Abstract
Here we report the case of a 30-year-old woman with relapsed acute myeloid leukemia (AML) who was treated with all-trans retinoic acid (ATRA) as part of investigational therapy (NCT02273102). The patient died from rapid disease progression following eight days of continuous treatment with ATRA. Karyotype analysis and RNA-Seq revealed the presence of a novel t(4;15)(q31;q22) reciprocal translocation involving the TMEM154 and RASGRF1 genes. Analysis of primary cells from the patient revealed the expression of TMEM154-RASGRF1 mRNA and the resulting fusion protein, but no expression of the reciprocal RASGRF1-TMEM154 fusion. Consistent with the response of the patient to ATRA therapy, we observed a rapid proliferation of t(4;15) primary cells following ATRA treatment ex vivo. Preliminary characterization of the retinoid response of t(4;15) AML revealed that in stark contrast to non-t(4;15) AML, these cells proliferate in response to specific agonists of RARα and RARγ. Furthermore, we observed an increase in the levels of nuclear RARγ upon ATRA treatment. In summary, the identification of the novel t(4;15)(q31;q22) reciprocal translocation opens new avenues in the study of retinoid resistance and provides potential for a new biomarker for therapy of AML.
Keywords: acute myeloid leukemia, t(4, 15)(q31, q22), TMEM154-RASGRF1, ATRA, retinoid, NCT02273102
1. Introduction
Acute myeloid leukemia (AML) is the most commonly occurring leukemia in adults, accounting for an estimated 33% of all cases and more that 80% of acute cases in 2016 [1]. AML is a disease whose incidence increases with age and—in line with an aging population—this has increased from 3.4 per 100,000 in 2004 to 5.1 per 100,000 in 2013 [2]. Advances in the treatment of AML have dramatically improved treatment outcomes for younger patients, with a relative 5-year survival rate (2005–2011) of approximately 67% (0–14 years) [1]. In the elderly, however (who account for the majority of new cases), the prognosis remains poor. Indeed, AML accounts for an estimated 43% of all leukemia deaths, with an overall average 5-year survival rate of only 27% [2].
The use of combination therapy based on the use all-trans retinoic acid (ATRA) and arsenic trioxide (As2O3) has revolutionized the clinical outcome of acute promyelocytic leukemia (APL), with complete remission rates of 90% and cure rates of around 80% [3]. However, the success of ATRA-based therapy in AML has not been translated into other non-APL subtypes of AML. Based on in vitro data showing efficacy with cytotoxic chemotherapy agents such as Ara-C and anthracyclines, several clinical studies have investigated the impact of adding ATRA to chemotherapy in patients with non-APL AML [4]. Unfortunately, however, these studies have yielded inconsistent and conflicting results. One possible reason for a lack of response to ATRA treatment in non-APL AML patients is epigenetic repression of the retinoic acid receptor pathway, especially the RARA gene [5]. Indeed, combination therapy of ATRA with epi-drugs targeting the histone H3 lysine 4 demethlyase LSD1 (KDM1A) has shown promise in in vivo models of AML [6]. This approach is now being evaluated in a phase I clinical trial where patients are given escalating doses of the LSD1 inhibitor Parnate (tranylcypromine, TCP) with fixed continuous doses of ATRA.
Acute myeloid leukemia is characterized by clonal or oligoclonal hematopoiesis that varies amongst patients. On average, at least four-to-five oncogenic mutations or chromosomal abnormalities are present at diagnosis, along with passenger mutations of unclear significance [7]. Clonal evolution occurs after exposure to chemotherapy, and the secondary mutations that give rise to these clones are more likely to be random, as opposed to recurrent primary abnormalities [8]. Here we report a case of relapsed AML characterized by a previously unidentified chromosomal translocation.
