockdown were rescued by the forced expression of Apc2, suggesting that APC2 may be a critical downstream gene of NSD1 in human neuronal cells.
Beckwith-Wiedemann syndrome (BWS; OMIM 130650) is another distinct overgrowth disorder with a broad clinical spectrum including hypoglycemia, ear creases/pits, cleft palate, and predisposition to embryonal tumors [124]. Martinez-y-Martinez et al. documented that mental retardation was observed in 6 of the 39 BWS cases [125]. It is well known that a major cause of BWS is the dysregulation of imprinted growth regulatory genes on chromosome 11p15 [126]. Interestingly, mutations in the NSD1 gene have been identified in 2 patients among 52 individuals clinically diagnosed with BWS, which suggests the involvement of NSD1 in imprinting of the 11p15 region [127].
3.4.2. NSD2 and Wolf-Hirshhorn Syndrome
NSD2 is one of the major genes associated with Wolf-Hirshhorn syndrome (WHS; OMIM 194190), of which key features include severe growth and mental retardation, microcephaly, “Greek helmet” facies, and closure defects [128]. Like patients with WHS, mice with Nsd2 gene deletions were growth-retarded, showed midline, craniofacial, and ocular anomalies [129]. However, these mice did not show any learning deficits [129]. Although the downstream effectors of NSD2, such as RUNX2 and p300, which are known to play a role in bone development [130], have been identified, the mechanism by which NSD2 deficiency causes neurological disorders in patients with WHS is still unknown.
3.5. H4K20 Methylation
Siderius X-Linked Syndromic Mental Retardation and Meier-Gorlin Syndrome 1
Thus far, two developmental diseases associated with dysregulated H4K20 methylation have been reported in human. As described above, one is MRXSSD (OMIM 300263) caused by mutations in PHF8, which encodes an eraser of H3K9 and H4K20 methylation. The other is Meier-Gorlin syndrome 1 (MGORS1; OMIM 224690) caused by homozygous or compound heterozygous mutation in the ORC1 gene [131], which encodes a specific reader of H4K20me2 [46]. MGORS1 is a rare disorder characterized by severe intrauterine, postnatal growth retardation, and microcephaly [132]. Interestingly, however, despite the presence of microcephaly, intellects of patients with MGORS1 are usually normal [133].
4. Perspectives
As reviewed above, the pathogenesis of various neurodevelopmental disorders is closely associated with alterations in histone methylation status, which, in many cases, can be primarily attributed to loss-of-function mutations in related factors. Given that histone methylation status is meticulously regulated by the balance between two opposing enzymes (i.e., KMTs and KDMs), pharmaceutical inhibition of specific targets counteracting the loss-of-function mutations responsible for diseases can be a possible therapeutic option. Interestingly, a subset of currently available psychotherapeutic drugs, such as the atypical antipsychotic Clozapine, the mood-stabilizer Valproate, and the antidepressant Phenelzine are known to interfere with histone methylation in the brain [134], although the relative contribution of this interference to their psychotherapeutic effects remains to be elucidated. In principle, an estimated 100 lysine and arginine residue-specific histone methyltransferases and demethylases [135] can be reasonable therapeutic targets, since they are considered more specific than HDACs [134]. Of note, histone methylation has been the most flourishing area of epigenetics research recently, and in line with this, huge efforts have been made to develop several potential therapeutic molecules, which specifically regulate histone methyltransferases and methylation reader proteins, particularly for cancer treatment [136]. For example, selective inhibitors, such as EPZ005687, GSK126, and EI1, which target EZH2 of PRC2, were recently reported by three independent groups to inhibit proliferation of B-cell lymphomas harboring EZH2-activating mutations [137,138,139]. In addition, tranylcypromine derivatives and polyaminoguanidine derivatives were designed and characterized to inhibit histone demethylases with potential anti-cancer activity [136]. Several epigenetic compounds, such as ORY-1001 and GSK2879552, are currently undergoing clinical trials for cancer treatment. If they meet the required biosafety standards, they could potentially be strong candidates for treating neurodevelopmental disorders, by correcting the impaired histone methylation status. Moreover, a microRNA-based gene silencing strategy targeting a specific histone methyltransferase or demethylase can be an alternative therapeutic option to consider in this regard. Indeed, several studies have reported the important roles of miRNA in histone methylation and following transcriptional gene silencing in various model systems [140,141,142]. Although further research is warranted, it will be interesting to establish whether these epigenetic compounds and/or microRNA-based specific gene silencing approaches have obvious therapeutic benefits for the patients with the neurodevelopmental disorders outlined in this review.
Acknowledgments
This work was supported by the DGIST R&D and MIREBraiN program, Basic Science Research Program through the ministry of science, ICT & future planning of Korea (17-BD-0402, 17-BT-02, and 17-01-HRSS-02); the Development of Platform Technology for Innovative Medical Measurements Program from the Korea Research Institute of Standards and Science (KRISS-2017-GP2017-0020) (Sung Bae Lee); the Basic Science Research Program of the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology (NRF-2014R1A1A2056768 and NRF-2017R1A2B4004241 Kyoung Sang Cho), and NRF-2014R1A1A3051462) (Im-Soon Lee); the Korea Health Technology Research & Development Project, Ministry of Health & Welfare, Republic of Korea (HI12C1472) (Kyoung Sang Cho); and KRIBB initiative program (Jeong-Hoon Kim).
