Abstract
Nucleus pulposus (NP) cells are derived from the notochord and differ from neighboring cells of the intervertebral disc in phenotypic marker expression and morphology. Adult human NP cells lose this phenotype and morphology with age in a pattern that contributes to progressive disc degeneration and pathology. Select laminin-mimetic peptide ligands and substrate stiffnesses were examined for their ability to regulate human NP cell phenotype and biosynthesis through the expression of NP-specific markers aggrecan, N-cadherin, collagen types I and II, and GLUT1. Peptide-conjugated substrates demonstrated an ability to promote expression of healthy NP-specific markers, as well as increased biosynthetic activity. We show an ability to re-express markers of the juvenile NP cell and morphology through control of peptide presentation and stiffness on well-characterized polyacrylamide substrates. NP cells cultured on surfaces conjugated with α3 integrin receptor peptides P4 and P678, and on α2, α5, α6, β1 integrin-recognizing peptide AG10, show increased expression of aggrecan, N-cadherin, and types I and II collagen, suggesting a healthier, more juvenile-like phenotype. Multi-cell cluster formation was also observed to be more prominent on peptide-conjugated substrates. These findings indicate a critical role for cell-matrix interactions with specific ECM-mimetic peptides in supporting and maintaining a healthy NP cell phenotype and bioactivity.
Graphical abstract
1. Introduction
Nucleus pulposus (NP) cells are derived from the embryonic notochord and are responsible for the original synthesis and maintenance of the ECM of the intervertebral disc. An early decrease in their cell number, loss of this developmental phenotype, and infiltration of alternate cell types are considered critical events in the alterations in mechanical function associated with intervertebral disc degeneration [1-3]. NP cells may interact with collagens, fibronectin and laminins of the extracellular matrix (ECM) through integrin and non-integrin mediated mechanisms [4-10] with profound effects on cellular biosynthesis, attachment and morphology. Studies have shown the α5β1 integrin heterodimer regulates NP cell interactions with fibronectin [8], and are also involved in the onset of cell pathobiology following exposure to degraded fragments of fibronectin [11]. Studies of rat NP cells have shown that attachment to type II collagen is mediated by the α2 integrin subunit in a process that involves activation of extracellular signal-regulated kinase-1 (ERK) [5], while porcine NP cells were instead shown to use the α1 integrin subunit to attach to type II collagen [8]. While collagens and fibronectin are compositionally abundant in the intervertebral disc, NP cell interactions with laminin proteins may be a key feature that distinguishes juvenile from aged, degenerate NP cells. Multiple isoforms of laminin are present in the juvenile NP, but not adjacent anulus fibrosus (AF) regions, as identified by immunohistochemical staining for the γ1 and other laminin chains [9, 12]. Porcine NP cells have been shown to interact with laminins LM-111 and LM-511 through integrins α6 and β1 [8, 13], while human NP cells derived from aged, degenerate tissues rely upon integrins α3, α5 and β1 for binding to these same laminins [10]. Together, these findings begin to reveal a role for specific integrin subunits in mediating NP cell-ECM interactions.
Peptides derived from ECM molecules may act as cell recognition motifs and can be used to increase cell attachment and elicit specific cell responses [14]. As compared to full-length proteins, peptides have many beneficial properties, such as receptor specificity, increased stability, ease of coupling, and cost effectiveness. The most commonly used cell recognition peptide, RGD (Arg-Gly-Asp), was derived from fibronectin [15] and has been shown to interact with integrins α5β1 and αVβ5, and up to 12 additional integrin subunits [16-18]. Of relevance to NP cells, a mechanically stimuli-driven increase in ECM production for NP cells can be attenuated when incubating cells with the RGD peptide, given evidence of functional interactions for NP cells with RGD [7]. To date, there is no information on peptide effects on NP cells other than RGD. In our prior work, we have demonstrated that surfaces coupled with full-length laminins (LM-111, LM-511), or basement membrane extract rich in laminins, can promote healthier, more biosynthetically active NP cells. These cells cultured on laminin-presenting substrates show elevated glycosaminoglycan (GAG) synthesis, a prototypical rounded and clustered cellular morphology, and elevated expression of healthy NP molecular markers including N-cadherin, type II collagen, and brachyury [13, 19-21]. In particular, we have identified laminin-coupled substrates of polyethylene glycol (PEG) or polyacrylamide (PAAm) that are uniquely well-suited to promote these features when engineered with stiffness less than 0.5 kPa [21-23]. Thus, cell recognition peptides derived from laminins may play a unique role in regulating NP cell interactions and behaviors. Nomizu and co-workers have been screening laminin chains since 1995 for active peptide sequences, resulting in a library of laminin-derived bioactive peptides [24-27]. Due to these and other similar studies, there is a large selection of laminin-derived peptides that have been shown to engage specific integrin and non-integrin cell surface receptors and to modulate the behaviors of multiple cell types (e.g. [28-31]). The objective of this study was to identify a subset of ECM mimetic peptides that can regulate human NP cell attachment, morphology and behaviors, and to reveal if peptide-coupled substrates of varying stiffness and peptide specificity can maintain the healthy NP-specific cell phenotype.