2. Results
2.1. Clinical Presentation
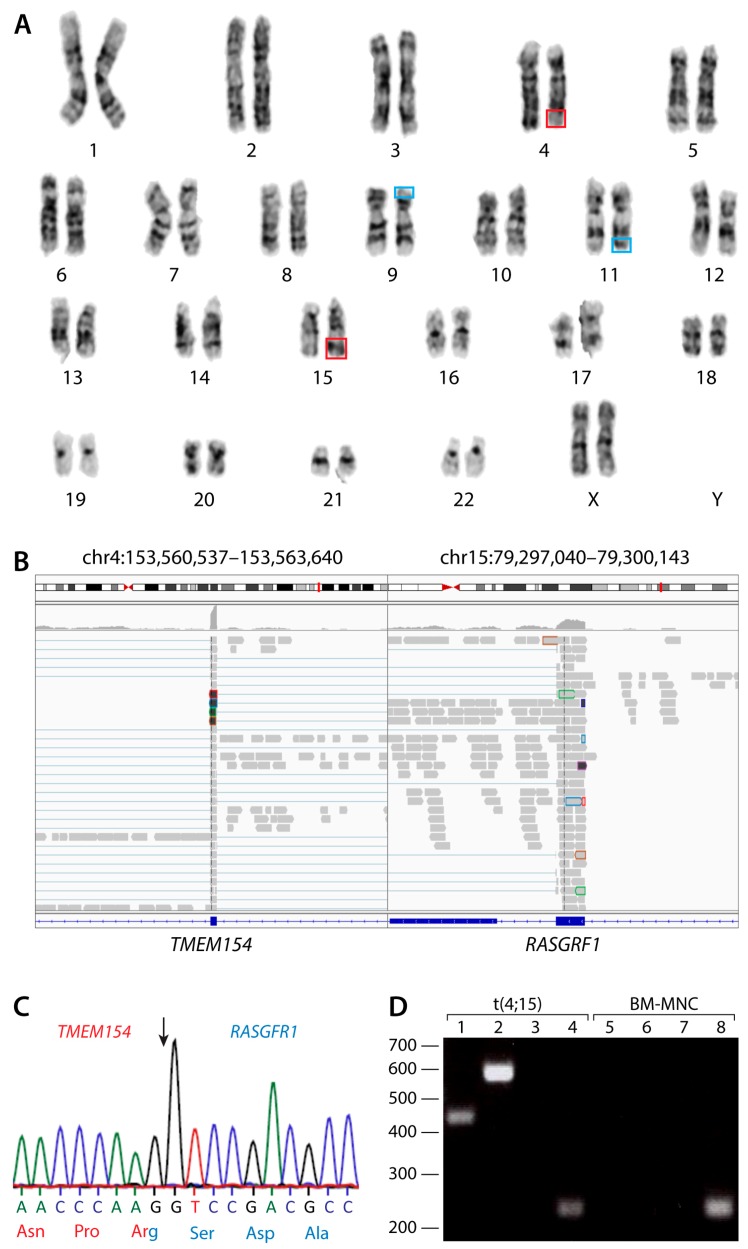
A 30-year-old previously healthy African-American female presented with a white blood cell count (WBC) of 180,000/µL, hemoglobin 7.6 g/dL, platelets 93,000/µL. Physical examination was unremarkable. Bone marrow aspiration and biopsy were consistent with AML with monocytic differentiation. Blasts amounted to 95% of nucleated elements and expressed CD34, CD117, CD13, CD33, CD64 (weak), CD38, HLA-DR, and were negative for terminal deoxynucleotidyl transferase (TdT) and myeloperoxidase (MPO) by flow cytometry. Cytogenetic analysis revealed an abnormal female karyotype: 46,XX,t(9;11)(p22;q23)[13]/46,XX[7]. Additionally, mutational analysis revealed an activating mutation in NRAS gene (c.34G > A; p.G12S). Following treatment with standard therapy, the patient achieved complete cytogenetic remission. Post remission therapy followed, with sequential cycles of high-dose cytarabine in the absence of an optimal donor. Following a disease-free interval of 8 months, the patient relapsed. Karyotype at relapse was consistent with clonal evolution: 46,XX,t(4;15)(q31;q22), t(9;11)(p22;q23)[20] (Figure 1A). Mutational analysis confirmed the presence of an NRAS mutation as before. The patient failed two lines of salvage chemotherapy. In the setting of chemotherapy refractory disease, the patient was enrolled in the phase I clinical trial (NCT02273102) and given escalating doses of TCP with fixed continuous doses of ATRA. However, the patient died from rapid disease progression after eight days of treatment (WBC at screening was 400/µL and at withdrawal from study WBC was 99,300/µL).