Abbreviations
| APC2 | Adenomatosis polyposis coli 2 |
| ARX | Aristaless related homeobox |
| ASH1L | ASH1 like histone lysine methyltransferase |
| BHC complex | BRAF35/histone deacetylase complex |
| BWS | Beckwith-Wiedemann syndrome |
| DOT1L | DOT1 like histone lysine methyltransferase |
| EED | Embryonic ectoderm development |
| EHMT1 | Euchromatic histone lysine methyltransferase 1 |
| EHMT2 | Euchromatic histone lysine methyltransferase 2 |
| ERK | Extracellular signal-regulated kinase |
| ESC | Embryonic stem cells |
| EZH1 | Enhancer of zeste 1 polycomb repressive complex 2 subunit |
| EZH2 | Enhancer of zeste 2 polycomb repressive complex 2 subunit |
| FAD | Flavin adenosine dinucleotide |
| H3K4 | Histone H3 lysine 4 |
| H3K4me | Methylation on histone H3 lysine 4 |
| H3K9 | Histone H3 lysine 9 |
| H3K9me | Methylation on histone H3 lysine 9 |
| H3K27 | Histone H3 lysine 27 |
| H3K27me | Methylation on histone H3 lysine 27 |
| H3K36 | Histone H3 lysine 36 |
| H3K36me | Methylation on histone H3 lysine 36 |
| H3K79 | Histone H3 lysine 79 |
| H3K79me | Methylation on histone H3 lysine 79 |
| H3R2 | Histone H3 arginine 2 |
| H3R8 | Histone H3 arginine 8 |
| H3R17 | Histone H3 arginine 17 |
| H3R26 | Histone H3 arginine 26 |
| H4K20 | Histone H4 lysine 20 |
| H4K20me | Methylation on histone H4 lysine 20 |
| H4R3 | Histone H4 arginine 3 |
| HDAC | Histone deacetylase |
| HP1 | Heterochromatin protein 1 |
| JHDM | JmjC-domain containing histone demethylases |
| JmjC | Jumonji C |
| KABUK1 | Kabuki syndrome 1 |
| KABUK2 | Kabuki syndrome 2 |
| KBGS | KBG syndrome |
| KS | Kleefstra syndrome |
| LSD1n | Lysine-specific demethylase 1 variant |
| KDM | Lysine demethylase |
| KMT | Lysine methyl transferase |
| MAPK | Mitogen-activated protein kinase |
| MEIS2 | Myeloid ecotropic viral integration site 1 homolog 2 |
| MEK | Mitogen-activated protein kinase kinase |
| MGORS1 | Meier-Gorlin syndrome 1 |
| MRXSCJ | Mental retardation, X-linked, syndromic, Claes-Jensen type |
| MRXSSD | Siderius X-linked mental retardation syndrome |
| NFIX | Nuclear factor I X |
| NSD1 | Nuclear receptor-binding SET domain protein 1 |
| NSD2 | Nuclear receptor-binding SET domain protein 2 |
| NSD3 | Nuclear receptor-binding SET domain protein 3 |
| ORC1 | Origin recognition complex subunit 1 |
| PHF2 | PHD finger protein 2 |
| PHF21A | PHD finger protein 21A |
| PHF8 | PHD finger protein 8 |
| PRC1 | Polycomb repressive complex 1 |
| PRC2 | Polycomb repressive complex 2 |
| PRDM | PR/SET domain family |
| PSS | Potocki-Shaffer syndrome |
| RAP1A/B | RAS-related protein 1A/B |
| RIOX1 | Ribosomal oxygenase 1 |
| RUNX2 | Runt related transcription factor 2 |
| SCZD | Schizophrenia |
| SCN3A | Sodium voltage-gated channel alpha subunit 3 |
| SETD1A | SET domain containing 1A |
| SETD1B | SET domain containing 1B |
| SETD2 | SET domain containing 2 |
| SETD3 | SET domain containing 3 |
| SETDB1 | SET domain bifurcated 1 |
| SETMAR | SET domain and mariner transposase fusion gene |
| SMYD2 | SET and MYND domain containing 2 |
| SOTOS1 | Sotos syndrome 1 |
| SOTOS2 | Sotos syndrome 2 |
| SOTOS3 | Sotos syndrome 3 |
| SUV39H1 | Suppressor of variegation 3-9 homolog 1 |
| SUV39H2 | Suppressor of variegation 3-9 homolog 2 |
| UTY | Ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
| WDSTS | Wiedemann-Steiner syndrome |
| WHS | Wolf-Hirshhorn syndrome |
| WVS | Weaver syndrome |
Author Contributions
Sung Bae Lee and Kyoung Sang Cho designed the review; and Jeong-Hoon Kim, Jang Ho Lee, Im-Soon Lee, Sung Bae Lee and Kyoung Sang Cho wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Martin C., Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 4.Greer E.L., Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 6.Di Lorenzo A., Bedford M.T. Histone arginine methylation. FEBS Lett. 2011;585:2024–2031. doi: 10.1016/j.febslet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuchs J., Demidov D., Houben A., Schubert I. Chromosomal histone modification patterns–from conservation to diversity. Trends Plant Sci. 2006;11:199–208. doi: 10.1016/j.tplants.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Hyun K., Jeon J., Park K., Kim J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017;49:e324. doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bale T.L., Baram T.Z., Brown A.S., Goldstein J.M., Insel T.R., McCarthy M.M., Nemeroff C.B., Reyes T.M., Simerly R.B., Susser E.S. Early life programming and neurodevelopmental disorders. Biol. Psychiatry. 2010;68:314–319. doi: 10.1016/j.biopsych.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millan M.J. An epigenetic framework for neurodevelopmental disorders: From pathogenesis to potential therapy. Neuropharmacology. 2013;68:2–82. doi: 10.1016/j.neuropharm.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Gilissen C., Hoischen A., Brunner H.G., Veltman J.A. Unlocking Mendelian disease using exome sequencing. Genome Biol. 2011;12:228. doi: 10.1186/gb-2011-12-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ronan J.L., Wu W., Crabtree G.R. From neural development to cognition: Unexpected roles for chromatin. Nat. Rev. Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vissers L.E., Gilissen C., Veltman J.A. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 14.Tatton-Brown K., Loveday C., Yost S., Clarke M., Ramsay E., Zachariou A., Elliott A., Wylie H., Ardissone A., Rittinger O. Mutations in epigenetic regulation genes are a major cause of overgrowth with intellectual disability. Am. J. Hum. Genet. 2017;100:725–736. doi: 10.1016/j.ajhg.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 16.Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allis C.D., Berger S.L., Cote J., Dent S., Jenuwien T., Kouzarides T., Pillus L., Reinberg D., Shi Y., Shiekhattar R. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 18.Shilatifard A. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 19.Rao R.C., Dou Y. Hijacked in cancer: The KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer. 2015;15:334–346. doi: 10.1038/nrc3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner E.J., Carpenter P.B. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 22.Lachner M., O’carroll D., Rea S., Mechtler K., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama J.-I., Rice J.C., Strahl B.D., Allis C.D., Grewal S.I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 24.Grewal S.I., Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 25.Schultz D.C., Ayyanathan K., Negorev D., Maul G.G., Rauscher F.J. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metzger E., Wissmann M., Yin N., Müller J.M., Schneider R., Peters A.H., Günther T., Buettner R., Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 27.Kim H.J., Park J.W., Lee K.H., Yoon H., Shin D.H., Ju U.I., Seok S.H., Lim S.H., Lee Z.H., Kim H.H. Plant homeodomain finger protein 2 promotes bone formation by demethylating and activating Runx2 for osteoblast differentiation. Cell Res. 2014;24:1231–1249. doi: 10.1038/cr.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plath K., Fang J., Mlynarczyk-Evans S.K., Cao R., Worringer K.A., Wang H., Cecile C., Otte A.P., Panning B., Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 29.Wiles E.T., Selker E.U. H3K27 methylation: A promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 2017;43:31–37. doi: 10.1016/j.gde.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 31.Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K.-I. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harikumar A., Meshorer E. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep. 2015;16:1609–1619. doi: 10.15252/embr.201541011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margueron R., Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kizer K.O., Phatnani H.P., Shibata Y., Hall H., Greenleaf A.L., Strahl B.D. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3K36 methylation with transcript elongation. Mol. Cell. Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S., Kim H., Fong N., Erickson B., Bentley D.L. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc. Natl. Acad. Sci. USA. 2011;108:13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bell O., Conrad T., Kind J., Wirbelauer C., Akhtar A., Schübeler D. Transcription-coupled methylation of histone H3 at lysine 36 regulates dosage compensation by enhancing recruitment of the MSL complex in Drosophila melanogaster. Mol. Cell. Biol. 2008;28:3401–3409. doi: 10.1128/MCB.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fnu S., Williamson E.A., De Haro L.P., Brenneman M., Wray J., Shaheen M., Radhakrishnan K., Lee S.-H., Nickoloff J.A., Hromas R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. USA. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strahl B.D., Grant P.A., Briggs S.D., Sun Z.-W., Bone J.R., Caldwell J.A., Mollah S., Cook R.G., Shabanowitz J., Hunt D.F. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 2002;22:1298–1306. doi: 10.1128/MCB.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edmunds J.W., Mahadevan L.C., Clayton A.L. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cloos P.A., Christensen J., Agger K., Helin K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li G.-M. Decoding the histone code: Role of H3K36me3 in mismatch repair and implications for cancer susceptibility and therapy. Cancer Res. 2013;73:6379–6383. doi: 10.1158/0008-5472.CAN-13-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinha K.M., Yasuda H., Coombes M.M., Dent S.Y., De Crombrugghe B. Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J. 2010;29:68–79. doi: 10.1038/emboj.2009.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farooq Z., Banday S., Pandita T.K., Altaf M. The many faces of histone H3K79 methylation. Mutat. Res. Rev. Mutat. Res. 2016;768:46–52. doi: 10.1016/j.mrrev.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Leeuwen F., Gafken P.R., Gottschling D.E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/S0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z., Zang C., Rosenfeld J.A., Schones D.E., Barski A., Cuddapah S., Cui K., Roh T.