Abstract
Many sequences in eukaryotic genomes have the potential to adopt a left-handed Z-DNA conformation. We used a previously described assay based on the binding of a mAb to Z-DNA to inquire whether Z-DNA is formed in the rat nucleolin (Ncl) gene in metabolically active, permeabilized nuclei. Using real-time PCR to measure Z-DNA formation, the potential Z-DNA sequence element Z1 [(CA)10(CG)8] in the promoter region was found to be enriched 571- to 4,040-fold in different cell lines, whereas Z2 [AC(GC)5CCGT(CG)2] in the first intron was enriched 12- to 34-fold. Ncl promoter activity was 1.5- to 16-fold stronger than that of the simian virus 40 promoter and enhancer. This activity was further increased 36–54% when Z1 was deleted. The inhibitory effect of Z1 on Ncl promoter activity was independent of location and orientation. The Ncl Z1 element is identical to the genetic marker D9Arb5. Five allelic variants of Z1 were identified by sequence analysis of genomic DNA from various rats. The two most common alleles differed significantly (up to 27%) in their capacity to inhibit Ncl promoter activity. This finding suggests that differences in Z-DNA formation by polymorphic dinucleotide repeats may be one of the factors contributing to genetic variation.
Keywords: gene regulation, polymorphism, microsatellites, left-handed DNA
Dinucleotide repeats are found throughout the genome of eukaryotes. (CA/TG)n is the most abundant one in mammals, found on average every 20–30 kb (1). These microsatellites often are polymorphic and have been used extensively in genetic mapping and the identification of susceptibility loci in genetic diseases (2). Evidence is accumulating that such elements also can be of functional importance. Dinucleotide repeats have the potential to form alternative DNA structures such as Z-DNA, H-DNA, and cruciform DNA (3, 4). One of the best-characterized alternative DNA conformations is left-handed Z-DNA, which can be adopted by stretches of alternating purine-pyrimidine nucleotides.
Z-DNA formation of many sequences can be induced in vitro (5). (CG)n repeats have the highest potential to form Z-DNA, followed by (CA/TG)n and (TA)n (5). Sequences containing alternating purine-pyrimidine nucleotides have been shown to modulate promoter activity. They may either enhance (6, 7) or repress (8) promoter activity. Because of the polymorphic nature of dinucleotide repeats, it seems possible that Z-DNA-forming sequences may provide a source of genetic variation if they occur in regions that are important for the regulation of gene activity.
An assay based on the preparation of metabolically active nuclei, introduced by Jackson and Cook (9), has been developed to investigate whether specific potential Z-DNA-forming sequences actually adopt this conformation in actively transcribing eukaryotic genes (10). These nuclei maintain transcription and show a DNA replication rate of 85%, compared with untreated cells (11). The permeable nuclei are incubated with a biotinylated mAb specific for Z-DNA. A 10-ns laser pulse at 266 nm is used to specifically crosslink proteins to DNA, followed by the extraction of the antibody with its bound DNA restriction fragments. The isolated DNA then is analyzed.
Using this method Z-DNA formation has been demonstrated in the human c-myc gene (10), the corticotropin-releasing hormone gene (12), and DNA sequences of the β-globin cluster (13). An association of Z-DNA formation with transcriptional activity has been established in all investigated genes. Physiological down-regulation of c-myc gene transcription is accompanied by a loss of Z-DNA formation (10). An explanation for the tight correlation between transcription and Z-DNA formation is that Z-DNA is stabilized by negative supercoiling (5), which is generated behind a moving RNA polymerase (14). When polymerases stop moving, topoisomerases then can relax the supercoiling and facilitate the transition from Z-DNA to B-DNA. Indeed inhibition of topoisomerase I by camptothecin increased the amount of Z-DNA detected (10). Potential Z-DNA sequences in human genes tend to be located near transcription start sites (15), making it possible that these sequences play a role in the transcriptional control of these genes.
In this report, we ask whether Z-DNA is formed by two sequences located in the rat nucleolin (Ncl) gene, the first in the promoter region and the second in the first intron. Ncl is a multifunctional protein with interesting properties (reviewed in refs. 16 and 17). It is the most abundant protein in the nucleolus, is constitutively expressed at a high level, and is a key player in ribosome biogenesis. We investigated whether potential Z-DNA forming sequences in the Ncl gene actually adopt this conformation and whether one of these sequences (Ncl Z1) located in the upstream region plays a role in transcriptional regulation. We further identified allelic variants of this dinucleotide repeat and tested the effect of polymorphisms in this element on promoter activity.
Materials and Methods
Animals and Cell Lines.
DNA of inbred rat strains BDE, BH, BN, DA, E3, F344, LE, LEW, WF, and WKY was kindly provided by D. Wedekind (Hannover Medical School, Hannover, Germany). LEW.6B rats were kindly provided by K. Wonigeit (Hannover Medical School, Hannover, Germany). BB/DR and BB/DP rats were purchased from Møllegaard (Skensved, Denmark). BUF, PVG, and SHR rats were purchased from Harlan (Horst, The Netherlands). Two wild rats (Rattus norvegicus) were caught in the Hamburg harbor (rnHH1 and rnHH2), and one rat was caught in the Buchholzer Moor (Schleswig Holstein, rnBM). Rattus rattus (black rat) from outbred populations were kindly provided by W. Schuster (Hygieneinstitut, Magdeburg, Germany) and J. Lodal (Technical University, Lyngby, Denmark). C58NT, EpSM30, EpD3 and rat1 cells were maintained as described (18).
