Abstract
The brain-derived neurotrophic factor (BDNF) Val66Met single nucleotide polymorphism (SNP) has been associated with individual differences in brain structure and function, and cognition. Research on BDNF’s influence on brain and cognition has largely been limited to adults, and little is known about the association of this gene, and specifically the Val66Met polymorphism, with developing brain structure and emerging cognitive functions in children.
We performed a targeted genetic association analysis on cortical thickness, surface area, and subcortical volume in 78 children (ages 6–10) who were Val homozygotes (homozygous Val/Val carriers) or Met carriers (Val/Met, Met/Met) for the Val66Met locus using Atlas-based brain segmentation. We observed greater cortical thickness for Val homozygotes in regions supporting declarative memory systems (anterior temporal pole/entorhinal cortex), consistent with adult findings. Met carriers had greater surface area in the prefrontal and parietal cortices and greater cortical thickness in lateral occipital/parietal cortex in contrast to prior adult findings that may relate to performance on cognitive tasks supported by these regions in Met carriers. Finally, we found larger right hippocampal volume in Met carriers, although inconsistent with adult findings (generally reports larger volumes for Val homozygotes), is consistent with a recent finding in children.
Gene expression levels vary across different brain regions and across development and our findings highlight the need to consider this developmental change in explorations of BDNF-brain relationships. The impact of the BDNF Val66Met polymorphism on the structure of the developing brain therefore reflects regionally-specific developmental changes in BDNF expression and cortical maturation trajectories.
Keywords: BDNF, rs6265, neurogenetics, sMRI neuroimaging, grey matter volume, cortical thickness, surface area, development
Introduction
Multiple genes and gene by environment interactions regulate the emerging cognitive abilities in the child’s developing brain. The BDNF gene (located on chromosome 11p14.1) encodes for the brain-derived neurotrophic factor, a member of the nerve growth family of proteins, and has been implicated in brain development, maturation and cognition (Alfimova et al., 2012, Bath & Lee, 2006, Dincheva et al., 2012, Egan et al., 2003, Hariri et al., 2003, Harrisberger et al., 2014, Molendijk et al., 2012, Yin et al., 2015). More specifically, BDNF influences the proliferation, differentiation and survival of neurons, neural morphology and function, synaptic changes (i.e. long-term potentiation [LTP] in the hippocampus), and, correspondingly, neuroplasticity (Frielingsdorf et al., 2010). The listed pathways and processes depend on the amount of neurotrophin present, its appropriate release, and its binding affinity to target cell membranes (Bath & Lee, 2006). Both animal and human studies suggested that common genetic polymorphisms in bdnf/BDNF are related to the structure and function of the developing brain; see Bath and Lee (2006) for a review of human and animal findings and see Hanover et al. (1999), Huang et al. (1999) for studies of mouse visual cortex. Despite these studies having established a link between BDNF and synaptic changes over development, few studies have examined the relationship between BDNF and brain function or structure in young children (Hashimoto et al., 2016, Marusak et al., 2015, Thomason et al., 2009). Here, we examine the association between the BDNF Val66Met polymorphism (dbSNP rs6265) and individual differences in brain structure in a sample of neurotypical children.
A common missense (i.e., leading to the alteration of the amino acid composition of the protein) single nucleotide polymorphism (SNP) in the BDNF gene, rs6265 or Val66Met, affects production of the BDNF protein (Egan et al., 2003). Specifically, this polymorphism results in a single amino acid substitution (valine to methionine) in the proBDNF peptide [a precursor peptide to BDNF (Hariri et al., 2003, Mowla et al., 2001)], at codon 66. Two alternative alleles (G/A) are possible at this locus, with G being the ancestral allele, and A – the derived one. Thus, three genotypes are possible: GG, AG, and AA, corresponding to Val/Val, Val/Met or Met/Met, respectively. Activity-dependent BDNF release is highest in Val/Val individuals relative to Val/Met and Met/Met individuals (however as discussed below this may vary with development). Moreover, differences in the activity-dependent BDNF release between Val/Val, Val/Met and Met/Met carriers have been observed to be associated with behavioral differences as well as differences in brain structure (e.g., white matter architecture; Ziegler et al., 2013) and function, in animals (Bath & Lee, 2006, Hanover et al., 1999, Huang et al., 1999) and human adults (Egan et al., 2003, Hariri et al., 2003).
Behaviorally, the BDNF Val66Met polymorphism has been associated with multiple aspects of cognition and, most notably, memory function. In studies with adult samples, Val homozygotes generally perform better than Met carriers on measures of declarative and working memory (Brooks et al., 2014, Galloway et al., 2008, Hansell et al., 2007), as well as other aspects of cognition, including attention (Scassellati et al., 2014, Shim et al., 2008, Zeni et al., 2012), and executive function (Alfimova et al., 2012, Smith et al., 2004). Working memory (WM) capacity involves a system for combining information storage and manipulation to involved in cognitive activities. This is often measured using an n-back task, where the participant is asked to monitor a series of stimuli (e.g., numbers, letters) and make a response when a stimulus presented is same as that presented n trials previously. During such an n-back working memory task, Met carriers were found to show reduced brain activation, compared to Val homozygotes, in brain regions including the frontal lobe (Chen et al., 2015). Compared to Val homozygotes, Met carriers also demonstrate reduced declarative memory, which includes long term storage of episodic and semantic knowledge (Kambeitz et al., 2012). However, Dodds et al. (2013) suggests that the Val66Met polymorphisms may have an effect on memory retrieval, and not on memory encoding.
However, the relationships between the BDNF gene and cognition have been almost exclusively documented in samples of healthy adults (Hashimoto et al., 2016, see Thomason et al., 2009) and psychiatric patient populations (Dias et al., 2009, Gatt et al., 2009, Lau et al., 2010, Scassellati et al., 2014, Shim et al., 2008, Smith et al., 2004, Song et al., 2015, Zeni et al., 2012), but see Hashimoto et al. (2016) for relevant research on the Val66Met polymorphism in child and adolescent brain structure. The impact of this polymorphism on the healthy development of the brain and cognition in children is not fully understood.
At the neural level, studies with adults have revealed that the Val66Met polymorphism is associated with structural variation in both cortical and subcortical grey and white matter structure, including volume, cortical thickness, and white matter integrity (Chen et al., 2007, Egan et al., 2003, Hariri et al., 2003, Pezawas et al., 2004, Szeszko et al., 2005), and brain function (Dodds et al., 2013, Egan et al., 2003, Hariri et al., 2003, Kauppi et al., 2013, Lau et al., 2010). Most prominently, this polymorphism has a well-documented effect on hippocampal structure and function, with most studies finding that the Val66Met polymorphism has an atrophic effect on the hippocampus. Indeed, murine studies have found that homozygous carriers of the Met allele have reduced hippocampal volumes, less dendritic arbors, and a 30% reduction in activity-dependent release of bdnf protein (c.f. Chen et al., 2007). In human neuroimaging studies, reduced hippocampal volume has also been observed in Met carriers, compared to Val homozygotes (Egan et al., 2003, Hariri et al., 2003, Kambeitz et al., 2012, Pezawas et al., 2004, Szeszko et al., 2005).