-Y., Peng W., Zhang M.Q. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuo A.J., Song J., Cheung P., Ishibe-Murakami S., Yamazoe S., Chen J.K., Patel D.J., Gozani O. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484:115–119. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Botuyan M.V., Lee J., Ward I.M., Kim J.-E., Thompson J.R., Chen J., Mer G. Structural basis for the methylation state-specific recognition of histone H4K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jørgensen S., Schotta G., Sørensen C.S. Histone H4 lysine 20 methylation: Key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013;41:2797–2806. doi: 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishioka K., Rice J.C., Sarma K., Erdjument-Bromage H., Werner J., Wang Y., Chuikov S., Valenzuela P., Tempst P., Steward R. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell. 2002;9:1201–1213. doi: 10.1016/S1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 50.Yang H., Pesavento J.J., Starnes T.W., Cryderman D.E., Wallrath L.L., Kelleher N.L., Mizzen C.A. Preferential dimethylation of histone H4 lysine 20 by Suv4–20. J. Biol. Chem. 2008;283:12085–12092. doi: 10.1074/jbc.M707974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu W., Tanasa B., Tyurina O.V., Zhou T.Y., Gassmann R., Liu W.T., Ohgi K.A., Benner C., Garcia-Bassets I., Aggarwal A.K. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J., Telese F., Tan Y., Li W., Jin C., He X., Basnet H., Ma Q., Merkurjev D., Zhu X. LSD1n is an H4K20 demethylase regulating memory formation via transcriptional elongation control. Nat. Neurosci. 2015;18:1256–1264. doi: 10.1038/nn.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stender J.D., Pascual G., Liu W., Kaikkonen M.U., Do K., Spann N.J., Boutros M., Perrimon N., Rosenfeld M.G., Glass C.K. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol. Cell. 2012;48:28–38. doi: 10.1016/j.molcel.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strom S.P., Lozano R., Lee H., Dorrani N., Mann J., O’Lague P.F., Mans N., Deignan J.L., Vilain E., Nelson S.F. De Novo variants in the KMT2A (MLL) gene causing atypical Wiedemann-Steiner syndrome in two unrelated individuals identified by clinical exome sequencing. BMC Med. Genet. 2014;15:49. doi: 10.1186/1471-2350-15-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta S., Kim S.Y., Artis S., Molfese D.L., Schumacher A., Sweatt J.D., Paylor R.E., Lubin F.D. Histone methylation regulates memory formation. J. Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim S.Y., Levenson J.M., Korsmeyer S., Sweatt J.D., Schumacher A. Developmental regulation of Eed complex composition governs a switch in global histone modification in brain. J. Biol. Chem. 2007;282:9962–9972. doi: 10.1074/jbc.M608722200. [DOI] [PubMed] [Google Scholar]
- 57.Jakovcevski M., Ruan H., Shen E.Y., Dincer A., Javidfar B., Ma Q., Peter C.J., Cheung I., Mitchell A.C., Jiang Y. Neuronal Kmt2a/Mll1 histone methyltransferase is essential for prefrontal synaptic plasticity and working memory. J. Neurosci. 2015;35:5097–5108. doi: 10.1523/JNEUROSCI.3004-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Niikawa N., Matsuura N., Fukushima Y., Ohsawa T., Kajii T. Kabuki make-up syndrome: A syndrome of mental retardation, unusual facies, large and protruding ears, and postnatal growth deficiency. J. Pediatr. 1981;99:565–569. doi: 10.1016/S0022-3476(81)80255-7. [DOI] [PubMed] [Google Scholar]
- 59.Ng S.B., Bigham A.W., Buckingham K.J., Hannibal M.C., McMillin M.J., Gildersleeve H.I., Beck A.E., Tabor H.K., Cooper G.M., Mefford H.C. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paulussen A.D., Stegmann A., Blok M.J., Tserpelis D., Posma-Velter C., Detisch Y., Smeets E.E., Wagemans A., Schrander J.J., van den Boogaard M.J.H. MLL2 mutation spectrum in 45 patients with Kabuki syndrome. Hum. Mutat. 2011;32:E2018–E2025. doi: 10.1002/humu.21416. [DOI] [PubMed] [Google Scholar]
- 61.Bögershausen N., Wollnik B. Unmasking Kabuki syndrome. Clin. Genet. 2013;83:201–211. doi: 10.1111/cge.12051. [DOI] [PubMed] [Google Scholar]
- 62.Lederer D., Grisart B., Digilio M.C., Benoit V., Crespin M., Ghariani S.C., Maystadt I., Dallapiccola B., Verellen-Dumoulin C. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am. J. Hum. Genet. 2012;90:119–124. doi: 10.1016/j.ajhg.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benjamin J.S., Pilarowski G.O., Carosso G.A., Zhang L., Huso D.L., Goff L.A., Vernon H.J., Hansen K.D., Bjornsson H.T. A ketogenic diet rescues hippocampal memory defects in a mouse model of Kabuki syndrome. Proc. Natl. Acad. Sci. USA. 2017;114:125–130. doi: 10.1073/pnas.1611431114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bjornsson H.T., Benjamin J.S., Zhang L., Weissman J., Gerber E.E., Chen Y.