Figure 1.
Analysis of the t(4;15)(q31;q22) reciprocal chromosomal translocation. (A) Conventional karyotyping showing 46,XX,t(4;15)(q31;q22), outlined in red and t(9;11)(p22;q23), outlined in blue. Chromosome analysis was performed on 20 G-banded metaphase cells from multiple unstimulated cultures. Both translocations were present in all cells examined. (B) Breakpoint analysis of the novel fusion transcript produced by the t(4;15) chromosomal translocation. TMEM154 and RASGRF1 form a chimeric mRNA transcript with the breakpoint indicated by the black arrow. (C) Sanger sequencing chromatogram showing the transcribed sequence surrounding the breakpoint. (D) PCR analysis of and TMEM154-RASGRF1, RASGRF1-TMEM154, and TMEM154 transcripts. PCR was performed on t(4;15) acute myeloid leukemia (AML) cells (lanes 1–4) and bone marrow mononuclear cells (BM-MNCs) from a healthy donor (lanes 5–8). Bands correspond to a 445 bp product amplified fromTMEM154-RASGRF1 mRNA (lanes 1 and 5), a 589 bp product amplified from TMEM154-RASGRF1 mRNA (lanes 2 and 6), and a 445 bp product amplified fromTMEM154 mRNA (lanes 4 and 8). No product was amplified using primers corresponding to RASGRF1-TMEM154 mRNA (lanes 3 and 7).
2.2. Molecular and Functional Characterization of the t(4;15) Fusion Gene
To characterize the translocation partners and their breakpoints, we used RNA-Seq and Sanger sequencing, confirming the t(9;11) MLL(KMT2A)-AF9(MLLT3) reciprocal translocation present at diagnosis (data not shown). We also defined a novel fusion gene comprising exons 1–6 of the TMEM154 gene (4q31.3) and exons 15–24 of the RASGRF1 gene (15q24.2) (Figure 1B). This generates a putative 830 amino acid TMEM154-RASGRF1 fusion protein comprising amino acids 1–178 of TMEM154 and 622–1273 of RASGRF1 (Figure 1C). RT-PCR analysis confirmed the presence of TMEM154-RASGRF1 mRNA in t(4;15) AML cells but not normal controls (Figure 1D, lanes 1–2 and 5–6). By contrast, controls for TMEM154 mRNA were present in both t(4;15) AML cells and normal controls (Figure 1D, lanes 4 and 8). Consistent with the RNA-Seq results, expression of mRNA transcripts arising from the reciprocal RASGRF1-TMEM154 fusion gene were not detected in t(4;15) AML cells (Figure 1D, lane 3).