BDNF is also abundantly expressed in the frontal lobes, the occipital lobe and in the temporal lobe, which connect with the hippocampus through its afferent connections (Baquet et al., 2004, Bartsch & Arzy, 2014). Correspondingly, the Val66Met polymorphism has also been associated with cortical structure in these regions, specifically, Met carriers have generally demonstrated reduced gray matter volume in the superior and middle frontal gyri, including the dorsolateral prefrontal cortex (Forde et al., 2014, Matsuo et al., 2009, Pezawas et al., 2004) anterior cingulate cortex (Gerritsen et al., 2012, Matsuo et al., 2009, Mueller et al., 2013), lingual gyri (Forde et al., 2014), and fusiform gyri (Montag et al., 2009), compared to the Val homozygotes.
In addition to these structural brain differences, there are also differences in functional brain activation observed between Val homozygotes and Met carriers. Hariri et al. (2003) found reduced activation of the hippocampus during a declarative memory task among Met carriers as compared with Val homozygotes. Further, Chen et al. (2007) found that Met allele carriers showed consistently lower brain activation in the right superior frontal gyrus (SFG) and the middle occipital gyrus during a N-back working memory task. Using resting state MRI, Wei et al. (2012) also found reduced functional connectivity between the right hippocampus and left parahippocampal gyrus with cortical regions (middle temporal gyrus, inferior and middle frontal gyrus, fusiform gyrus) in Met carriers compared to Val homozygotes. These differences in brain activation between Met carriers and Val homozygotes generally correspond to observed differences in memory and general cognitive performance in adults (Brooks et al., 2014, Galloway et al., 2008, Hansell et al., 2007, Kambeitz et al., 2012). Generally, reduced activation and smaller volume of the hippocampus is associated with poorer memory performance (Rahman et al., 2016, Riggins et al., 2015, Van Strien et al., 2009, Yonelinas, 2013). Indeed, Kambeitz et al. (2012)’s meta-analysis of relation between BDNF and memory performance and both brain structure and function observed this relationship.
With respect to neural development in humans, there is currently very limited research that has explored this polymorphism in children, and findings to date are not completely consistent with the adult data. For example, in a cohort of Japanese children aged 5–18, Hashimoto et al. (2016) found greater grey matter volume in the right cuneus for Met homozygotes (Met/Met) relative to Val homozygotes (a finding not previously observed in adults), whereas they observed greater left insula and left ventromedial prefrontal cortex volumes in Val homozygotes relative to Met homozygotes which is consistent with the adult studies. Further, Marusak et al. (2015) reported larger right hippocampal volume among children and adolescents aged 7–15 who were Met carriers, whereas adult studies report larger volumes for Val carriers. Finally, Thomason et al. (2009) found that 11–12-year-old children who were Met carriers showed reduced resting state connectivity between the hippocampus and parahippocampal gyrus with cortical regions (middle temporal lobe, posterior cingulate, inferior parietal lobule, precuneus), compared to Val homozygotes, which is consistent with the adult literature and may reflect less robust hippocampal-cortical projections among children who are Met carriers.
In sum, there are demonstrated relationships between the BDNF Val66Met polymorphism, and brain structure and function; yet, the effects of this polymorphism on the developing brain are poorly characterized given the limited research on children to date. In the present study, we aimed to bridge this gap in the literature by performing a targeted genetic association study focusing on the role of the BDNF Val66Met polymorphism on the structure (cortical thickness and surface area, and subcortical volume) of developing brain systems in a sample of school-aged children. To this goal, we investigated cortical and subcortical structure in children who were homozygous for the Val allele (Val/Val), as compared with children who were Met allele carriers (Val/Met and Met/Met). We examined cortical surface area, cortical thickness, and subcortical volume differences related to the BDNF Val66Met polymorphism because these indices of brain structure are heritable, yet, reflect distinct components of brain maturation (Chouinard-Decorte et al., 2014, Panizzon et al., 2009, Winkler et al., 2010). The present study investigates whether this common genetic variant impacts brain structure in the human developing brain in ways that are relevant for children’s emerging cognitive abilities, with the goal of better understanding the complex pathways that link variation in genetic and brain structure during the course of development.
Materials and Methods
Experimental Subjects
Seventy-eight children between the ages of 6 and 10 (47 males, 31 females, mean age = 8.1, SD = 1.2) participated in this study. Participants were part of a larger longitudinal study investigating the genetic underpinnings of structural and functional brain changes over a period in development corresponding to rapid acquisition of reading and other academic skills (Landi et al., 2013, Palejev et al., 2011). Participants were divided into two groups based on their BDNF genotype: Val homozygotes (Val/Val; n=49) and Met carriers (n=29; this group included Val/Met [n=26] and Met/Met [n=3] individuals). The minor allele frequency (MAF, here for the Met allele) in the sample was 23%. As noted above, given the low frequency of the Met allele, we collapsed Val/Met and Met/Met individuals into one group: Met carriers (Petryshen et al., 2010, Tost et al., 2013, Yang et al., 2012, Ziegler et al., 2013), which is a common practice in the studies of this polymorphism in specific and low-MAF variants in general. But see recent work on differences between Val/Met and Met/Met animal models (Notaras et al., 2016).
There were no significant differences between the Val66Met genotype groups with respect to age, F(1,76) =.311, p = .578, or gender distribution, χ2 (1) = 2.098, p = .07. Table 1 presents summary of the participants’ demographics and genotypic status. Further, given reported associations between the Val66Met polymorphism and cognitive ability in adults, children in our study completed the Weschler Abbreviated Scales of Intelligence (WASI) IQ. Children in this study all had IQ scored in the normal range (Performance IQ: 79–146; Verbal IQ: 70–141) and there were no significant differences in verbal or performance IQ between the two genotype groups (Verbal IQ: F(1,76) =0.001, p>.05; Performance IQ: F(1,76)=0.705, p>.05).
Table 1.
Summary of participants
| Val homozygotes | Met carriers | |
|---|---|---|
| n | 49 | 29 |
| Gender Ratio (M:F) | 26:23 | 21:8 |
| Mean Age (SD) | 8.2 (1.2) | 8.0 (1.1) |
| Verbal IQ | 107.2 (14.5) | 107.1 (13.7) |
| Performance IQ | 109.2 (16.3) | 106.1 (15.9) |
The study received ethical approval from the Yale University Human Research Protection Program. All child subjects provided informed assent and all parents provided informed consent.
Procedure
MRI Processing and Analysis
Structural MRI data were acquired using a Siemens 1.5T Sonata scanner with 8-channel receiver array head coil. We employed the 3D magnetization prepared rapid acquisition gradient echo (MPRAGE) sequence (TR=2000ms, TE=3.65ms, flip angle=8 degrees, 160 slices, 256×256 matrix), most data were acquired using a 1mmˆ3 resolution, however, 14% of the data were acquired using the same sequence under the 1.3mmˆ3 resolution, which was resampled to 1mmˆ3 for the analysis. Each participant’s anatomical scan and resulting segmentation were visually inspected. Image processing and statistical analyses were completed with the software packages Freesurfer version 5.3.0 (Fischl et al., 2002) and R (The R Core Team, 2013).
Using Freesurfer, each high-resolution anatomical scan was registered to the Talairach space and voxels were classified as gray or white matter based on intensity and neighborhood constraints. Images were parcellated into specific regions by white and grey matter based on Desikan-Killiany Atlas (Destrieux et al., 2010) to examine volumetric, surface area, and cortical thickness differences between groups.