-C., Vaurio R.G., Potter M.C., Hansen K.D., Dietz H.C. Histone deacetylase inhibition rescues structural and functional brain deficits in a mouse model of Kabuki syndrome. Sci. Transl. Med. 2014;6:256ra135. doi: 10.1126/scitranslmed.3009278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takata A., Xu B., Ionita-Laza I., Roos J.L., Gogos J.A., Karayiorgou M. Loss-of-function variants in schizophrenia risk and SETD1A as a candidate susceptibility gene. Neuron. 2014;82:773–780. doi: 10.1016/j.neuron.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh T., Kurki M.I., Curtis D., Purcell S.M., Crooks L., McRae J., Suvisaari J., Chheda H., Blackwood D., Breen G. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat. Neurosci. 2016;19:571–577. doi: 10.1038/nn.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takata A., Ionita-Laza I., Gogos J.A., Xu B., Karayiorgou M. De novo synonymous mutations in regulatory elements contribute to the genetic etiology of autism and schizophrenia. Neuron. 2016;89:940–947. doi: 10.1016/j.neuron.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Najmabadi H., Hu H., Garshasbi M., Zemojtel T., Abedini S.S., Chen W., Hosseini M., Behjati F., Haas S., Jamali P. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- 69.Jensen L.R., Amende M., Gurok U., Moser B., Gimmel V., Tzschach A., Janecke A.R., Tariverdian G., Chelly J., Fryns J.-P. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 2005;76:227–236. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tahiliani M., Mei P., Fang R., Leonor T., Rutenberg M., Shimizu F., Li J., Rao A., Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 71.Poeta L., Fusco F., Drongitis D., Shoubridge C., Manganelli G., Filosa S., Paciolla M., Courtney M., Collombat P., Lioi M.B. A regulatory path associated with X-linked intellectual disability and epilepsy links KDM5C to the polyalanine expansions in ARX. Am. J. Hum. Genet. 2013;92:114–125. doi: 10.1016/j.ajhg.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strømme P., Mangelsdorf M.E., Scheffer I.E., Gécz J. Infantile spasms, dystonia, and other X-linked phenotypes caused by mutations in Aristaless related homeobox gene, ARX. Brain Dev. 2002;24:266–268. doi: 10.1016/S0387-7604(02)00079-7. [DOI] [PubMed] [Google Scholar]
- 73.Kitamura K., Yanazawa M., Sugiyama N., Miura H., Iizuka-Kogo A., Kusaka M., Omichi K., Suzuki R., Kato-Fukui Y., Kamiirisa K. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat. Genet. 2002;32:359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- 74.Kato M., Das S., Petras K., Kitamura K., Morohashi K.I., Abuelo D.N., Barr M., Bonneau D., Brady A.F., Carpenter N.J. Mutations of ARX are associated with striking pleiotropy and consistent genotype–phenotype correlation. Hum. Mutat. 2004;23:147–159. doi: 10.1002/humu.10310. [DOI] [PubMed] [Google Scholar]
- 75.Tunovic S., Barkovich J., Sherr E.H., Slavotinek A.M. De novo ANKRD11 and KDM1A gene mutations in a male with features of KBG syndrome and Kabuki syndrome. Am. J. Med. Genet. 2014;164:1744–1749. doi: 10.1002/ajmg.a.36450. [DOI] [PubMed] [Google Scholar]
- 76.Shi Y. Histone lysine demethylases: Emerging roles in development, physiology and disease. Nat. Rev. Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 77.Lan F., Collins R.E., De Cegli R., Alpatov R., Horton J.R., Shi X., Gozani O., Cheng X., Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim H.-G., Kim H.-T., Leach N.T., Lan F., Ullmann R., Silahtaroglu A., Kurth I., Nowka A., Seong I.S., Shen Y. Translocations disrupting PHF21A in the Potocki-Shaffer-syndrome region are associated with intellectual disability and craniofacial anomalies. Am. J. Hum. Genet. 2012;91:56–72. doi: 10.1016/j.ajhg.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Labonne J.D., Vogt J., Reali L., Kong I.K., Layman L.C., Kim H.G. A microdeletion encompassing PHF21A in an individual with global developmental delay and craniofacial anomalies. Am. J. Med. Genet. 2015;167:3011–3018. doi: 10.1002/ajmg.a.37344. [DOI] [PubMed] [Google Scholar]
- 80.Hakimi M.-A., Bochar D.A., Chenoweth J., Lane W.S., Mandel G., Shiekhattar R. A core–BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. USA. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kleefstra T., Smidt M., Banning M., Oudakker A., Van Esch H., De Brouwer A., Nillesen W., Sistermans E., Hamel B., De Bruijn D. Disruption of the gene Euchromatin Histone Methyl Transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J. Med. Genet. 2005;42:299–306. doi: 10.1136/jmg.2004.028464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kleefstra T., Brunner H.G., Amiel J., Oudakker A.R., Nillesen W.M., Magee A., Geneviève D., Cormier-Daire V., Van Esch H., Fryns J.-P. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am. J. Hum. Genet. 2006;79:370–377. doi: 10.1086/505693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harada N., Visser R., Dawson A., Fukamachi M., Iwakoshi M., Okamoto N., Kishino T., Niikawa N., Matsumoto N. A 1-Mb critical region in six patients with 9q34. 3 terminal deletion syndrome. J. Hum. Genet. 2004;49:440–444. doi: 10.1007/s10038-004-0166-z. [DOI] [PubMed] [Google Scholar]