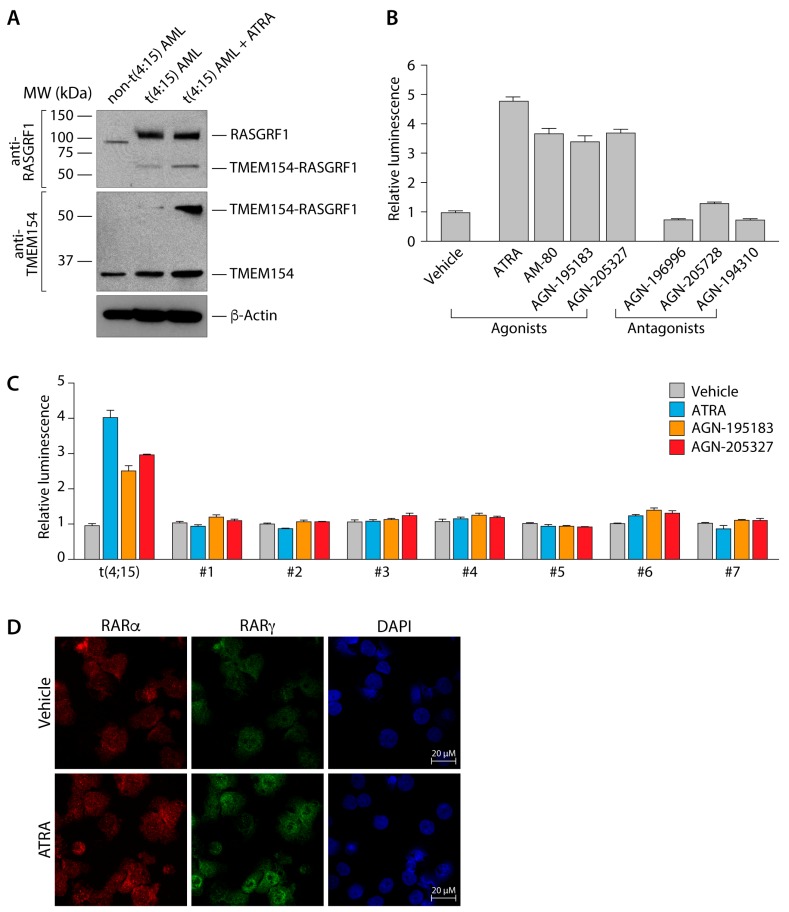
In agreement with our analysis of mRNA expression, Western blot analysis revealed the expression of endogenous TMEM154 and RASGRF1 proteins, as well as TMEM154-RASGRF1 fusion protein, which could be detected using antibodies directed against both a N-terminal region of TMEM154 as well as a C-terminal region of RASGRF1 (Figure 2A). Interestingly, treatment with ATRA led to an increase in the expression of TMEM154-RASGRF1 fusion protein, but not endogenous TMEM154 or RASGRF1 (Figure 2A). Supporting the clinical history, we observed a rapid proliferation of the patient’s blasts following treatment with ATRA ex vivo (Figure 2B). The same phenotype was seen when the cells were exposed to other agonists of the retinoic acid pathway. Primary cells lacking a t(4;15)(q31;q22) rearrangement displayed no growth alteration compared to controls when treated with either ATRA (a pan-RAR isotype agonist) or isotype-selective agonists specific for RARα or RARγ (Figure 2C). To further examine the effects of ATRA on primary t(4;15) AML cells, we performed immunofluorescence analysis of RARα or RARγ. Here, we found that treatment of t(4;15) AML cells with ATRA led to an increase in nuclear RARγ (Figure 2D).
Figure 2.
Characterization of t(4;15) AML primary cells. (A) Immunoblot analysis of TEM154-RASGRF1 expression. After 72 h treatment, vehicle control non-t(4;15) AML cells (lane 1), vehicle control t(4;15) AML cells (lane 2) and all-trans retinoic acid (ATRA) t(4;15) AML cells (lane 3) were subjected to immunoblot analysis. Samples were probed with rabbit monoclonal antibody against a C-terminal region of RASGRF1 and mouse monoclonal antibody against an N-terminal region of TMEM154. β-actin was used as a loading control. Molecular weight standards (left) and the identities of bands (right) are indicated. (B) Proliferation of t(4;15) AML cells in response to retinoids. Cells were treated with vehicle control, ATRA, RARα agonist (AM-80 or AGN-195183), or RARγ agonist (AGN-205327). Cells were also treated with RARα antagonist (AGN-196996), RARγ antagonist (AGN-205728), or RARα/β/γ antagonist (AGN-194310). All retinoids were used at a concentration of 1 µM. Cell proliferation was determined by CellTiter-Glo luminescent cell viability assay (Promega) after 72 h of treatment. Values shown are normalized to vehicle control. (C) Proliferation of t(4;15) and non-t(4;15) AML cells in response to retinoid agonists. Samples from t(4;15) and 7 non-t(4;15) AML patients were incubated with vehicle control, ATRA, RARα agonist (AGN-195183), or RARγ agonist (AGN-205327) at a concentration of 1 µM. Cells were analyzed and normalized as described for (B). (D) Treatment of t(4;15) cells with ATRA increases levels of nuclear RARγ. t(4;15) cells were treated with vehicle control or 1 µM ATRA and incubated for 72 h before staining with RARα mouse monoclonal and RARγ rabbit polyclonal antibodies.