Preparation of Metabolically Active Nuclei and Isolation of Z-DNA Fragments.
Encapsulation of cells, permeabilization of nuclei, and crosslinking of Z-DNA- specific antibody Z22 was carried out as described (10). Briefly, after crosslinking the biotinylated Z-DNA-specific antibody Z22 to DNA by a 10-ns pulse of the fourth harmonic wavelength at 266 nm of a Ns-YAG laser, unbound proteins were removed by high-salt treatment (2 M NaCl). Genomic DNA was digested overnight with restriction enzymes PvuII and BanII. Supernatants were incubated with streptavidin-coated magnetic beads (Dynabeads, Dynal, Oslo) for 30 min. The unbound fraction was kept for control reactions. Dynabeads were washed 10 times to remove nonspecifically bound DNA and then were treated with proteinase K to release the Z-DNA-specific fragments. DNA was precipitated with ethanol after phenol extraction.
Detection of Z-DNA-Specific Fragments.
Primers for amplification of PvuII and BanII restriction fragments were as follows: Ncl Z1F, 5′-CTGATGAGGGCACCCGTTTGCTAC-3′; Ncl Z1R, 5′-AAACAGTCCATTTAATCTCTGACCTCACG-3′; Ncl Z2F, 5′-GCTGGCAGGCGGTTGTACGTG-3′; Ncl Z2R, 5′-TCTCAGAAAGCAGGCCTAGTCGTC-3′; aldehyde reductase (Aldr) Z1F, 5′-CTGGACTACCTGGACCTCTACC-3′; Aldr Z1R, 5′-GCAGGCTTCACCTACAGCCGTG-3′; RT6 Ne1F, 5′-CTGATTTCTGGTGGATATTTTGAACATCC-3′; RT6 Ne1R, 5′-GCTCCTTGGGAGAGCCTGGGTA-3′; RT6 Ne2F, 5′-CTGGTTTATTTTGGGTGATAGTGATG-3′; RT6 Ne3R, 5′-ATGAGAGCTAGCTCAGGGTCAC-3′; RT6 Ne3F, 5′-CAGCAAGCAGGCTATTCTGAC-3′; and RT6 Ne3R, 5′-GGCTCCTCTCTTGCCATGTCC-3′.
PCR amplification was performed in a 40-μl reaction volume containing 1% of the Z-DNA-specific fraction (or 0.03% of the unbound supernatant) using 1 unit of AmpliTaq Gold polymerase (Perkin–Elmer). An initial heat activation for 8 min at 94°C was followed by 32 or 38 cycles (1 min 94°C, 1 min 60°C, 40 sec 72°C). Reaction products were visualized on 1.6% ethidium bromide containing agarose gels.
Real-time PCR for segments containing Ncl Z1, Ncl Z2, and RT6 Ne1 was carried out by using an Applied Biosystems Prism 5700 Sequence Detector according to the manufacturer's instructions. Amplification reactions (50 μl) were performed in triplicate and contained the SYBR Green PCR Mastermix (Perkin–Elmer), 0.30 μM of each primer, and 75 nM of probe. The thermal cycling conditions were 50°C for 2 min, 95°C for 12 min, and 45 cycles of 95°C for 15 sec, 60°C for 30 sec, and 72° for 40 sec. The amount of a specific fragment of the bound (Z-DNA specific) fraction was related to that of the unbound (control) fraction by the ΔΔCt method as described by K. Livak (Perkin–Elmer Sequence Detector User Bulletin 2) and ref. 19. Briefly, the number of amplification cycles necessary to generate a signal above a given threshold was determined as the Ct (threshold cycle) value. The relative amount of Z-DNA was determined as ΔCt values by subtracting the Ct values of the control reaction from those of the Z-DNA-specific fraction (Table 1). Low ΔCt values imply that high amounts of Z-DNA have been detected. Relative enrichment was calculated with the formula: relative enrichment = 2ΔΔCt (see Fig. 2C).
Table 1.
Quantification of the relative DNA amounts
| Segment | C58NT/ bound | C58NT/ unbound | ΔCT | ΔΔCT | EpSM30/ bound | EpSM30/ unbound | ΔCT | ΔΔCT | EpD3/ bound | EpD3/ unbound | ΔCT | ΔΔCT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nu Z1 | 24.58 ± 0.24 | 24.39 ± 0.13 | 0.19 | 11.98 | 24.26 ± 0.09 | 24.10 ± 0.09 | 0.16 | 10.78 | 27.52 ± 0.13 | 26.03 ± 0.02 | 1.49 | 9.16 |
| Nu Z2 | 32.46 ± 0.46 | 25.36 ± 0.23 | 7.1 | 5.08 | 32.80 ± 0.42 | 25.48 ± 0.20 | 7.32 | 3.61 | 36.16 ± 1.36 | 26.33 ± 0.02 | 9.84 | 0.81 |
| RT6 Ne1 | 37.42 ± 0.26 | 25.24 ± 0.08 | 12.18 | 0 | 36.91 ± 0.07 | 25.98 ± 0.18 | 10.93 | 0 | 37.65 ± 0.08 | 27.00 ± 0.01 | 10.65 | 0 |
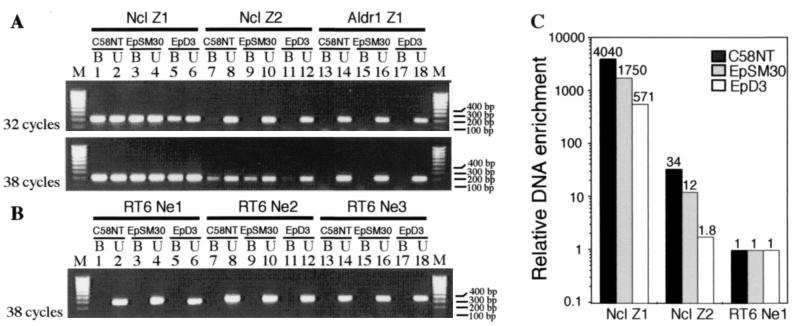
Figure 2.