Surface based analysis: Individual cortical reconstructions were smoothed using a Gaussian kernel of 10 mm FWHM. Differences in cortical thickness at each vertex and differences in surface area between Val homozygotes and Met carriers were determined using Freesurfer’s QDEC (Query, Design, Estimate, Contrast) and a general linear model (GLM) with a different offset different slope (DODS) design matrix that controlled for age, and for mean cortical thickness and mean surface area by hemisphere respectively. We performed a Monte Carlo simulation with a 2-sided vertex-wise threshold of p < .05 to correct for multiple comparisons (Hagler et al., 2006). Effect size maps were calculated using Cohen’s d formula whereby the contrast effect size (Freesurfer gamma) was divided by the square root of product of the contrast variance (Freesurfer gammavar) and sample N. Surface areas were reported as number of voxels in mm2. Cortical thickness was computed as the distance between the white and plial surfaces at each vertex as described in Fischl and Dale (2000).
Subcortical volume analysis: Subcortical volumes were exported using Freesurfer’s “asegstats2table” function for additional analyses in R. We compared subcortical values between Val homozygotes and Met carriers in R using a linear regression model with participant age and overall cortical volume as covariates. Nonparametric permutation testing (1000 permutations) was performed to estimate the significance of each linear model and adjust for multiple statistical tests. Subcortical gray matter volumes were reported as the number of voxels in mm3 within each segmented region.
DNA Collection and Analysis
We obtained biological samples from all participants using sterile Oragene™ saliva collection kits (DNA Genotek, Inc). DNA was collected, extracted, and stored according to the manufacturer’s protocols. We used the Applied Biosystems Inc. (ABI) TaqMan protocol for SNP genotyping. Specifically, the Assays-on-Demand™ SNP Genotyping Product containing forward and reverse primers as well as the probe for the SNP of interest was utilized. In order to amplify the region of interest, a polymerase chain reaction (PCR) was carried out using MJ Research Tetrad Thermocycler on a 384-well plate format. TaqMan reactions included 100 ng of genomic DNA, 2.5 μl of ABI Taqman® Universal PCR Master Mix, 0.2 μl of ABI 40X Assays-on-Demand™ SNP Genotyping Assay Mix (assay ID C_11592758_10), 2.0 μl of sterile H2O and 0.5 μl of Bovine Serum Albumin (BSA). The genotyping call rate was 92%; quality was controlled by regenotyping.
Results
Cortical Thickness
With respect to cortical thickness, children who were Val homozygotes showed greater thickness than Met carriers in a cortical region encompassing the left superior temporal gyrus (STG), anterior temporal pole (ATP) and the entorhinal cortex (EC). On the other hand, children who were Met carriers showed greater cortical thickness relative to Val homozygotes in bilateral lateral occipital/superior parietal gyri, and in a region encompassing the left para-, post-, and precentral gyri (see Figure 1 and Table 2).
Figure 1.
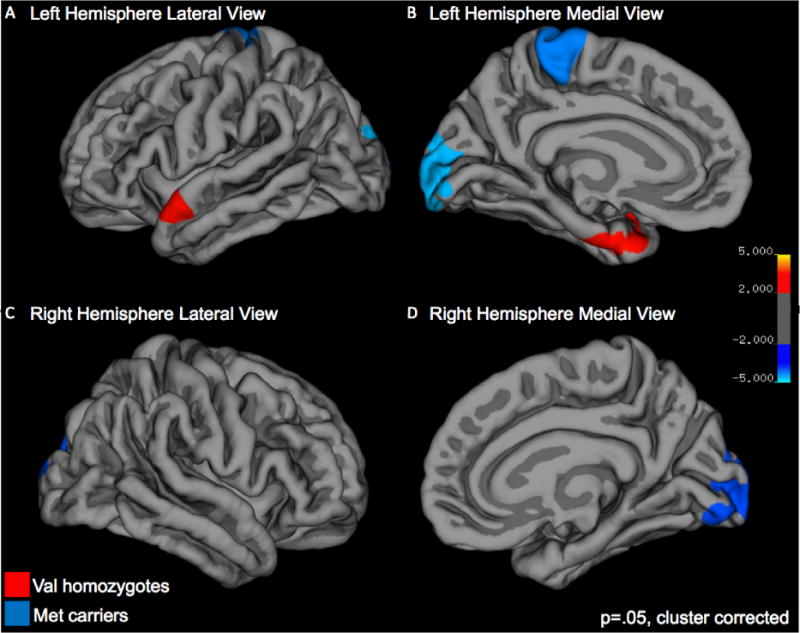
Group differences in cortical thickness. Greater cortical thickness for Val homozygotes versus Met carriers is noted in red, greater cortical thickness for Met carriers versus Val homozygotes is noted in blue.
Table 2.
Group mean differences in regional cortical thickness and surface area.
| Brain Region | Val vs Met | Difference | X | Y | Z | Cohen’s d |
|---|---|---|---|---|---|---|
| Cortical Thickness | ||||||
| L. Lateral Occipital, Superior Parietal, Pericalcarine, Cuneus | Met>Val | 1516 | −11.5 | −95 | 7 | 0.275 |
| L. Paracentral, Postcentral, Precentral | Met>Val | 1427 | −5 | −30 | 68 | 0.336 |
| L. Superior Temporal, Temporal Pole, Entorhinal Cortex | Val>Met | 869 | −54 | 12 | −19 | 0.303 |
| R. Lateral Occipital, Superior Parietal, Lingual, Pericalcarine | Met>Val | 1361 | 13.4 | −87 | 25.8 | 0.266 |
| Surface Area | ||||||
| R. Superior Parietal, Lateral Occipital | Met>Val | 1591 | 24 | −61 | 52 | 0.276 |
| R. Rostral Middle Frontal, Pars Orbitalis | Met>Val | 1482 | 40 | 41 | 24 | 0.276 |
| L. Rostral Middle Frontal, Pars Opercularis | Met>Val | 1317 | −56 | 23 | 16.7 | 0.267 |
Centroid of significant clusters are reported in MNI coordinates. Cohen’s d effect sizes are reported. Val and Met notation refer to Val homozygotes (Val/Val) and Met carriers (Val/Met, Met/Met), respectively. Differences in structure between groups for surface areas are reported as number of voxels in mm2. Cortical thickness was computed as the distance between the white and plial surfaces at each vertex as described in Fischl and Dale (2000).
Cortical Surface Area
Children who were Met carriers showed greater surface area than Val homozygotes in the right lateral occipital and superior parietal gyri, in bilateral rostral middle frontal gyri, left pars opercularis, and right pars orbitalis (see Figure 2 and Table 2).
Figure 2.
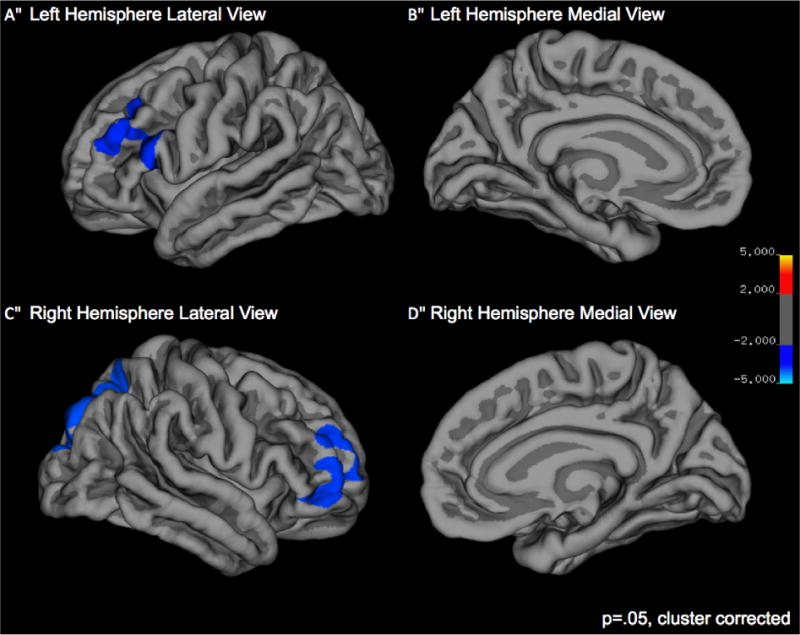
Group differences in surface area. Greater cortical surface area for Met carriers versus Val homozygotes is noted in blue.