3. Discussion
Our data suggest that the t(4;15)(q31;q22) reciprocal translocation confers a growth-stimulatory response to retinoid-based therapy. However, as we did not measure levels of apoptosis at the time the proliferation assays were performed, it is possible that t(4;15) cells also exhibited improved survival ex vivo. Nevertheless, an ex vivo growth-stimulatory response is consistent with the increased WBC in the patient following ATRA treatment. The basis for this response requires further investigation, but could lie—at least in part—in an increase in nuclear RARγ following ATRA treatment. While RARα is required for myeloid differentiation, RARγ has been shown to promote hematopoietic stem and progenitor cell self-renewal and proliferation [9]. Additionally, it is likely that the ATRA-mediated increase in TMEM154-RASGRF1 fusion protein (which is consistent with established ATRA regulation of TMEM154 [10]) cooperates with mutant NRAS G12S (which results in decreased GTPase activity and constitutive RAS signaling). Between 12–19% of AML patients possess gain-of-function mutated RAS genes [7], with NRAS being most frequently affected [11]. Furthermore, other components of the RAS pathway signaling cascade are often mutated [12]. RASGRF1 (RAS protein specific nucleotide releasing factor 1) is a guanine nucleotide exchange factor (GEF) similar to the Saccharomyces cerevisiae CDC25 gene product, and functions to activate RAS by catalyzing the exchange of RAS-bound GDP for GTP [13]. While we have not yet investigated the activity of TMEM154-RASGRF1 towards NRAS, it is noteworthy that the fusion protein (which lacks regulatory domains contained in N-terminal RASGRF1) contains the membrane anchoring domain of TMEM154 and functional domains of RASGRF1. These include the GDP/GTP RAS exchanger and CDC25H motifs of RASGRF1, which have been shown to be sufficient for functional protein [14].
Another aspect of the consequences of the t(4;15) translocation that merits further consideration is the fact that RASGRF1 is a paternally expressed imprinted gene. Thus, under normal conditions, only the paternal allele of the gene is translated into protein. In t(4;15) AML, both the TMEM154-RASGRF1 fusion protein as well endogenous RASGRF1 are expressed, leading to possible gene dosage effects. Conversely, the lack of expression of the reciprocal RASGRF1-TMEM154 transcript indicates that the differentially methylated domain (DMD) located 30 kb 5′ of the RASGRF1 transcription initiating site [15] is intact, resulting in imprinted methylation. A functionally important consequence of this could be inappropriate silencing of genes downstream of TMEM154. Here, it is noteworthy that the gene immediately downstream of TMEM154 is FBXW7, which encodes an F-box family protein that is a component of the SCF (SKP1-cullin-F-box) ubiquitin protein ligase complex. In the context of AML, FBXW7 has clinically relevant targets including MYC, MCL-1, and Cyclin E [16,17].
4. Materials and Methods
4.1. Patient Samples
Primary AML samples were collected by the Tissue Banking Core Facility (TBCF) at the University of Miami according to an institutional review board approved protocol (20060858). In order to purify the mononuclear fraction, bone marrow aspirates were subjected to density gradient centrifugation using Ficoll Paque Plus (GE Healthcare Life Sciences, Pittsburgh, PA, USA).
4.2. DNA/RNA Isolation
DNA and RNA were isolated from mononuclear cells from patients’ bone marrow aspirate. DNA was isolated using QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. RNA was extracted with PureLink RNA kit (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s recommendations.