Enrichment of Z-DNA-forming sequences by mAb Z22. (A) Bound Z-DNA enriched (B) and unbound control (U) fractions were used as PCR templates. Amplifications were performed for fragments containing potential Z-DNA-forming sequences Ncl Z1 and Ncl Z2 from the Ncl gene and for Aldr Z1. Three DNA segments from the RT6 gene RT6 Ne1, RT6 Ne2, and RT6 Ne3, which lack the potential to form Z-DNA, were used as controls. Amplification products for fragments Ncl Z1, Ncl Z2, and Aldr Z1 were visualized after 32 (Upper) and 38 cycles (Lower). (B) Products for control fragments RT6 Ne1, RT6 Ne2, and RT6 Ne3 were seen after 38 cycles. (C) The amount of Z-DNA containing fragments Ncl Z1 and Ncl Z2 was measured by real-time PCR and calculated relative to RT6 Ne1, which does not contain potential Z-DNA-forming sequences, as described in Materials and Methods. Relative enrichment is shown for cell lines C58NT, EpSM30, and EpD3. Values are plotted on a logarithmic scale.
Plasmid Construction.
Primers for amplification of the Ncl promoter region, designed after the published sequence (GenBank M55015), were Ncl pr1F, 5′-AGTCCCGGGGCAGCCAGGGCTACA-3′, and Ncl pr1R, 5′-CCATGATGGCGGATTACGGCGACGG-3′. PCR was performed with C58NT and EpSM30 DNA. Initial heat activation for 8 min at 94°C was followed by 34 cycles: 1 min at 96°C, 30 sec at 60°C, and 2 min at 72°C. One microliter of reaction product was used for nested PCR with primers Ncl pr2F, 5′-TCGGTACCAGCGGTAGAGCGCTTGCCTAGC-3′, containing a KpnI restriction site, and Ncl pr2R, 5′-TCAAGCTTACGGCGACGGCGTGTGTAGA-3′, containing a HindIII restriction site. Reaction products were digested with KpnI and HindIII, gel-extracted, and ligated into the pGL3-Basic luciferase vector (Promega), yielding pGL3 Ncl11f (−1,264/+139 relative to transcription start site).
A naturally occurring ApaLI restriction site immediately 5′ of the dinucleotide repeat was used to generate pGL3 NuΔZ1. The whole promoter region was PCR-amplified, using pGL3 Ncl11f as template, with primers RVprimer3 and GLprimer2 (Promega), which flank the cloning site of the pGL3 plasmid. The reaction product was digested with ApaLI, and the 530-bp band was recovered. A second PCR product was generated with primers GLprimer2 and pGL3 Δ Z1, 5′-TGTTTGTGCACCGAAATTGCTTTTATTAGGAGCTC-3′, which starts immediately 3′ to the dinucleotide repeat and has an ApaLI site integrated. This product also was digested with ApaLI and gel-purified. Ligation of both digests was performed overnight. After KpnI and HindIII digestion, the expected 1.3-kb ligation product was gel-purified and cloned into the pGL3-Basic luciferase vector.
pGL3 NclRe6 and pGL3 NclRe5 were created by combining primers Ncl Re6F, 5′-ATCTTGGTACCACACACACACACACACACACGCGCG-3′, or Ncl 5Re5F, 5′-GGGGGGGTACCAAATTGCTTTTATTAGGAGC-3′, with Ncl pr2R. PCR products were cloned into the pGL3-Basic luciferase vector after KpnI/HindIII digestion.
pGL3 NclRe7a was created by PCR with primers Ncl Re7F, 5′-AGAAGGGTACCTGTGCACTGCTCACACAC-3′, with Ncl pr2R. pGL3 NclRe7b was created by amplifying a cloned fragment from the LEW.6B rat with primers Ncl Re7F and Ncl 3R, 5′-GGTGGTCTGGGCATTTCTCGG-3′, digesting with KpnI and StuI and ligating into pGL3 NclRe7, which had been digested with the same enzymes.
To generate plasmids pGL3 NuΔZ1+NclZ1, pGL3 NuΔZ1+NclZ1 o.o. and pGL3 promoter+NclZ1, PCR was performed by using primers NclZ1 KpnI–KpnI, 5′-AAAAAGGTACCCACACACACACACACACACACGCGCGCGCGCGCGCGGGTACCAAATTGCTTTTATTAGGAGCTC-3′, where KpnI sites were introduced immediately adjacent to Ncl Z1 and Ncl pr2R by using pGL3 NclRe7a as template. The PCR product was digested with HindIII, and the 40-bp band was gel-purified. This fragment was ligated into the HindIII-digested plasmids pGL3 NuΔZ1 and pGL3 promoter. Orientation of Ncl Z1 was determined by DNA sequencing. The inserts in all plasmids were verified by DNA sequencing.
Transient Transfection.