Subcortical Volume
Children who were Met carriers showed greater right hippocampal volume than Val homozygotes (see Table 3). Additionally, we examined whether hippocampal size was related to either verbal or performance IQ and found significant relation with performance IQ (F(1,70)=5.653, p<.05); this finding is discussed below, with particular relevance for future research directions.
Table 3.
Group mean differences hippocampal volume.
| Group | Mean (mm3) | Std. Error | p value | Cohen’s d |
|---|---|---|---|---|
| Val homozygotes | 3967 | 50 | 0.018 | 0.504 |
| Met carriers | 4146 | 67 |
Discussion
In this study, we asked whether common genetic variation in the BDNF gene (specifically, the Val66Met polymorphism) is related to brain structure in children. To this end, we examined cortical surface area, cortical thickness and subcortical volume in children who were Val homozygotes (Val/Val) and Met carriers (Val/Met and Met/Met).
The majority of the published literature on the neural phenotypes associated with the Val66Met BDNF polymorphism has focused on hippocampal differences. However, BDNF is widely expressed in the brain – correspondingly, our investigation examined both cortical and subcortical structures, including the hippocampus. Differences in hippocampal and prefrontal regions related to the BDNF Val66Met polymorphism have previously been reported, but these findings were largely restricted to patient and adult populations (Matsuo et al., 2009), despite findings from the animal and human literatures documenting significant developmental changes in BDNF expression (Webster et al., 2006). Here, we provide one of the first accounts of BDNF-related neuroanatomical differences in school-aged children. Critically, our findings revealed a host of neuroanatomical differences associated with the BDNF Val66Met polymorphism, in particular in the prefrontal cortex, parietal lobes, lateral occipital area, and the hippocampus.
Cortical Thickness and Surface Area
Cortical Thickness
Previous research generally suggests that increased cortical thickness is associated with better cognitive function (Knierim et al., 2014). Here we find that children who were Val homozygotes have greater cortical thickness in the left anterior temporal lobe/entorhinal cortex – a large region that is involved in semantic processing and declarative memory (Bonner & Price, 2013), and is a primary sources of cortical projections to the hippocampus (Knierim et al., 2014). This finding is consistent with previous adult studies that report superior declarative memory performance in Val homozygotes (Egan et al., 2003, Hariri et al., 2003).
Interestingly, we also identified several regions where Met carriers showed greater cortical thickness, compared to Val homozygotes, including bilateral lateral occipital/superior parietal gyri, which are involved in comprehension, working memory, and reading (Binder et al., 2009, Chertkow et al., 1997, Ullman et al., 2014), and in a region encompassing the left para-, post-, and precentral gyri (somatosensory cortex and motor cortex).
Cortical Surface Area
With respect to surface area, all of our findings indicate larger surface area for Met carriers relative to Val homozygotes, which is somewhat unexpected given extant reports of poorer cognitive performance in adult Met carriers. Specifically, children who were Met carriers showed greater surface area relative to Val homozygotes in the right lateral occipital/superior parietal gyri (an overlapping region to that reported above), in bilateral rostral middle frontal gyri, and in a region of the inferior frontal cortex (encompassing IFG [left pars opercularis, and right pars orbitalis] near the dorsolateral prefrontal cortex: DLPFC). This region of inferior frontal cortex is involved in higher-level cognition including attention and executive function as well as language processing, task-switching, attention and working memory (Balconi, 2013, Bookheimer, 2002, Caplan, 2001, Petrides, 2005, Price, 2012).
Our analysis revealed some regions that showed similar genotype group effects for cortical thickness and cortical surface area (e.g., lateral occipital/superior parietal gyri), whereas other regions did not pattern consistently. Such differences across brain regions are unsurprising given that cortical surface area and cortical thickness reflect unique structural properties of the brain that have been found to be independent of each other and genetically uncorrelated (Panizzon et al., 2009, Winkler et al., 2010). Heritability estimates for cortical surface area and cortical thickness are high in prefrontal regions but heritability estimates for other regions vary considerably between the two measures (Hulshoff Pol et al., 2006, Joshi et al., 2011, Kremen et al., 2010, Lenroot et al., 2009, Rimol et al., 2010). Further, differing maturational rates of brain structures may interact with heritability, which is likely to influence both the overall pattern of gene-brain associations observed here, as well as the consistency between measures of thickness and surface area. Developmental changes in genetic variance offer support for this view: genetic variance increases in the frontal and temporal lobes until adulthood, which suggests higher initial heritability in early-maturing regions of the brain (e.g., occipital cortex) and higher heritability later in development in later-maturing regions (e.g., frontal cortex) such as those associated with higher cognitive functions (Chouinard-Decorte et al., 2014, Lenroot et al., 2009). This heritability pattern also matches findings from gene-expression studies showing greater variance in expression levels in childhood versus adulthood (Sterner et al., 2012). Thus, it is expected that changing genetic variability in brain structures and independent measures of those structures (thickness, surface area) may pattern differently over development.
With respect to extant cognitive-behavioral findings associated with the BDNF Val66Met polymorphism, our finding of greater cortical thickness for Val homozygotes in a region serving higher cognitive function was expected, given the oft observed superior performance of adult Val homozygotes on cognitive tasks that rely on these networks, and the fact that studies of cortical thickness and cognition typically find greater cortical thickness to be associated with better cognitive performance. On the other hand, regions where we observed greater cortical thickness for Met carriers are also involved in aspects of higher-level cognition, and these findings were unexpected based on existing adult data and relations between thickness and cognitive performance; however, these findings may be specific to children, given both different maturational levels of brain regions and levels of BDNF expression across these regions. For example, Shaw et al. (2008) found that thickness shows initial increases throughout the brain in childhood, followed by declines in adolescence until stabilization in early adulthood; higher-order areas (e.g. frontal) reach peak thickness last (late childhood). Likewise, BDNF expression levels follow different trajectories across brain regions. In the DLPFC, BDNF expression increases gradually until adulthood (Webster et al., 2002); in the temporal cortex, BDNF expression levels are highest in infancy and decline over childhood (Webster et al., 2006); in the occipital cortex, BDNF expression is stable over development (Webster et al., 2002). In the present study, Met carriers’ greater cortical thickness was observed in regions in parietal and occipital cortex, which show more stable BDNF expression over development. However, Val homozygotes’ greater cortical thickness was observed in the temporal cortex, which shows declining BDNF expression over development (Shaw et al., 2006, Sowell et al., 2004). Thus, these region-specific differences in cortical thickness between Val homozygotes and Met carriers may reflect a combination of developmental changes in BDNF expression and brain maturation across regions. Differences in brain development and BDNF expression highlight the need to interpret gene-brain relationships in a developmental context, as relations between brain structure and genotype are dynamic and developmentally-patterned.