2. Materials and Methods
2.1 Primary Human NP cell culture
Cells from the NP region of to-be-discarded surgical waste tissues (ages 33-77, degenerative scoliosis, non-human subjects, IRB exemption) from lumbar and thoracic regions were isolated using a pronase-collagenase digestion, as previously described [10]. Cells from each patient were collected as separate samples, but vertebral level and grade of pathology were not known for the non-human subjects research designation (only age, race and gender information were collected). Briefly, NP tissue was separated from the surgical sample and surrounding anulus fibrosus and cartilaginous tissue and digested 2-4 hours in 25mL digestion solution per gram of NP tissue (0.3% collagenase type II, 0.2% pronase). The cells were expanded up to passage 2 in F-12 media supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin under 5% CO2.
2.2 Peptide selection for NP cell attachment
Ten ECM mimetic peptides were chosen for their demonstrated role in regulating cell adhesion and spreading in other cell types [29, 31-40]. Peptide selection was additionally constrained by using peptides derived from laminins and/or peptides shown to engage integrin α3, α5, or α6 (Table 1), based on demonstrated interactions of NP cells with laminin through these integrin subunits. The final set of peptides were synthesized with a cysteine-glycine-glycine at the N-terminus (American Peptide Co., Sunnyvale, CA). The peptides were manufactured at over 95% purity and solubilized in ultrapure water at 1 mg/mL; peptides p678 and AG10 were the exception, requiring 20% acetonitrile to fully solubilize at 1 mg/mL. Stock peptides were stored at -80°C until use. Experimental peptide solutions were prepared from the stock by dilution with ultrapure water.
Table 1. Peptides selected for screening against NP cells.
| Peptide Name | Amino Acid Sequence | Reported Cell Receptors |
|---|---|---|
| GD6 | CGG(KQNCLSSRAS)FRGCVRNLRLSR | Integrin α3 [32,35,40,44,45] |
| P4 | CGGPPFLMLLKGSTR | Integrin α3 [32,35,44,45,71] |
| p678 | CGGFQGVLQNVRFVF | Integrin α3 [32,34,35,44,45,71] |
| cNGQ | CGGcMGQGEQc (cyclic) | Integrin α3 [32,35,44,45] |
| AG32 | CGGTWYKIAFQRNRK | Integrins α2, α6, β1 [27, 29,38] |
| AG10 | CGGNRWHSIYITRFG | Integrins α2, α5, α6, β1 [27,29,38] |
| T1 | CGGTTSWSQCSKS | Integrin α6 [31,46,47] |
| AG73 | CGGRKRLQVQLSIRT | Syndecans [25,27,37,49,53-56,72] |
| RGD | CGGRGDS | Integrins α5, αV, α8 [15-18,50,51] |
| IKVAV | CSRARKQAASIKVAVSADR | Integrins [33,39,48,49] |
Table 1. Laminin-mimetic peptides used for initial screening.