EpD3 (7 × 106 cells) and rat1 fibroblasts (2 × 106 cells) were transfected by electroporation (0.3 V/960 μF) with 10 μg plasmid DNA in 700 μl of appropriate medium containing 10% FCS by using a Gene pulser electroporator (Bio-Rad) and analyzed after 44–48 h for luciferase activity using the luciferase (Promega) assay according to the manufacturer's instructions in a luminometer (Berthold, Nashua, NH). Four or six independent transfections with plasmids from two plasmid preparations were performed for each construct, and normalized standard deviations were calculated. Statistical analysis was performed by using Student's t test.
Determination of Polymorphisms.
R. norvegicus DNA was PCR-amplified by using primers Ncl pr2F and Ncl 3R and from three R. rattus individuals by using primers Ncl 12f 5′-TTCTACTTATCTCCAAAGGCCCG-3′ and Ncl 3R. AmpliTaq polymerase was activated for 8 min at 94°C, followed by 32 cycles, 1 min at 96°C, 1 min at 65°C, and 50 sec at 72°C by using primers Ncl pr2F and Ncl 3R. Amplification products were gel-purified and directly sequenced with primer Ncl Z1F by using the dye-terminator protocol. Additionally, products from heterozygous and LEW.6B rats, as well as R. rattus were cloned into the pCR 2.1 vector (Invitrogen) and sequenced.
Results
Z-DNA Is Formed in the Promoter and First Intron of the Rat Ncl Gene.
We asked whether potential Z-DNA-forming sequences adopt this conformation in intact functional nuclei. To this end we used an established protocol (10) to isolate Z-DNA-forming sequences using the Z-DNA-specific mAb Z22, followed by comparative PCR analyses of antibody-bound vs. unbound DNA.
Permeabilized, metabolically active nuclei were prepared from rat thymoma and T cell hybridoma cell lines C58NT, EpSM30, and EpD3, as described (9, 20). Biotinylated monoclonal Z-DNA-specific antibody Z22 (mAb Z22) was incubated with the nuclei for 2 h to allow the binding of the antibody to Z-DNA. Proteins were crosslinked to DNA by a high-energy, 266-nm laser pulse for 10 ns. DNA was digested with BanII and PvuII, and the antibody with its crosslinked DNA fragments was recovered by using streptavidin-coated magnetic beads. DNA bound to mAb Z22 was released by Proteinase K digestion. DNA of bound and unbound fractions was phenol-extracted, precipitated with ethanol, and subjected to PCR analyses.
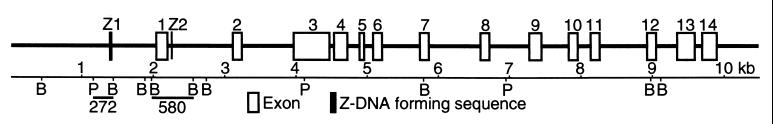
Primers were designed to amplify short (200–300 bp) DNA segments within the same BanII/PvuII restriction fragment that contained the potential Z-DNA-forming element. The potential Z-DNA-forming elements thus analyzed were Ncl Z1 [(CA)10(CG)8] and Ncl Z2 [(AC(GC)5CCGT(CG)2] from the promoter region and first intron of the Ncl gene (GenBank accession nos. M55015 and M55017) as well as Aldr Z1 [(GC)7ACGC(AC)22] from the third intron of Aldr (GenBank accession no. M60322). For control, PCR primers also were designed to amplify segments from restriction fragments lacking potential Z-DNA-forming elements. The control fragments, designated RT6 Ne1, RT6 Ne2, and RT6 Ne3, were from the promoter region and first and third introns of the gene for the T cell differentiation marker RT6, respectively (21). The structure of the rat Ncl gene, locations of Ncl Z1 and Ncl Z2, and restriction sites for BanII and PvuII are illustrated schematically in Fig. 1.
Figure 1.
Diagram of the rat Ncl gene. Location of exons (open boxes) and Z-DNA-forming sequences Z1 and Z2 (black boxes) are shown. Restriction sites for BanII (B) and PvuII (P) as well as restriction fragments that include Z-DNA-forming sequences are shown. The map is modified from Bourbon et al. (31). GenBank entries: M55015, M55017, M55020, and M55022.
Using these primers we performed comparative PCR analyses from mAb Z22 bound and unbound DNA fractions obtained from cell lines C58NT, EpSM30, or EpD3. Fig. 2 shows reaction products obtained after 32 or 38 PCR cycles. After 32 amplification rounds, prominent bands are observed in the mAb Z22-bound fractions in all three cell lines but only for Ncl Z1 (lanes 1, 3, and 5 in Fig. 2A Upper). However, none are observed in corresponding reactions for the other segments analyzed. After 38 cycles amplification products also are seen for Ncl Z2 in the mAb Z22-bound fractions of C58NT and EpSM30 cells (lanes 7 and 9 in Fig. 2A Lower). No bands were obtained from the bound fractions of any of the RT6 gene fragments used as negative controls (Fig. 2B). Control reactions from the unbound fractions show bands for all investigated fragments from all cell lines. These results demonstrate a strong enrichment of the Ncl Z1-containing fragment by Z-DNA-specific mAb Z22 and a weaker enrichment of the Ncl Z2-containing fragment.