Hippocampal Structure
The results of our study indicated that children who were Val homozygotes had smaller right hippocampi than Met carriers. The adult literature on both healthy/neurotypical and atypical populations typically reports on the opposite pattern: greater hippocampal volumes among Val homozygotes than Met carriers. Further, we observed volumetric differences only in the right hippocampus, which may be a feature of asymmetric hippocampal structure development as young infants have been previously reported to have a larger right hippocampus, an asymmetry not found in adults (Thompson et al., 2009). Indeed, our finding is consistent with (Marusak et al., 2015), who found larger right (only) hippocampal volume for children, who were Met carriers. Developmental changes in hippocampal volume may reflect general u-shaped developmental trajectories of subcortical volumes which peak in adolescence (Giedd et al., 1999, Giedd & Rapoport, 2010, Wierenga et al., 2014), as well as, variable expression of BDNF over the lifespan (Webster et al., 2006). Differences between our hippocampal volume findings and those of previous studies with adults may be linked to differences in BDNF expression and its consequent impact on brain structure over development. The significant relationship between performance IQ and hippocampal size also observed here indicates that the cognitive functions supported by this structure may be related to the BDNF Val66Met polymorphism and require further investigation using additional specific measures of children’s cognitive abilities (for example, n-back memory task).
In sum, the juxtaposition of our findings and those from the adult literature, in particular studies by Marusak et al. (2015) and Hashimoto et al. (2016), suggests that the BDNF Val66Met polymorphism exerts a significant influence on the cortical and subcortical structures of the brain, but does so differently in children and adults. These differences are likely driven by developmental differences in both structural brain development and BDNF expression, and suggest the need for more research on gene-brain relationships that take a developmental approach.
Conclusion
In the current study, we found that the BDNF gene Val66Met polymorphism was associated with hippocampal volume and cortical surface area and cortical thickness in frontal, temporal, and occipital cortices in school-aged children. Our findings were partially consistent with previous reports in adults and children but also revealed novel associations between this polymorphism and cortical thickness, surface area, and hippocampal volume. Thus, Val homozygotes showed greater cortical thickness in the anterior temporal pole/entorhinal cortex, consistent with better declarative memory performance observed in adult Val homozygotes. On the other hand, Met carriers showed greater cortical thickness in lateral occipital and somatosensory and motor cortex. Further, Met carriers showed increased surface area in a number of regions that also serve higher level cognitive processes including bilateral prefrontal cortex (left and right IFG and DLPFC), right superior parietal cortex/lateral occipital gyri, as well as greater right hippocampal volume. Although these findings were somewhat unexpected based on the adult literature, they implicate future regions to be explored in a developmental context and also suggest the need to look at both cortical thickness and surface area, as these neural features have different ontological trajectories and can be associated with different aspects of cognition. The Val/Val homozygotes and Met carriers in this study did show similar performance and verbal IQ scores, which were within a normal range. Future research will be directed at understanding whether the structural differences in observed between these groups have direct relationship to specific cognitive functions beyond IQ measures, particularly memory function. Finally, it is important to note that our findings are limited to associations between brain structure and one SNP on one gene. Complex developmental changes in brain structure, and their relationship to emerging cognition, is governed by multiple genes, gene-gene and gene-environment interactions, and future studies should explore the relation between brain structure and multiple SNPs and SNP interactions on BDNF as well as interactions with other genes. Our findings add to an increasingly complex pattern of the relationship between BDNF and brain development.
Acknowledgments
This study was supported by NIH grants R01 HD 048830 (K. Pugh, PI); P01 HD052120, (R. Wagner, PI); P01 HD 001994 (C. Fowler, PI); and R03 HD053409 (N. Landi, PI). We also want to thank Beth Eaton and Annie Stutzman for behavioral assessment of children, as well as Teri Hickey, Hedy Serofin and Cheryl McMurray for imaging participants, and Cheryl Lacadie for fMRI preprocessing. We are exceptionally grateful to the families and children who participated in this study, and extend a special thanks to the Windward School for their collaboration. Note that grantees undertaking such projects are encouraged to express freely their professional judgment. This article, therefore, does not necessarily reflect the position or policies of the National Institutes of Health and no official endorsement should be inferred.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alfimova MV, Korovaitseva GI, Lezheiko TV, Golimbet VE. Effect of BDNF Val66Met Polymorphism on Normal Variability of Executive Functions. Bulletin of Experimental Biology and Medicine. 2012;152:606–609. doi: 10.1007/s10517-012-1587-x. [DOI] [PubMed] [Google Scholar]
- Balconi M. Dorsolateral prefrontal cortex, working memory and episodic memory processes: insight through transcranial magnetic stimulation techniques. Neuroscience bulletin. 2013;29:381–389. doi: 10.1007/s12264-013-1309-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquet ZC, Gorski JA, Jones KR. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. Journal of Neuroscience. 2004;24:4250–4258. doi: 10.1523/JNEUROSCI.3920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch T, Arzy S. Human memory: insights into hippocampal networks in epilepsy. 2014 doi: 10.1093/brain/awu125. [DOI] [PubMed] [Google Scholar]
- Bath KG, Lee FS. Variant BDNF (Val66Met) impact on brain structure and function. Cognitive, affective & behavioral neuroscience. 2006;6:79–85. doi: 10.3758/cabn.6.1.79. [DOI] [PubMed] [Google Scholar]
- Binder JR, Desai RH, Graves WW, Conant LL. Where is the semantic system? A critical review and meta-analysis of 120 functional neuroimaging studies. Cerebral cortex. 2009;19:2767–2796. doi: 10.1093/cercor/bhp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner MF, Price AR. Where Is the Anterior Temporal Lobe and What Does It Do? The Journal of Neuroscience. 2013;33:4213–4215. doi: 10.1523/JNEUROSCI.0041-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookheimer S. Functional MRI of language: new approaches to understanding the cortical organization of semantic processing. Annual review of neuroscience. 2002;25:151–188. doi: 10.1146/annurev.neuro.25.112701.142946. [DOI] [PubMed] [Google Scholar]