- 84.Kramer J.M., Kochinke K., Oortveld M.A., Marks H., Kramer D., de Jong E.K., Asztalos Z., Westwood J.T., Stunnenberg H.G., Sokolowski M.B. Epigenetic regulation of learning and memory by Drosophila EHMT/G9a. PLoS Biol. 2011;9:e1000569. doi: 10.1371/journal.pbio.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Balemans M., Nadif Kasri N., Kopanitsa M.V., Afinowi N.O., Ramakers G., Peters T.A., Beynon A.J., Janssen S.M., van Summeren R.C., Eeftens J.M. Hippocampal dysfunction in the Euchromatin histone methyltransferase 1 heterozygous knockout mouse model for Kleefstra syndrome. Hum. Mol. Genet. 2013;22:852–866. doi: 10.1093/hmg/dds490. [DOI] [PubMed] [Google Scholar]
- 86.Martens M.B., Frega M., Classen J., Epping L., Bijvank E., Benevento M., van Bokhoven H., Tiesinga P., Schubert D., Kasri N.N. Euchromatin histone methyltransferase 1 regulates cortical neuronal network development. Sci. Rep. 2016;6:35756. doi: 10.1038/srep35756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Benevento M., Iacono G., Selten M., Ba W., Oudakker A., Frega M., Keller J., Mancini R., Lewerissa E., Kleefstra T. Histone methylation by the Kleefstra syndrome protein EHMT1 mediates homeostatic synaptic scaling. Neuron. 2016;91:341–355. doi: 10.1016/j.neuron.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 88.Kleefstra T., Kramer J.M., Neveling K., Willemsen M.H., Koemans T.S., Vissers L.E., Wissink-Lindhout W., Fenckova M., van den Akker W.M., Kasri N.N. Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am. J. Hum. Genet. 2012;91:73–82. doi: 10.1016/j.ajhg.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Agha Z., Iqbal Z., Azam M., Ayub H., Vissers L.E., Gilissen C., Ali S.H.B., Riaz M., Veltman J.A., Pfundt R. Exome sequencing identifies three novel candidate genes implicated in intellectual disability. PLoS ONE. 2014;9:e112687. doi: 10.1371/journal.pone.0112687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Siderius L.E., Hamel B.C., van Bokhoven H., de Jager F., van den Helm B., Kremer H., Heineman-de Boer J.A., Ropers H.H., Mariman E.C. X-linked mental retardation associated with cleft lip/palate maps to Xp11.3-q21.3. Am. J. Med. Genet. 1999;85:216–220. doi: 10.1002/(SICI)1096-8628(19990730)85:3<216::AID-AJMG6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 91.Laumonnier F., Holbert S., Ronce N., Faravelli F., Lenzner S., Schwartz C., Lespinasse J., Van Esch H., Lacombe D., Goizet C. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J. Med. Genet. 2005;42:780–786. doi: 10.1136/jmg.2004.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Abidi F., Miano M., Murray J., Schwartz C. A novel mutation in the PHF8 gene is associated with X-linked mental retardation with cleft lip/cleft palate. Clin. Genet. 2007;72:19–22. doi: 10.1111/j.1399-0004.2007.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koivisto A., Ala-Mello S., Lemmelä S., Komu H., Rautio J., Järvelä I. Screening of mutations in the PHF8 gene and identification of a novel mutation in a Finnish family with XLMR and cleft lip/cleft palate. Clin. Genet. 2007;72:145–149. doi: 10.1111/j.1399-0004.2007.00836.x. [DOI] [PubMed] [Google Scholar]
- 94.Feng W., Yonezawa M., Ye J., Jenuwein T., Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat. Struct. Mol. Biol. 2010;17:445–450. doi: 10.1038/nsmb.1778. [DOI] [PubMed] [Google Scholar]
- 95.Kleine-Kohlbrecher D., Christensen J., Vandamme J., Abarrategui I., Bak M., Tommerup N., Shi X., Gozani O., Rappsilber J., Salcini A.E. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol. Cell. 2010;38:165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qi H.H., Sarkissian M., Hu G.-Q., Wang Z., Bhattacharjee A., Gordon D.B., Gonzales M., Lan F., Ongusaha P.P., Huarte M. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Riveiro A.R., Mariani L., Malmberg E., Amendola P.G., Peltonen J., Wong G., Salcini A.E. JMJD-1.2/PHF8 controls axon guidance by regulating Hedgehog-like signaling. Development. 2017;144:856–865. doi: 10.1242/dev.142695. [DOI] [PubMed] [Google Scholar]
- 98.Walsh R.M., Shen E.Y., Bagot R.C., Anselmo A., Jiang Y., Javidfar B., Wojtkiewicz G.J., Cloutier J., Chen J.W., Sadreyev R. Phf8 loss confers resistance to depression-like and anxiety-like behaviors in mice. Nat. Commun. 2017;8:15142. doi: 10.1038/ncomms15142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Weaver D.D., Graham C.B., Thomas I., Smith D.W. A new overgrowth syndrome with accelerated skeletal maturation, unusual facies, and camptodactyly. J. Pediatr. 1974;84:547–552. doi: 10.1016/S0022-3476(74)80675-X. [DOI] [PubMed] [Google Scholar]
- 100.Cohen M.M. Mental deficiency, alterations in performance, and CNS abnormalities in overgrowth syndromes. Am. J. Med. Genet. 2003;117C:49–56. doi: 10.1002/ajmg.c.10013. [DOI] [PubMed] [Google Scholar]