To quantify the relative enrichment of the Z-DNA-forming sequences in the mAb Z22-bound fraction, we performed real-time PCR assays (22) with primers specific for Ncl Z1, Ncl Z2, and RT6 Ne1. The results are shown in Fig. 2C and Table 1. Relative quantification was performed according to the ΔΔCt method (19) as described in Materials and Methods. Threshold cycle values were determined for mAb Z22 bound and unbound fractions, and ΔCT values were calculated as the difference between the threshold cycle value for the bound and unbound fraction. After ≈37 PCR cycles the threshold values were reached for the control segment that is unable to form Z-DNA. This fraction contains DNA that was not specifically bound and thus represents the background. ΔΔCT values were calculated by subtracting ΔCT values for segments Ncl Z1 and Ncl Z2 from that of RT6 Ne1. The threshold was set at values where PCRs were in exponential growth phases. This was determined by standard dilution series for each primer pair (not shown). Relative enrichment of the sequences that formed Z-DNA was calculated by the formula 2ΔΔCt and is shown in Fig. 2C. Segment Ncl Z1 was enriched in mAb Z22-bound fractions of all three cell lines by factors ranging from 571- to 4,040-fold. Ncl Z2 was enriched 12- to 34-fold in mAb Z22-bound fractions of cell lines C58NT and EpSM30.
The Z-DNA-Forming Sequence Ncl Z1 Inhibits Ncl Promoter Activity.
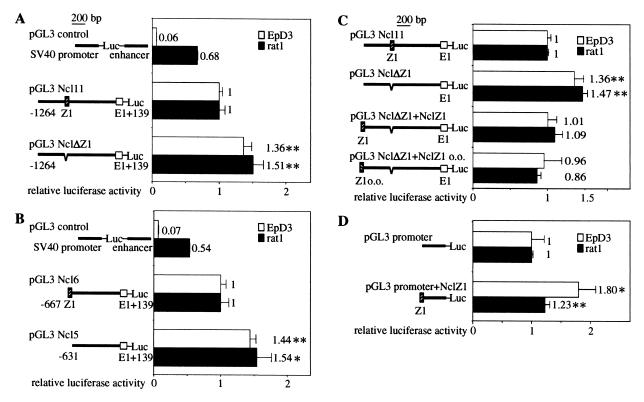
Because the location of the Z-DNA-forming sequence Ncl Z1 is 631 bp upstream of the transcription start site, we tested whether this sequence might play a regulatory role in Ncl transcription. To this end, we cloned a 1.3-kb fragment from a region containing Ncl Z1 and the first part of exon 1 into a luciferase reporter vector, yielding pGL3 Ncl11. Transient transfections of this construct into rat1 and EpD3 cells revealed a very strong promoter activity of this fragment, i.e., 1.5- and 16-fold stronger than that of the simian virus 40 (SV40) promoter and enhancer contained in the pGL3 control vector (Fig. 3A). We next deleted the Z-DNA-forming sequence Ncl Z1 and 4 bp 5′ of it from pGL3 Ncl11, yielding pGL3 NclΔZ1. Deletion of Ncl Z1 increases promoter activity by 36–51%. As a second approach we built shorter constructs beginning either exactly before or after the Z-DNA-forming sequence, termed pGL3 Ncl6 and pGL3 Ncl5, respectively. Promoter activity of the plasmid lacking the Z-DNA-forming sequence was 44–54% stronger than the plasmid containing it (Fig. 3B). These results demonstrate a repressive effect of the Z-DNA-forming element Ncl Z1 on the activity of the Ncl promoter in these cell lines.
Figure 3.
Effects of Ncl Z1 on promoter activity. EpD3 and rat1 cells were transiently transfected with the indicated reporter plasmids. Luciferase activities are shown relative to those of constructs containing Ncl Z1 (the standard deviation for each construct was calculated from four independent transfections). The pGL3 control vector, which contains the SV40 promoter and enhancer, was used as a control. (A) Construct pGL3 Ncl11 contains the wild-type sequence −1,264 to +139 relative to the transcription start site. In construct pGL3 NclΔZ1, the motif TGCT(CA)10(CG)8 is deleted, while surrounding sequences are identical to pGL3 Ncl11. (B) Promoter activities of pGL3 Ncl6 (−667/+139), which starts precisely with the Z-DNA sequence, and pGL3 Ncl5 (−631/+139), which begins just 3′ of the Z-DNA sequence, are compared. These sequences are identical except for the Z-DNA sequence. Asterisks denote significant differences in luciferase activity compared with the constructs containing the Z-DNA sequences. *, P < 0.05; **, P < 0.01. (C) The influence of the location and orientation of Ncl Z1 on Ncl promoter activity is shown. Results are shown with constructs pGL3 Ncl11 (wild type), pGL3 NclΔZ1 (Z-DNA sequence deleted), pGL3 ΔZ1+NclZ1 (Z-DNA sequence moved upstream) and pGL3 ΔZ1+NclZ1 o.o. (Z-DNA sequence moved upstream, other orientation). (D) The influence of Ncl Z1 on SV40 promoter activity is shown. Construct pGL3 promoter+NclZ1, containing Ncl Z1 upstream of the SV40 promoter differs from pGL3 promoter only by the insertion of Ncl Z1 in the KpnI site. Asterisks denote significant differences in luciferase activity compared with the wild-type construct pGL3 Ncl11 or wild-type SV40 promoter. *, P < 0.05; **, P < 0.01.
Ncl Z1 Inhibits Ncl Promoter Activity Independent of Location and Orientation but Enhances SV40 Promoter Activity.