- Brooks SJ, Nilsson EK, Jacobsson JA, Stein DJ, Fredriksson R, Lind L, Schioth HB. BDNF Polymorphisms Are Linked to Poorer Working Memory Performance, Reduced Cerebellar and Hippocampal Volumes and Differences in Prefrontal Cortex in a Swedish Elderly Population. PloS one. 2014;9 doi: 10.1371/journal.pone.0082707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan D. Functional neuroimaging studies of syntactic processing. Journal of Psycholinguistic Research. 2001;30:297–320. doi: 10.1023/a:1010495018484. [DOI] [PubMed] [Google Scholar]
- Chen CC, Chen CJ, Wu D, Chi NF, Chen PC, Liao YP, Chiu HW, Hu CJ. BDNF Val66Met Polymorphism on Functional MRI During n-Back Working Memory Tasks. Medicine. 2015;94:e1586. doi: 10.1097/MD.0000000000001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kitanishi T, Ikeda T, Matsuki N, Yamada MK. Contextual learning induces an increase in the number of hippocampal CA1 neurons expressing high levels of BDNF. Neurobiol Learn Mem. 2007;88:409–415. doi: 10.1016/j.nlm.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Chertkow H, Bub D, Deaudon C, Whitehead V. On the status of object concepts in aphasia. Brain and language. 1997;58:203–232. doi: 10.1006/brln.1997.1771. [DOI] [PubMed] [Google Scholar]
- Chouinard-Decorte F, McKay DR, Reid A, Khundrakpam B, Zhao L, Karama S, Rioux P, Sprooten E, Knowles E, Kent JW, Jr, Curran JE, Goring HH, Dyer TD, Olvera RL, Kochunov P, Duggirala R, Fox PT, Almasy L, Blangero J, Bellec P, Evans AC, Glahn DC. Heritable changes in regional cortical thickness with age. Brain imaging and behavior. 2014;8:208–216. doi: 10.1007/s11682-014-9296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destrieux C, Fischl B, Dale A, Halgren E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. NeuroImage. 2010;53:1–15. doi: 10.1016/j.neuroimage.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias VV, Brissos S, Frey BN, Andreazza AC, Cardoso C, Kapczinski F. Cognitive function and serum levels of brain-derived neurotrophic factor in patients with bipolar disorder. Bipolar Disorders. 2009;11:663–671. doi: 10.1111/j.1399-5618.2009.00733.x. [DOI] [PubMed] [Google Scholar]
- Dincheva I, Glatt CE, Lee FS. Impact of the BDNF Val66Met polymorphism on cognition: implications for behavioral genetics. Neuroscientist. 2012;18:439–451. doi: 10.1177/1073858411431646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds CM, Henson RN, Suckling J, Miskowiak KW, Ooi C, Tait R, Soltesz F, Lawrence P, Bentley G, Maltby K, Skeggs A, Miller SR, McHugh S, Bullmore ET, Nathan PJ. Effects of the BDNF Val66Met polymorphism and met allele load on declarative memory related neural networks. PloS one. 2013;8:e74133. doi: 10.1371/journal.pone.0074133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Forde NJ, Ronan L, Suckling J, Scanlon C, Neary S, Holleran L, Leemans A, Tait R, Rua C, Fletcher PC, Jeurissen B, Dodds CM, Miller SR, Bullmore ET, McDonald C, Nathan PJ, Cannon DM. Structural neuroimaging correlates of allelic variation of the BDNF val66met polymorphism. NeuroImage. 2014;90:280–289. doi: 10.1016/j.neuroimage.2013.12.050. [DOI] [PubMed] [Google Scholar]
- Frielingsdorf H, Bath KG, Soliman F, Difede J, Casey BJ, Lee FS. Variant brain-derived neurotrophic factor Val66Met endophenotypes: implications for posttraumatic stress disorder. Annals of the New York Academy of Sciences. 2010;1208:150–157. doi: 10.1111/j.1749-6632.2010.05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway EM, Woo NH, Lu B. Persistent neural activity in the prefrontal cortex: a mechanism by which BDNF regulates working memory? Essence of Memory. 2008;169:251–266. doi: 10.1016/S0079-6123(07)00015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatt JM, Nemeroff CB, Dobson-Stone C, Paul RH, Bryant RA, Schofield PR, Gordon E, Kemp AH, Williams LM. Interactions between BDNF Val66Met polymorphism and early life stress predict brain and arousal pathways to syndromal depression and anxiety. Molecular psychiatry. 2009;14:681–695. doi: 10.1038/mp.2008.143. [DOI] [PubMed] [Google Scholar]
- Gerritsen L, Tendolkar I, Franke B, Vasquez AA, Kooijman S, Buitelaar J, Fernandez G, Rijpkema M. BDNF Val66Met genotype modulates the effect of childhood adversity on subgenual anterior cingulate cortex volume in healthy subjects. Molecular psychiatry. 2012;17:597–603. doi: 10.1038/mp.2011.51. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, Paus T, Evans AC, Rapoport JL. Brain development during childhood and adolescence: A longitudinal MRI study. Nature neuroscience. 1999;2:861–863. doi: 10.1038/13158. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Rapoport JL. Structural MRI of Pediatric Brain Development: What Have We Learned and Where Are We Going? Neuron. 2010;67:728–734. doi: 10.1016/j.neuron.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagler DJ, Jr, Saygin AP, Sereno MI. Smoothing and cluster thresholding for cortical surface-based group analysis of fMRI data. NeuroImage. 2006;33:1093–1103. doi: 10.1016/j.neuroimage.2006.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JL, Huang ZJ, Tonegawa S, Stryker MP. Brain-derived neurotrophic factor overexpression induces precocious critical period in mouse visual cortex. Journal of Neuroscience. 1999;19:Rc40. doi: 10.1523/JNEUROSCI.19-22-j0003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansell NK, James MR, Duffy DL, Birley AJ, Luciano M, Geffen GM, Wright MJ, Montgomery GW, Martin NG. Effect of the BDNF V166M polymorphism on working memory in healthy adolescents. Genes Brain and Behavior. 2007;6:260–268. doi: 10.1111/j.1601-183X.2006.00254.x. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. Journal of Neuroscience. 2003;23:6690–6694. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrisberger F, Spalek K, Smieskova R, Schmidt A, Coynel D, Milnik A, Fastenrath M, Freytag V, Gschwind L, Walter A, Vogel T, Bendfeldt K, de Quervain DJ, Papassotiropoulos A, Borgwardt S. The association of the BDNF Val66Met polymorphism and the hippocampal volumes in healthy humans: a joint meta-analysis of published and new data. Neurosci Biobehav Rev. 2014;42:267–278. doi: 10.1016/j.neubiorev.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Fukui K, Takeuchi H, Yokota S, Kikuchi Y, Tomita H, Taki Y, Kawashima R. Effects of the BDNF Val66Met Polymorphism on Gray Matter Volume in Typically Developing Children and Adolescents. Cerebral cortex. 2016;26:1795–1803. doi: 10.1093/cercor/bhw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, Kirkwood A, Pizzorusso T, Porciatti V, Morales B, Bear MF, Maffei L, Tonegawa S. BDNF regulates the maturation of inhibition and the critical period of plasticity in mouse visual cortex. Cell. 1999;98:739–755. doi: 10.1016/s0092-8674(00)81509-3. [DOI] [PubMed] [Google Scholar]