4.3. RNA-Seq for Breakpoint Identification
Preparation of RNA libraries for sequencing on the Illumina HiSeq2500 (Illumina, San Diego, CA, USA) platform was carried out in the John P. Hussman Institute for Human Genomics Center for Genome Technology at the University of Miami. Briefly, total RNA was quantified and qualified by Agilent Bioanalyzer (Agilent, Santa Clara, CA, USA) to have an RNA integrity score (RIN) of 7. For each of the two samples, 1000 ng of total RNA was used as input for the Illumina TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero to create ribosomal RNA-depleted sequencing libraries. Each sample was barcoded to allow for multiplexing and was sequencing to ~30 million raw reads in a 2 × 125 paired end sequencing run on an Illumina HiSeq2500. Raw sequence data from the Illumina HiSeq2500 was processed by the on-instrument Real Time Analysis software (v1.8, Illumina, San Diego, CA, USA) to basecall files. These were converted to demultiplexed FASTQ files with the Illumina supplied scripts in the BCL2FASTQ software (v1.8.4, Illumina, San Diego, CA, USA). The quality of the reads was determined with FastQC software for per-base sequence quality, duplication rates, and overrepresented kmers (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Illumina adapters were trimmed from the ends of the reads using Trim Galore! resulting in two trimmed FASTQ files per sample (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Reads were aligned to the human reference genome (hg19) with the STAR aligner (v2.5.0a) [18]. Gene fusions were detected in the aligned BAM files using the Manta software [19].
4.4. Accession Numbers
The Genbank accession number for TMEM154-RASGRF1 mRNA is MF175878. RNA-Seq FASTQ files were submitted to the sequence read archive (SRA) with the Genbank accession number SRR5681084.
4.5. RT-PCR
Reverse transcription and PCR amplification was performed using One-Step RT-PCR System (Life Technologies) according to manufacturer’s instructions. Primers used to amplify TMEM154-RASGRF1 mRNA were TMEM154-Fwd333: 5′-TAGCCAAGGATCTCAGAGTG-3′; and RASGRF1-Rev778: 5′-GAATGGCACTGATAGGCTTC-3′ or RASGRF1-Rev922: 5′-GTGACGATGTCTTGGTGATG-3′. Primers used to amplify TMEM154 were TMEM154-Fwd1762: 5′-TAGCCAAGGATCTCAGAGTG-3′ and TMEM154-Rev2272: 5′-TGAGCGCCATTCAGGTTTAG-3′. Primers to amplify RASGRF1-TMEM154 were RASGRF1-Fwd1762: 5′-CTCATTCAGGTGCCCATGTC-3′ and TMEM154-Rev2272: 5′-TGAGCGCCATTCAGGTTTAG-3′. Amplified PCR fragments were subjected to DNA sequence (Eurofins, Louisville, KY, USA) to validate the translocation breakpoint.
4.6. Western Blot
Protein sample concentrations were normalized using DC Protein Assay (Bio-Rad, Hercules, CA, USA). Fifty micrograms of protein were separated by SDS-PAGE using 4–20% Mini-PROTEAN TGX pre-cast gels with Precision Plus ProteinDual Color Standards (Bio-Rad). Primary antibodies used were as follows: rabbit monoclonal antibody against the C-terminal region of RASGRF1 (ab118830, Abcam, Cambridge, UK) and mouse monoclonal antibody against the N-terminal region of TMEM154 (sc-398802, Santa Cruz Biotechnology, Dallas, TX, USA). β-actin was used as a loading control (A5441, Sigma, St. Louis, MO, USA,). Protein bands were detected using HRP-linked Anti-rabbit IgG (#7074, Cell Signaling Technology, Danvers, MA, USA) and HRP-linked Anti-mouse IgG (Cell Signaling Technology, #7076) secondary antibodies.