To investigate whether the observed inhibition of the Ncl promoter by Ncl Z1 depends on location and orientation we cloned it 458 bp further upstream in both orientations and performed transfections. Both variants showed a comparable repressive effect as the wild-type construct, pGL3 Ncl11 (Fig. 3C). To investigate whether the modulatory effect of Ncl Z1 is specific for the Ncl promoter, we introduced the element into the SV40 promoter. Transfection of the corresponding construct into rat1 and EpD3 cells showed that the presence of Ncl Z1 enhanced rather than repressed the transcriptional activity of the SV40 promoter. Ncl Z1 augmented the activity of the SV40 promoter 1.2- to 1.8-fold (Fig. 3D).
Allelic Variations of the Z-DNA-Forming Sequence.
The Z-DNA-forming sequence Ncl Z1 consists of dinucleotide repeats. Such sequences often are polymorphic. To investigate polymorphism of Ncl Z1, we sequenced this element from various inbred rat strains, cell lines, and three wild rats. Five allelic variants were identified that differ with respect to the repeat number of CA or CG units (Fig. 4A). Allele a, consisting of [(CA)10(CG)8], was found in inbred rat strains BB/DR, BB/DP, BN, BUF, DA, WF, and WKY as well as in the cell lines C58NT (from which we cloned the Ncl promoter region) and EpSM30. Allele b consists of [(CA)7(CG)9] and was found in rat strains BDE, BH, E3, F344, LEW.6B, and PVG as well as in cell line rat1. Allele c, consisting of [(CA)10(CG)7] was found in SHR rats, as well in three wild rats that were caught in Hamburg and Schleswig Holstein (Germany). These wild rats were heterozygous, also carrying allele d, that consists of [(CA)9(CG)8]. Allele e, consisting of [(CA)11(CG)7], was found only in LE rats. None of the investigated sequences matched the one deposited in the database, consisting of [(CA)7(CG)8]. We also analyzed whether the Z- DNA-forming element Ncl Z1 is conserved in the closely related species R. rattus (black rat). A single allele, consisting of [(CA)7(CG)5(TG)], was found in three investigated rats from two distinct colonies.
Figure 4.
Allelic variants of Ncl Z1 show differences in the modulation of promoter activity. (A) Sequence analysis of Ncl Z1 from various strains of R. norvegicus reveals the existence of five allelic variants (a–e). The sequence M55015 from the database was designated allele f. Ncl Z1 also is found in the closely related species R. rattus. (B) Promoter activities of two constructs, which are identical except for their Z-DNA-forming sequence, are shown. Construct pGL3 Ncl7a contains the dinucleotide repeat [(CA)10(CG)8] from allele a, pGL3 Ncl7b contains [(CA)7(CG)9] from allele b. Standard deviations from six independent transfections are shown. The significant difference is indicated by asterisks (**, P < 0.01).
Allele-Specific Modulation of Promoter Activity.
Promoter fragments containing the two major allelic variants a and b were cloned into the pGL3 vector and transiently transfected to look for differences in their capability to modulate the activity of the Ncl promoter. The two constructs are identical except for the Z-DNA-forming sequence. The results shown in Fig. 4B reveal that the activity of the fragment containing allele b was significantly stronger (27%) in rat1 cells than its counterpart, demonstrating that the repressive effect on promoter activity exerted by allele a was greater than that of allele b.
Discussion
An important step toward elucidating the biological role of left-handed Z-DNA was made in the assay developed by Wittig et al. (10) that identifies Z-DNA formation in specific genes in metabolically active nuclei. Under physiological conditions Z-DNA formation is a dynamic process. In all cases where Z-DNA formation has been shown, a tight correlation between transcriptional activity and detection of Z-DNA has been observed (10, 12, 13). Because formation of Z-DNA in vitro is influenced by experimental changes in the environment (5), the assay developed by Wittig et al. is important for determining whether a potential Z-DNA-forming sequence actually adopts this conformation under physiological conditions.
We assayed Z-DNA formation in two potential elements of the rat Ncl gene, Ncl Z1 [(CA)10(CG)8] and Ncl Z2 [AC(GC)5CCGT(CG)2], and by one element in the rat Aldr1 gene, Z1 [(GC)7ACGC(AC)22]. Based on the composition and length of their dinucleotide repeats (5), Ncl Z1 and Aldr1 Z1 are predicted to have a higher potential for Z-DNA formation than Ncl Z2. Three segments of the RT6 gene without the potential to form Z-DNA also were used as controls. In a first set of experiments designed to semiquantitatively assess Z-DNA formation by these elements, we found Ncl Z1 to be highly enriched in all three cell lines using Z-DNA-specific mAb Z22. This finding indicates that large amounts of Z-DNA were formed in each case. Ncl Z2 was enriched to a lesser degree in two of the three cell lines investigated. No Z-DNA formation was observed for Aldr1 Z1, even though it contains a sequence whose Z-DNA-forming potential is comparable to that of Ncl Z1. We then quantified the relative enrichment of Ncl Z1 and Ncl Z2 by using real-time PCR. This method yields reliable quantitation of even tiny amounts of target DNA. The signals obtained for Ncl Z1 and Ncl Z2 were compared with that obtained for the non-Z-DNA-forming segment RT6 Ne1, which was used to define the background caused by nonspecific binding of DNA in the assay system. Using these cell lines, we found 571- to 4,040-fold enrichment for Ncl Z1 and 12 to 34-fold enrichment for Ncl Z2 relative to RT6 Ne1, confirming and extending the results obtained from the semiquantitative experiments.