- Hulshoff Pol HE, Schnack HG, Posthuma D, Mandl RCW, Baaré WF, van Oel C, van Haren NE, Collins DL, Evans AC, Amunts K, Bürgel U, Zilles K, de Geus E, Boomsma DI, Kahn RS. Genetic Contributions to Human Brain Morphology and Intelligence. The Journal of Neuroscience. 2006;26:10235–10242. doi: 10.1523/JNEUROSCI.1312-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Lepore N, Joshi SH, Lee AD, Barysheva M, Stein JL, McMahon KL, Johnson K, de Zubicaray GI, Martin NG, Wright MJ, Toga AW, Thompson PM. The contribution of genes to cortical thickness and volume. Neuroreport. 2011;22:101–105. doi: 10.1097/WNR.0b013e3283424c84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambeitz JP, Bhattacharyya S, Kambeitz-Ilankovic LM, Valli I, Collier DA, McGuire P. Effect of BDNF val(66)met polymorphism on declarative memory and its neural substrate: A meta-analysis. Neuroscience and Biobehavioral Reviews. 2012;36:2165–2177. doi: 10.1016/j.neubiorev.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Kauppi K, Nilsson LG, Adolfsson R, Lundquist A, Eriksson E, Nyberg L. Decreased medial temporal lobe activation in BDNF (66)Met allele carriers during memory encoding. Neuropsychologia. 2013;51:2462–2468. doi: 10.1016/j.neuropsychologia.2012.11.028. [DOI] [PubMed] [Google Scholar]
- Knierim JJ, Neunuebel JP, Deshmukh SS. Functional correlates of the lateral and medial entorhinal cortex: objects, path integration and local–global reference frames. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2014;369 doi: 10.1098/rstb.2013.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremen WS, Prom-Wormley E, Panizzon MS, Eyler LT, Fischl B, Neale MC, Franz CE, Lyons MJ, Pacheco J, Perry ME, Stevens A, Schmitt JE, Grant MD, Seidman LJ, Thermenos HW, Tsuang MT, Eisen SA, Dale AM, Fennema-Notestine C. Genetic and environmental influences on the size of specific brain regions in midlife: The VETSA MRI study. NeuroImage. 2010;49:1213–1223. doi: 10.1016/j.neuroimage.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi N, Frost SJ, Mencl WE, Sandak R, Pugh KR. Neurobiological bases of reading comprehension: Insights from neuroimaging studies of word-level and text-level processing in skilled and impaired readers. Reading & Writing Quarterly: Overcoming Learning Difficulties. 2013;29:145–167. doi: 10.1080/10573569.2013.758566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau JY, Goldman D, Buzas B, Hodgkinson C, Leibenluft E, Nelson E, Sankin L, Pine DS, Ernst M. BDNF gene polymorphism (Val66Met) predicts amygdala and anterior hippocampus responses to emotional faces in anxious and depressed adolescents. NeuroImage. 2010;53:952–961. doi: 10.1016/j.neuroimage.2009.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenroot RK, Schmitt JE, Ordaz SJ, Wallace GL, Neale MC, Lerch JP, Kendler KS, Evans AC, Giedd JN. Differences in genetic and environmental influences on the human cerebral cortex associated with development during childhood and adolescence. Human brain mapping. 2009;30:163–174. doi: 10.1002/hbm.20494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusak HA, Kuruvadi N, Vila AM, Shattuck DW, Joshi SH, Joshi AA, Jella PK, Thomason ME. Interactive effects of BDNF Val66Met genotype and trauma on limbic brain anatomy in childhood. Eur Child Adolesc Psychiatry. 2015 doi: 10.1007/s00787-015-0759-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Walss-Bass C, Nery FG, Nicoletti MA, Hatch JP, Frey BN, Monkul ES, Zunta-Soares GB, Bowden CL, Escamilla MA, Soares JC. Neuronal correlates of brain-derived neurotrophic factor Val66Met polymorphism and morphometric abnormalities in bipolar disorder. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2009;34:1904–1913. doi: 10.1038/npp.2009.23. [DOI] [PubMed] [Google Scholar]
- Molendijk ML, Bus BA, Spinhoven P, Kaimatzoglou A, Oude Voshaar RC, Penninx BW, van IMH, Elzinga BM. A systematic review and meta-analysis on the association between BDNF val(66)met and hippocampal volume–a genuine effect or a winners curse? American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2012;159b:731–740. doi: 10.1002/ajmg.b.32078. [DOI] [PubMed] [Google Scholar]
- Montag C, Weber B, Fliessbach K, Elger C, Reuter M. The BDNF Val66Met polymorphism impacts parahippocampal and amygdala volume in healthy humans: incremental support for a genetic risk factor for depression. Psychological medicine. 2009;39:1831–1839. doi: 10.1017/S0033291709005509. [DOI] [PubMed] [Google Scholar]
- Mowla SJ, Farhadi HF, Pareek S, Atwal JK, Morris SJ, Seidah NG, Murphy RA. Biosynthesis and post-translational processing of the precursor to brain-derived neurotrophic factor. Journal of Biological Chemistry. 2001;276:12660–12666. doi: 10.1074/jbc.M008104200. [DOI] [PubMed] [Google Scholar]
- Mueller SC, Aouidad A, Gorodetsky E, Goldman D, Pine DS, Ernst M. Gray matter volume in adolescent anxiety: an impact of the brain-derived neurotrophic factor Val(66)Met polymorphism? Journal of the American Academy of Child and Adolescent Psychiatry. 2013;52:184–195. doi: 10.1016/j.jaac.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaras MJ, Hill RA, Gogos JA, van den Buuse M. BDNF Val66Met Genotype Interacts With a History of Simulated Stress Exposure to Regulate Sensorimotor Gating and Startle Reactivity. Schizophrenia bulletin. 2016 doi: 10.1093/schbul/sbw077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palejev D, Hwang W, Landi N, Eastman M, Frost SJ, Fulbright RK, Kidd JR, Kidd KK, Mason GF, Mencl WE, Yrigollen C, Pugh KR, Grigorenko EL. An application of the elastic net for an endophenotype analysis. Behavior genetics. 2011;41:120–124. doi: 10.1007/s10519-011-9443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizzon MS, Fennema-Notestine C, Eyler LT, Jernigan TL, Prom-Wormley E, Neale M, Jacobson K, Lyons MJ, Grant MD, Franz CE, Xian H, Tsuang M, Fischl B, Seidman L, Dale A, Kremen WS. Distinct genetic influences on cortical surface area and cortical thickness. Cerebral cortex. 2009;19:2728–2735. doi: 10.1093/cercor/bhp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrides M. Lateral prefrontal cortex: architectonic and functional organization. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2005;360:781–795. doi: 10.1098/rstb.2005.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryshen TL, Sabeti PC, Aldinger KA, Fry B, Fan JB, Schaffner SF, Waggoner SG, Tahl AR, Sklar P. Population genetic study of the brain-derived neurotrophic factor (BDNF) gene. Molecular psychiatry. 2010;15:810–815. doi: 10.1038/mp.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, Egan MF, Meyer-Lindenberg A, Weinberger DR. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. Journal of Neuroscience. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CJ. A review and synthesis of the first 20 years of PET and fMRI studies of heard speech, spoken language and reading. NeuroImage. 2012;62:816–847. doi: 10.1016/j.neuroimage.2012.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MM, Callaghan CK, Kerskens CM, Chattarji S, O’Mara SM. Early hippocampal volume loss as a marker of eventual memory deficits caused by repeated stress. Sci Rep. 2016;6:29127. doi: 10.1038/srep29127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggins T, Blankenship SL, Mulligan E, Rice K, Redcay E. Developmental Differences in Relations Between Episodic Memory and Hippocampal Subregion Volume During Early Childhood. Child development. 2015;86:1710–1718. doi: 10.1111/cdev.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimol LM, Panizzon MS, Fennema-Notestine C, Eyler LT, Fischl B, Franz CE, Hagler DJ, Lyons MJ, Neale MC, Pacheco J, Perry ME, Schmitt JE, Grant MD, Seidman LJ, Thermenos HW, Tsuang MT, Eisen SA, Kremen WS, Dale AM. Cortical Thickness Is Influenced by Regionally Specific Genetic Factors. Biological psychiatry. 2010;67:493–499. doi: 10.1016/j.biopsych.2009.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scassellati C, Zanardini R, Tiberti A, Pezzani M, Valenti V, Effedri P, Filippini E, Conte S, Ottolini A, Gennarelli M, Bocchio-Chiavetto L. Serum brain-derived neurotrophic factor (BDNF) levels in attention deficit-hyperactivity disorder (ADHD) European Child & Adolescent Psychiatry. 2014;23:173–177. doi: 10.1007/s00787-013-0447-1. [DOI] [PubMed] [Google Scholar]