4.7. Cell Proliferation
Cell proliferation was measured using CellTiter-Glow luminescent cell viability assay (Promega, Madison, WI, USA) following the manufacturer’s recommendations. Primary AML mononuclear cells were seeded at a density of 10,000 cells/well in 96-well plate for 24 h, followed by retinoid or vehicle (DMSO) treatment for 72 h. Cell were cultured in RPMI supplemented with 5% Charcoal-dextran fetal bovine serum (FBS, Life Technologies). ATRA was purchased from Sigma. AM-80 was purchased from Tocris (Bio-Techne, Minneapolis, MN, USA). AGN195183, AGN196996, AGN205327, AGN205728, and AGN194310 were manufactured under contract at the Chinese National Center for Drug Screening, Shanghai Institute of Materia Medica, Shanghai, China.
4.8. Immunofluorescence
AML cells were fixed with 4% paraformaldehyde in 1X phosphate-buffered saline (PBS) for 20 min at room temperature. For immunostaining, cells were permeabilized with 1% Triton X-100 in 1X PBS and blocked with 2.5% bovine serum albumin (BSA) and 1% Triton X-100 in 1X PBS for 1 h shaking at room temperature. Primary antibodies (dilution 1:100) were incubated for 2 h at room temperature in blocking buffer and washed five times with wash buffer (0.1% Triton X-100 in 1X PBS). Primary antibodies used were RARα-9a (mouse monoclonal) and RARγ-453 (rabbit polyclonal) (gift from Cécile Rochette-Egly, L’Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), France). Secondary antibodies (dilution 1:1000) were added for 1 h at room temperature in blocking buffer and then washed five times. Secondary antibodies used were Alexa Fluor 488-conjugated anti-mouse IgG (H + L), F(ab’)2 fragment (Cell Signaling Technology, #4408) and Alexa Fluor 555-conjugated anti-rabbit IgG (H + L), F(ab’)2 fragment (Cell Signaling Technology, #4413). Negative controls were performed using primary and secondary antibodies alone. Slides were then mounted with ProLong Gold antifade reagent with DAPI (Molecular Probes, Eugene, OR, USA), following the manufacturer’s instructions. Immunofluorescence microscopy was performed using a Leica DFC 310 FX microscope.
5. Conclusions
Here we report a case of relapsed AML characterized by a novel translocation associated with aggressive disease and increased proliferation in the presence of ATRA. We are currently generating a cell line bearing this translocation that will provide a new model system for the study of AML. Although the TMEM154-RASGRF1 translocation is likely to be a rare occurrence (as has been the case in the past with other translocations), it is predicted that further study of its biology will yield important insights into retinoic acid signaling and resistance to retinoid-based therapy in AML.
Acknowledgments
We thank Omar Lopez, Ana Rodrigues, Veronica Gil, Terrence Bradley, Fernando Vargas, Sandra Algaze and Jennifer Chapman for helpful advice and comments. This study was supported by Grant Number KL2TR000461, Miami Clinical and Translational Science Institute, from the National Center for Advancing Translational Sciences and the National Institute on Minority Health and Health Disparities. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Author Contributions
Justin M. Watts, Aymee Perez, Ronan T. Swords and Arthur Zelent conceived and designed the study. Justin M. Watts collected the clinical data. Aymee Perez and Lutecia Pereira performed laboratory experiments. Yao-Shan Fan and Francisco Vega performed cytogenetic analysis. Geoffrey Brown provided technical expertise on the manufacture and use of retinoid agonists and antagonists. Justin M. Watts, Aymee Perez and Kevin Petrie analyzed and interpreted the data and wrote the manuscript. Geoffrey Brown, Francisco Vega, Ronan T. Swords and Arthur Zelent analyzed and interpreted the data and edited the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society . Cancer Facts & Figures. American Cancer Society; Atlanta, GA, USA: 2017. [Google Scholar]