The stronger Z-DNA signal detected for Ncl Z1 relative to Ncl Z2 is consistent with the predicted higher potential of Ncl Z1 to form Z-DNA. Furthermore, the location of the sequences may be of importance. According to the theory of Liu and Wang (14) RNA polymerase plows through DNA that is being transcribed. This generates negative supercoils upstream of the polymerase, which would favor formation of left-handed Z-DNA, and positive supercoils downstream to it, which would favor the right-handed B-DNA form. The sequence Ncl Z2 located in the first intron would be only transiently in the Z-DNA form whereas Ncl Z1 in the promoter would be continuously stabilized as long as the gene is transcribed. Similarly, a position effect (in the third intron) might account for the lack of Z-DNA formation detected for Aldr1 Z1. However, it cannot be ruled out that Aldr1 Z1 formed Z-DNA, but that bound proteins with a high affinity to Z-DNA excluded the binding of antibody, thus preventing its detection.
Because Ncl Z1 is located upstream of the transcriptional start site we investigated whether it might exert a regulatory effect on Ncl gene transcription. Consistent with the high level of Ncl expression, we observed very strong activity in reporter gene assays of a 1.3-kb promoter fragment including the Z-DNA-forming sequence. Transient transfection experiments using two independent strategies showed that deletion of the Z-DNA sequence enhanced promoter activity by 36–54%, demonstrating a repressive effect of this element. Given that the Ncl gene is constitutively transcribed at a high level in all cell types under basal conditions, and therefore factors necessary for transcription are ubiquitously present, we consider an effect of this magnitude to be of potential biological relevance. Ncl Z1 repressed Ncl promoter activity in a position- and orientation-independent manner. Intriguingly, an enhancing rather than an inhibitory effect was observed when Ncl Z1 is cloned 5′ to the SV40 promoter (Fig. 3D). This may reflect the fact that the amount of negative supercoiling, produced by the moving polymerases during transcription of Ncl, may enhance the cooperation of transcription factors. In this scenario, Ncl Z1 would absorb supercoils and consequently repress transcription, thus serving as a negative feedback mechanism. On the other hand, the SV40 promoter might exhibit strongest promoter activity when sequences absorbing negative supercoils are present. This is consistent with the occurrence of Z-DNA-forming sequences in the SV40 enhancer, which are essential for enhancement (7).
Different effects on the transcriptional activity of different promoters also have been observed in the case of a dinucleotide repeat from the inducible promoter of the rat acetyl-CoA carboxylase (ACC) gene. Here a 76-bp fragment containing (CA)28 inhibited promoter activity of the inducible ACC promoter (PI). The same sequence also repressed activity of the TK promoter. However, no effect of this element was observed on the activity of the constitutively active ACC promoter (PII) (23).
Several studies suggest that naturally occurring potential Z-DNA-forming sequences have regulatory effects on transcription. In these experiments potential Z-DNA-forming sequences have been either deleted from or introduced into reporter plasmids, and inhibitory or enhancing effects were observed (24–26). The contribution of the potential Z-DNA-forming sequences to the observed effects is probable, but adjacent sequences also may play a role. The effect of such an element has been unequivocally demonstrated in the transcriptional control of the voltage-gated potassium channel Kv1.5. In Kv1.5 nonexpressing cells, the Kv1.5 gene repressor element (KRE) reduces promoter activity in reporter gene assays by approx. 50%. The repeat sequence [(GT)19GA(CA)15] is essential for the repression (27). It is interesting that KRE does not influence promoter activity in cells expressing Kv1.5 (8). However, KRE binding factor (KBF) is present only in Kv1.5-expressing cells. Thus, the KBF may protect these cells from the repressive effects of KRE (27).
Further support for a biological function of the Z-DNA-forming sequence Ncl Z1 is provided by its high degree of conservation between the rodent species R. norvegicus (Norway rat), mouse, hamster, and R. rattus (reported here). As Bourbon et al. (28) described it: “a most striking feature is a conserved stretch of alternating purine and pyrimidine nucleotides … it would seem that there has been a selective pressure to maintain the alternating order.”
We found Ncl Z1 to be polymorphic with respect to the number of dinucleotide repeats and identified five alleles. Because Ncl Z1 evidently affects promoter activity, we tested whether the two most predominant allelic variants might inhibit transcription to different extents. Indeed, discrete but significant differences were observed, as would be expected for the effects of allelic variants of a randomly chosen gene within a healthy population. It can be assumed that the other allelic variants also differ slightly in their capability to modulate promoter activity. One can speculate that even small differences in expression levels have phenotypic consequences if constitutive, highly expressed genes such as Ncl are affected.
Interestingly, the polymorphic nature of Ncl Z1 has been previously used in genetic mapping, and it is known as marker D9Arb5 on rat chromosome 9 (29). Many of the polymorphic dinucleotide repeats used in genetic mapping consist of alternating purine/pyrimidine repeats and thus have the potential to form Z-DNA. Our results show for one such element that Z-DNA formation occurs under physiological conditions and that this element has the capability to modulate promoter activity. The finding that allelic variants of this element show differences in their inhibitory potential demonstrates that such polymorphisms, besides being helpful tools in genetic studies, also can be of functional importance. Kashi et al. (30) proposed that these simple sequence repeats are an abundant source of mutations causing quantitative effects without reducing fitness. We speculate that polymorphisms in these elements might influence transcriptional activity, as shown in this paper for the Ncl gene, by affecting their ability to form alternative DNA structures such as Z-DNA, and thus contribute to genetic variation.