- Shaw P, Greenstein D, Lerch J, Clasen L, Lenroot R, Gogtay N, Evans A, Rapoport J, Giedd J. Intellectual ability and cortical development in children and adolescents. Nature. 2006;440:676–679. doi: 10.1038/nature04513. [DOI] [PubMed] [Google Scholar]
- Shaw P, Kabani NJ, Lerch JP, Eckstrand K, Lenroot R, Gogtay N, Greenstein D, Clasen L, Evans A, Rapoport JL, Giedd JN, Wise SP. Neurodevelopmental trajectories of the human cerebral cortex. Journal of Neuroscience. 2008;28:3586–3594. doi: 10.1523/JNEUROSCI.5309-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim SH, Hwangbo Y, Kwon YJ, Jeong HY, Lee BH, Lee HJ, Kim YK. Increased levels of plasma brain-derived neurotrophic factor (BDNF) in children with attention deficit-hyperactivity disorder (ADHD) Progress in Neuro-Psychopharmacology & Biological Psychiatry. 2008;32:1824–1828. doi: 10.1016/j.pnpbp.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Wilkie M, Smith G, Wolff R, Reid IC, Muir WJ, Blackwood DHR. The Val66Met polymorphism of BDNF is associated with impaired frontal executive function in young adults with recurrent depression. Journal of Psychopharmacology. 2004;18:A18–A18. [Google Scholar]
- Song X, Quan M, Lv L, Li X, Pang L, Kennedy D, Hodge S, Harrington A, Ziedonis D, Fan X. Decreased cortical thickness in drug naive first episode schizophrenia: In relation to serum levels of BDNF. Journal of psychiatric research. 2015;60:22–28. doi: 10.1016/j.jpsychires.2014.09.009. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Leonard CM, Welcome SE, Kan E, Toga AW. Longitudinal mapping of cortical thickness and brain growth in normal children. Journal of Neuroscience. 2004;24:8223–8231. doi: 10.1523/JNEUROSCI.1798-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner KN, Weckle A, Chugani HT, Tarca AL, Sherwood CC, Hof PR, Kuzawa CW, Boddy AM, Abbas A, Raaum RL, Grégoire L, Lipovich L, Grossman LI, Uddin M, Goodman M, Wildman DE. Dynamic Gene Expression in the Human Cerebral Cortex Distinguishes Children from Adults. PloS one. 2012;7:e37714. doi: 10.1371/journal.pone.0037714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeszko PR, Lipsky R, Mentschel C, Robinson D, Gunduz-Bruce H, Sevy S, Ashtari M, Napolitano B, Bilder RM, Kane JM, Goldman D, Malhotra AK. Brain-derived neurotrophic factor val66met polymorphism and volume of the hippocampal formation. Molecular psychiatry. 2005;10:631–636. doi: 10.1038/sj.mp.4001656. [DOI] [PubMed] [Google Scholar]
- The R Core Team. R; A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- Thomason ME, Yoo DJ, Glover GH, Gotlib IH. BDNF genotype modulates resting functional connectivity in children. Frontiers in human neuroscience. 2009;3:55. doi: 10.3389/neuro.09.055.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DK, Wood SJ, Doyle LW, Warfield SK, Egan GF, Inder TE. MR-determined hippocampal asymmetry in full-term and preterm neonates. Hippocampus. 2009;19:118–123. doi: 10.1002/hipo.20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tost H, Alam T, Geramita M, Rebsch C, Kolachana B, Dickinson D, Verchinski BA, Lemaitre H, Barnett AS, Trampush JW, Weinberger DR, Marenco S. Effects of the BDNF Val66Met polymorphism on white matter microstructure in healthy adults. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2013;38:525–532. doi: 10.1038/npp.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman H, Almeida R, Klingberg T. Structural maturation and brain activity predict future working memory capacity during childhood development. Journal of Neuroscience. 2014;34:1592–1598. doi: 10.1523/JNEUROSCI.0842-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Strien NM, Cappaert NL, Witter MP. The anatomy of memory: an interactive overview of the parahippocampal-hippocampal network. Nature reviews. Neuroscience. 2009;10:272–282. doi: 10.1038/nrn2614. [DOI] [PubMed] [Google Scholar]
- Webster MJ, Herman MM, Kleinman JE, Shannon Weickert C. BDNF and trkB mRNA expression in the hippocampus and temporal cortex during the human lifespan. Gene Expression Patterns. 2006;6:941–951. doi: 10.1016/j.modgep.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Webster MJ, Weickert CS, Herman MM, Kleinman JE. BDNF mRNA expression during postnatal development, maturation and aging of the human prefrontal cortex. Developmental Brain Research. 2002;139:139–150. doi: 10.1016/s0165-3806(02)00540-0. [DOI] [PubMed] [Google Scholar]
- Wei SM, Eisenberg DP, Kohn PD, Kippenhan JS, Kolachana BS, Weinberger DR, Berman KF. Brain-derived neurotrophic factor Val(6)(6)Met polymorphism affects resting regional cerebral blood flow and functional connectivity differentially in women versus men. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:7074–7081. doi: 10.1523/JNEUROSCI.5375-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga L, Langen M, Ambrosino S, van Dijk S, Oranje B, Durston S. Typical development of basal ganglia, hippocampus, amygdala and cerebellum from age 7 to 24. NeuroImage. 2014;96:67–72. doi: 10.1016/j.neuroimage.2014.03.072. [DOI] [PubMed] [Google Scholar]
- Winkler AM, Kochunov P, Blangero J, Almasy L, Zilles K, Fox PT, Duggirala R, Glahn DC. Cortical Thickness or Grey Matter Volume? The Importance of Selecting the Phenotype for Imaging Genetics Studies. NeuroImage. 2010;53:1135–1146. doi: 10.1016/j.neuroimage.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Liu P, Sun J, Wang G, Zeng F, Yuan K, Liu J, Dong M, von Deneen KM, Qin W, Tian J. Impact of brain-derived neurotrophic factor Val66Met polymorphism on cortical thickness and voxel-based morphometry in healthy Chinese young adults. PloS one. 2012;7:e37777. doi: 10.1371/journal.pone.0037777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Hou Z, Wang X, Sui Y, Yuan Y. The BDNF Val66Met polymorphism, resting-state hippocampal functional connectivity and cognitive deficits in acute late-onset depression. Journal of affective disorders. 2015;183:22–30. doi: 10.1016/j.jad.2015.04.050. [DOI] [PubMed] [Google Scholar]
- Yonelinas AP. The hippocampus supports high-resolution binding in the service of perception, working memory and long-term memory. Behavioural brain research. 2013;254:34–44. doi: 10.1016/j.bbr.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeni CP, Tramontina S, Aguiar BW, Salatino-Oliveira A, Pheula GF, Hutz M, Kapczinski F, Rohde LA. Brain-Derived Neurotrophic Factor in Juvenile Bipolar Disorder and Attention-Deficit Hyperactivity Disorder: differentiation using BDNF serum levels, and Val66Met. Bipolar Disorders. 2012;14:48–48. [Google Scholar]
- Ziegler E, Foret A, Mascetti L, Muto V, Le Bourdiec-Shaffii A, Stender J, Balteau E, Dideberg V, Bours V, Maquet P, Phillips C. Altered white matter architecture in BDNF met carriers. PloS one. 2013;8:e69290. doi: 10.1371/journal.pone.0069290. [DOI] [PMC free article] [PubMed] [Google Scholar]