- 3.Coombs C.C., Tavakkoli M., Tallman M.S. Acute promyelocytic leukemia: Where did we start, where are we now, and the future. Blood Cancer J. 2015;5:e304. doi: 10.1038/bcj.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forghieri F., Bigliardi S., Quadrelli C., Morselli M., Potenza L., Paolini A., Colaci E., Barozzi P., Zucchini P., Riva G., et al. All-trans retinoic acid (ATRA) in non-promyelocytic acute myeloid leukemia (AML): Results of combination of ATRA with low-dose Ara-C in three elderly patients with NPM1-mutated AML unfit for intensive chemotherapy and review of the literature. Clin. Case Rep. 2016;4:1138–1146. doi: 10.1002/ccr3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glasow A., Barrett A., Petrie K., Gupta R., Boix-Chornet M., Zhou D.C., Grimwade D., Gallagher R., von Lindern M., Waxman S., et al. DNA methylation-independent loss of RARA gene expression in acute myeloid leukemia. Blood. 2008;111:2374–2377. doi: 10.1182/blood-2007-05-088344. [DOI] [PubMed] [Google Scholar]
- 6.Schenk T., Chen W.C., Gollner S., Howell L., Jin L., Hebestreit K., Klein H.U., Popescu A.C., Burnett A., Mills K., et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat. Med. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J., Jr., Laird P.W., et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klco J.M., Miller C.A., Griffith M., Petti A., Spencer D.H., Ketkar-Kulkarni S., Wartman L.D., Christopher M., Lamprecht T.L., Helton N.M., et al. Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. Jama. 2015;314:811–822. doi: 10.1001/jama.2015.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrie K., Zelent A., Waxman S. Differentiation therapy of acute myeloid leukemia: Past, present and future. Curr. Opin. Hematol. 2009;16:84–91. doi: 10.1097/MOH.0b013e3283257aee. [DOI] [PubMed] [Google Scholar]
- 10.Simandi Z., Balint B.L., Poliska S., Ruhl R., Nagy L. Activation of retinoic acid receptor signaling coordinates lineage commitment of spontaneously differentiating mouse embryonic stem cells in embryoid bodies. FEBS Lett. 2010;584:3123–3130. doi: 10.1016/j.febslet.2010.05.052. [DOI] [PubMed] [Google Scholar]
- 11.Bacher U., Haferlach T., Schoch C., Kern W., Schnittger S. Implications of NRAS mutations in AML: A study of 2502 patients. Blood. 2006;107:3847–3853. doi: 10.1182/blood-2005-08-3522. [DOI] [PubMed] [Google Scholar]
- 12.Scholl C., Gilliland D.G., Frohling S. Deregulation of signaling pathways in acute myeloid leukemia. Semin. Oncol. 2008;35:336–345. doi: 10.1053/j.seminoncol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Medarde A., Santos E. The RasGrf family of mammalian guanine nucleotide exchange factors. Biochim. Biophys. Acta. 2011;1815:170–188. doi: 10.1016/j.bbcan.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Lenzen C., Cool R.H., Prinz H., Kuhlmann J., Wittinghofer A. Kinetic analysis by fluorescence of the interaction between Ras and the catalytic domain of the guanine nucleotide exchange factor Cdc25Mm. Biochemistry. 1998;37:7420–7430. doi: 10.1021/bi972621j. [DOI] [PubMed] [Google Scholar]
- 15.Shibata H., Yoda Y., Kato R., Ueda T., Kamiya M., Hiraiwa N., Yoshiki A., Plass C., Pearsall R.S., Held W.A., et al. A methylation imprint mark in the mouse imprinted gene Grf1/Cdc25Mm locus shares a common feature with the U2afbp-rs gene: An association with a short tandem repeat and a hypermethylated region. Genomics. 1998;49:30–37. doi: 10.1006/geno.1998.5218. [DOI] [PubMed] [Google Scholar]
- 16.Gores G.J., Kaufmann S.H. Selectively targeting Mcl-1 for the treatment of acute myelogenous leukemia and solid tumors. Genes Dev. 2012;26:305–311. doi: 10.1101/gad.186189.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeishi S., Nakayama K.I. Role of Fbxw7 in the maintenance of normal stem cells and cancer-initiating cells. Br. J. Cancer. 2014;111:1054–1059. doi: 10.1038/bjc.2014.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Kallberg M., Cox A.J., Kruglyak S., Saunders C.T. Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]