Acknowledgments
We thank Maike Eiben for excellent technical assistance, Kevin L. Cunningham for operating the laser, Stefan Wölfl, Ky Lowenhaupt, Thomas Schwartz, and Alan Herbert for helpful discussions, and Thomas Schwartz for critical reading of this manuscript. This work was supported by a stipend of the Boehringer Ingelheim Fonds to S.R. and grants from the National Institutes of Health (to A.R.) and Deutsche Forschungsgemeinschaft Ha2369/1 (to F.H.).
Abbreviations
- Ncl
nucleolin
- Aldr
aldehyde reductase
- SV40
simian virus 40
- Ct
threshold cycle
References
- 1.Stallings R L, Ford A F, Nelson D, Torney D C, Hildebrand C E, Moyzis R K. Genomics. 1991;10:807–815. doi: 10.1016/0888-7543(91)90467-s. [DOI] [PubMed] [Google Scholar]
- 2.Hearne C M, Ghosh S, Todd J A. Trends Genet. 1992;8:288–294. doi: 10.1016/0168-9525(92)90256-4. [DOI] [PubMed] [Google Scholar]
- 3.Wells R D. J Biol Chem. 1988;263:1095–1098. [PubMed] [Google Scholar]
- 4.Rich A. Gene. 1993;135:99–109. doi: 10.1016/0378-1119(93)90054-7. [DOI] [PubMed] [Google Scholar]
- 5.Rich A, Nordheim A, Wang A H. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- 6.Hamada H, Seidman M, Howard B H, Gorman C M. Mol Cell Biol. 1984;4:2622–2630. doi: 10.1128/mcb.4.12.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nordheim A, Rich A. Nature (London) 1983;303:674–679. doi: 10.1038/303674a0. [DOI] [PubMed] [Google Scholar]
- 8.Mori Y, Folco E, Koren G. J Biol Chem. 1995;270:27788–27796. doi: 10.1074/jbc.270.46.27788. [DOI] [PubMed] [Google Scholar]
- 9.Jackson D A, Cook P R. EMBO J. 1985;4:913–918. doi: 10.1002/j.1460-2075.1985.tb03718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wittig B, Wàlfl S, Dorbic T, Vahrson W, Rich A. EMBO J. 1992;11:4653–4663. doi: 10.1002/j.1460-2075.1992.tb05567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson D A, Cook P R. J Mol Biol. 1986;192:65–76. doi: 10.1016/0022-2836(86)90464-x. [DOI] [PubMed] [Google Scholar]
- 12.Wölfl S, Martinez C, Rich A, Majzoub J A. Proc Natl Acad Sci USA. 1996;93:3664–3668. doi: 10.1073/pnas.93.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müller V, Takeya M, Brendel S, Wittig B, Rich A. Proc Natl Acad Sci USA. 1996;93:780–784. doi: 10.1073/pnas.93.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L F, Wang J C. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroth G P, Chou P J, Ho P S. J Biol Chem. 1992;267:11846–11855. [PubMed] [Google Scholar]
- 16.Ginisty H, Sicard H, Roger B, Bouvet P. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 17.Srivastava M, Pollard H B. FASEB J. 1999;13:1911–1922. [PubMed] [Google Scholar]
- 18.Rothenburg S, Koch-Nolte F, Thiele H-G, Haag F. Immunogenetics. 2001;52:231–241. doi: 10.1007/s002510000267. [DOI] [PubMed] [Google Scholar]
- 19.Winer J, Jung C K, Shackel I, Williams P M. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 20.Wittig B, Dorbic T, Rich A. J Cell Biol. 1989;108:755–764. doi: 10.1083/jcb.108.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haag F A, Kuhlenbaumer G, Koch-Nolte F, Wingender E, Thiele H G. J Immunol. 1996;157:2022–2030. [PubMed] [Google Scholar]
- 22.Heid C A, Stevens J, Livak K J, Williams P M. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 23.Tae H J, Luo X, Kim K H. J Biol Chem. 1994;269:10475–10484. [PubMed] [Google Scholar]
- 24.Naylor L H, Clark E M. Nucleic Acids Res. 1990;18:1595–1601. doi: 10.1093/nar/18.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu T, Ikezono T, Angus C W, Shelhamer J H. Nucleic Acids Res. 1994;22:5093–5098. doi: 10.1093/nar/22.23.5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Y H, Shin E J, Campbell M J, Niederhuber J E. J Biol Chem. 1995;270:25968–25975. doi: 10.1074/jbc.270.43.25968. [DOI] [PubMed] [Google Scholar]
- 27.Valverde P, Koren G. Circ Res. 1999;84:937–944. doi: 10.1161/01.res.84.8.937. [DOI] [PubMed] [Google Scholar]
- 28.Bourbon H M, Prudhomme M, Amalric F. Gene. 1988;68:73–84. doi: 10.1016/0378-1119(88)90600-2. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe T K, Bihoreau M T, McCarthy L C, Kiguwa S L, Hishigaki H, Tsuji A, Browne J, Yamasaki Y, Mizoguchi-Miyakita A, Oga K, et al. Nat Genet. 1999;22:27–36. doi: 10.1038/8737. [DOI] [PubMed] [Google Scholar]
- 30.Kashi Y, King D, Soller M. Trends Genet. 1997;13:74–78. doi: 10.1016/s0168-9525(97)01008-1. [DOI] [PubMed] [Google Scholar]
- 31.Bourbon H M, Amalric F. Gene. 1990;88:187–196. doi: 10.1016/0378-1119(90)90031-l. [DOI] [PubMed] [Google Scholar]