- 101.Tatton-Brown K., Murray A., Hanks S., Douglas J., Armstrong R., Banka S., Bird L.M., Clericuzio C.L., Cormier-Daire V., Cushing T. Weaver syndrome and EZH2 mutations: Clarifying the clinical phenotype. Am. J. Med. Genet. 2013;161:2972–2980. doi: 10.1002/ajmg.a.36229. [DOI] [PubMed] [Google Scholar]
- 102.Tatton-Brown K., Hanks S., Ruark E., Zachariou A., Duarte S.D.V., Ramsay E., Snape K., Murray A., Perdeaux E.R., Seal S. Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget. 2011;2:1127–1133. doi: 10.18632/oncotarget.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gibson W.T., Hood R.L., Zhan S.H., Bulman D.E., Fejes A.P., Moore R., Mungall A.J., Eydoux P., Babul-Hirji R., An J. Mutations in EZH2 cause Weaver syndrome. Am. J. Hum. Genet. 2012;90:110–118. doi: 10.1016/j.ajhg.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 105.Cohen A.S., Tuysuz B., Shen Y., Bhalla S.K., Jones S.J., Gibson W.T. A novel mutation in EED associated with overgrowth. J. Hum. Genet. 2015;60:339–342. doi: 10.1038/jhg.2015.26. [DOI] [PubMed] [Google Scholar]
- 106.Pereira J.D., Sansom S.N., Smith J., Dobenecker M.-W., Tarakhovsky A., Livesey F.J. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc. Natl. Acad. Sci. USA. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Feng X., Juan A.H., Wang H.A., Ko K.D., Zare H., Sartorelli V. Polycomb Ezh2 controls the fate of GABAergic neurons in the embryonic cerebellum. Development. 2016;143:1971–1980. doi: 10.1242/dev.132902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Akizu N., García M.A., Estarás C., Fueyo R., Badosa C., de la Cruz X., Martínez-Balbás M.A. EZH2 regulates neuroepithelium structure and neuroblast proliferation by repressing p21. Open Biol. 2016;6:150227. doi: 10.1098/rsob.150227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang J., Ji F., Liu Y., Lei X., Li H., Ji G., Yuan Z., Jiao J. Ezh2 regulates adult hippocampal neurogenesis and memory. J. Neurosci. 2014;34:5184–5199. doi: 10.1523/JNEUROSCI.4129-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang J., Taylor R.J., La Torre A., Wilken M.S., Cox K.E., Reh T.A., Vetter M.L. Ezh2 maintains retinal progenitor proliferation, transcriptional integrity, and the timing of late differentiation. Dev. Biol. 2015;403:128–138. doi: 10.1016/j.ydbio.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hirabayashi Y., Suzki N., Tsuboi M., Endo T.A., Toyoda T., Shinga J., Koseki H., Vidal M., Gotoh Y. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 112.Yu Y.-L., Chou R.-H., Chen L.-T., Shyu W.-C., Hsieh S.-C., Wu C.-S., Zeng H.-J., Yeh S.-P., Yang D.-M., Hung S.-C. EZH2 regulates neuronal differentiation of mesenchymal stem cells through PIP5K1C-dependent calcium signaling. J. Biol. Chem. 2011;286:9657–9667. doi: 10.1074/jbc.M110.185124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hwang W.W., Salinas R.D., Siu J.J., Kelley K.W., Delgado R.N., Paredes M.F., Alvarez-Buylla A., Oldham M.C., Lim D.A. Distinct and separable roles for EZH2 in neurogenic astroglia. Elife. 2014;3:e02439. doi: 10.7554/eLife.02439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Di Meglio T., Kratochwil C.F., Vilain N., Loche A., Vitobello A., Yonehara K., Hrycaj S.M., Roska B., Peters A.H., Eichmann A. Ezh2 orchestrates topographic migration and connectivity of mouse precerebellar neurons. Science. 2013;339:204–207. doi: 10.1126/science.1229326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao Q., Rank G., Tan Y.T., Li H., Moritz R.L., Simpson R.J., Cerruti L., Curtis D.J., Patel D.J., Allis C.D. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tien C.-L., Jones A., Wang H., Gerigk M., Nozell S., Chang C. Snail2/Slug cooperates with Polycomb repressive complex 2 (PRC2) to regulate neural crest development. Development. 2015;142:722–731. doi: 10.1242/dev.111997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kurotaki N., Imaizumi K., Harada N., Masuno M., Kondoh T., Nagai T., Ohashi H., Naritomi K., Tsukahara M., Makita Y. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 2002;30:365–366. doi: 10.1038/ng863. [DOI] [PubMed] [Google Scholar]
- 118.Sotos J.F., Dodge P.R., Muirhead D., Crawford J.D., Talbot N.B. Cerebral gigantism in childhood: A syndrome of excessively rapid growth with acromegalic features and a nonprogressive neurologic disorder. N. Engl. J. Med. 1964;271:109–116. doi: 10.1056/NEJM196407162710301. [DOI] [PubMed] [Google Scholar]
- 119.Žilina O., Reimand T., Tammur P., Tillmann V., Kurg A., Õunap K. Patient with dup (5) (q35.2-q35.3) reciprocal to the common Sotos syndrome deletion and review of the literature. Eur. J. Med. Genet. 2013;56:202–206. doi: 10.1016/j.ejmg.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 120.Berdasco M., Ropero S., Setien F., Fraga M.F., Lapunzina P., Losson R., Alaminos M., Cheung N.-K., Rahman N., Esteller M. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc. Natl. Acad. Sci. USA. 2009;106:21830–21835. doi: 10.1073/pnas.0906831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Visser R., Landman E.B., Goeman J., Wit J.M., Karperien M. Sotos syndrome is associated with deregulation of the MAPK/ERK-signaling pathway. PLoS ONE. 2012;7:e49229. doi: 10.1371/journal.pone.0049229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Malan V., Rajan D., Thomas S., Shaw A.C., Louis Dit Picard H., Layet V., Till M., van Haeringen A., Mortier G., Nampoothiri S., et al. Distinct effects of allelic NFIX mutations on nonsense-mediated mRNA decay engender either a Sotos-like or a Marshall-Smith syndrome. Am. J. Hum. Genet. 2010;87:189–198. doi: 10.1016/j.ajhg.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Almuriekhi M., Shintani T., Fahiminiya S., Fujikawa A., Kuboyama K., Takeuchi Y., Nawaz Z., Nadaf J., Kamel H., Kitam A.K. Loss-of-function mutation in APC2 causes Sotos syndrome features. Cell Rep. 2015;10:1585–1598. doi: 10.1016/j.celrep.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 124.Engström W., Lindham S., Schofield P. Wiedemann-Beckwith syndrome. Eur. J. Pediatr. 1988;147:450–457. doi: 10.1007/BF00441965. [DOI] [PubMed] [Google Scholar]
- 125.Martínez-y-Martínez R., Martínez-Carboney R., Ocampo-Campos R., Rivera H., Cuevas A., Martín M.M. Wiedemann-Beckwith syndrome: Clinical, cytogenetical and radiological observations in 39 new cases. Genet. Couns. 1992;3:67–76. [PubMed] [Google Scholar]
- 126.Waziri M., Patil S.R., Hanson J.W., Bartley J.A. Abnormality of chromosome 11 in patients with features of Beckwith-Wiedemann syndrome. J. Pediatr. 1983;102:873–876. doi: 10.1016/S0022-3476(83)80014-6. [DOI] [PubMed] [Google Scholar]
- 127.Baujat G., Rio M., Rossignol S., Sanlaville D., Lyonnet S., Le Merrer M., Munnich A., Gicquel C., Cormier-Daire V., Colleaux L. Paradoxical NSD1 mutations in Beckwith-Wiedemann syndrome and 11p15 anomalies in Sotos syndrome. Am. J. Hum. Genet. 2004;74:715–720. doi: 10.1086/383093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Battaglia A., Carey J.C., Cederholm P., Viskochil D.H., Brothman A.R., Galasso C. Natural history of Wolf-Hirschhorn syndrome: Experience with 15 cases. Pediatrics. 1999;103:830–836. doi: 10.1542/peds.103.4.830. [DOI] [PubMed] [Google Scholar]
- 129.Näf D., Wilson L.A., Bergstrom R.A., Smith R.S., Goodwin N.C., Verkerk A., van Ommen G.J., Ackerman S.L., Frankel W.N., Schimenti J.C. Mouse models for the Wolf–Hirschhorn deletion syndrome. Hum. Mol. Genet. 2001;10:91–98. doi: 10.1093/hmg/10.2.91. [DOI] [PubMed] [Google Scholar]
- 130.Lee Y.F., Nimura K., Lo W.N., Saga K., Kaneda Y. Histone H3 lysine 36 methyltransferase Whsc1 promotes the association of RUNX2 and p300 in the activation of bone-related genes. PLoS ONE. 2014;9:e106661. doi: 10.1371/journal.pone.0106661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bicknell L.S., Walker S., Klingseisen A., Stiff T., Leitch A., Kerzendorfer C., Martin C.-A., Yeyati P., Al Sanna N., Bober M. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat. Genet. 2011;43:350–355. doi: 10.1038/ng.776. [DOI] [PubMed] [Google Scholar]
- 132.Shalev S.A., Hall J.G. Another adult with Meier-Gorlin syndrome-insights into the natural history. Clin. Dysmorphol. 2003;12:167–169. doi: 10.1097/01.mcd.0000065052.36236.32. [DOI] [PubMed] [Google Scholar]
- 133.Bicknell L.S., Bongers E.M., Leitch A., Brown S., Schoots J., Harley M.E., Aftimos S., Al-Aama J.Y., Bober M., Brown P.A. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat. Genet. 2011;43:356–359. doi: 10.1038/ng.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Peter C.J., Akbarian S. Balancing histone methylation activities in psychiatric disorders. Trends Mol. Med. 2011;17:372–379. doi: 10.1016/j.molmed.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Copeland R.A., Solomon M.E., Richon V.M. Protein methyltransferases as a target class for drug discovery. Nat. Rev. Drug Discov. 2009;8:724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 136.Liu Y., Liu K., Qin S., Xu C., Min J. Epigenetic targets and drug discovery: Part 1: Histone methylation. Pharmacol. Ther. 2014;143:275–294. doi: 10.1016/j.pharmthera.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 137.Knutson S.K., Wigle T.J., Warholic N.M., Sneeringer C.J., Allain C.J., Klaus C.R., Sacks J.D., Raimondi A., Majer C.R., Song J. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 138.McCabe M.T., Ott H.M., Ganji G., Korenchuk S., Thompson C., Van Aller G.S., Liu Y., Graves A.P., Diaz E., LaFrance L.V. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 139.Qi W., Chan H., Teng L., Li L., Chuai S., Zhang R., Zeng J., Li M., Fan H., Lin Y. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc. Natl. Acad. Sci. USA. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim D.H., Sætrom P., Snøve O., Rossi J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Varambally S., Cao Q., Mani R.-S., Shankar S., Wang X., Ateeq B., Laxman B., Cao X., Jing X., Ramnarayanan K. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hawkins P.G., Santoso S., Adams C., Anest V., Morris K.V. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]