2.3 NP cell attachment to peptide coated surfaces
Substrates (96 half-well assay plate, Corning) were prepared by adding 40 μl of each peptide solution to each well at 0.1, 1, 10, 100, and 200 μg/mL on the plates; controls included wells with no protein (PBS only), mouse LM-111 at 20 μg/mL (Millipore, Billerica, MA), or mouse LM-511 at 10 μg/mL (Sigma-Aldrich) (all values chosen based on prior work showing maximal NP cell attachment [8, 10]. Plates were incubated overnight at 4 °C to adsorb the peptides to the surface of the plastic. The wells were then blocked with 3.75% bovine serum albumin (BSA) for 3 hours at 37 °C followed by one rinse with 1× PBS. Wells without protein but blocked with BSA were negative controls to identify nonspecific cell binding.
NP cells (tissue ages 30-77, n=9 patients) were detached from tissue culture plastic, seeded in serum-free media in protein-coated wells (4,000 cells/well), allowed to attach for 1 hour, and washed twice with serum-free media to remove unattached cells. Attached cells were lysed and counted using the CellTiter-Glo luminescent cell viability assay (Promega). Cell attachment for each surface was performed in triplicate for cells from each sample and averaged. Differences in percent cell attachment amongst culture substrates were detected via one-factor ANOVA (substrate) and Tukey's HSD post hoc analysis for each plated peptide concentration. Data normality was determined by the Shapiro-Wilk test (α = 0.05).
2.4 Fabrication and characterization of peptide-functionalized substrates
Four peptides were selected for conjugation to polyacrylamide (PAAm) gel substrates of tunable mechanical properties, based on results from the initial NP cell attachment screen. These peptides were p4, p678, AG10 and AG73. Thin PAAm gels were created by polymerizing acrylamide (5% or 8% w/v, Bio-Rad) with bis-acrylamide crosslinker (0.03 – 0.15% w/v, Bio-Rad) in solution with 30 mM HEPES buffer (Gibco), 0.4% N,N,N′,N′-tetramethylethylene-diamine (TEMED, Bio-Rad), and sufficient 5N HCl to bring the pH to 6.0. Concerns about variations in peptide density led us to functionalize peptide to PAAm gels, through use of the N2-linker [41]. N2 is an amine-reactive N-hydroxysuccinimide ester that was added to acrylamide during co-polymerization (20 mM acrylic acid N-hydroxysuccinimide ester dissolved in ethanol, CovaChem). After the addition of 0.05% ammonium persulfate (Bio-Rad), the gel was allowed to polymerize for 15 minutes between a microscope slide and a 24 × 60 mm coverglass. The polymerized gel was partitioned into 8 distinct 0.7 cm2 gels by clamping an 8-well media chamber (Millicell EZ Slide, Millipore) onto the microscope slide. Gels were functionalized by immediately adding the desired ligand to each well (0.1 mM peptide or 0.13 mM LM-111 in cold 50 mM HEPES with 5 mM EDTA at pH 8.5) and incubated at 4 °C overnight. After ligand incubation, gels were rinsed with 1× PBS twice, sterilized in UV light for 20 minutes, and immediately used for experiments.
For characterization, the mechanical properties of PAAm gels functionalized with peptide AG73 were measured using atomic force microscopy (AFM) indentation (MFP-3D, Asylum Research), as described previously [42] Briefly, a cantilever and borosilicate spherical tipped probe (Novascan Technologies Inc.) were used to indent the PAAm gels of interest at a constant indentation rate of 15 μm/sec. The generated force-indentation curves were fit to the Hertz contact model for spherical indentation of a flat surface assuming a Poisson's ratio of 0.45 (MATLAB) [43]. A minimum of 216 indentations per gel were tested (at least three different gels, each with at least two separate locations and 36 indentations made in a 50 μm × 50 μm grid at each location). Elastic moduli were compared amongst gel formulations using the Mann-Whitney test. For subsequent experiments, only 14 kPa and 0.3 kPa PAAm gel formulations were used to test the effect of NP cell biosynthesis and phenotype upon substrate stiffness.
2.5 Characterization of peptide-PAAm conjugation
Polyacrylamide gels (8%/0.15% or 5%/0.03% acrylamide/bis-acrylamide) were functionalized with 0.1, 0.05, 0.01, or 0 mM peptide conjugated to the fluorophore FITC (CGG(KQNCLSSRAS)FRGCVRNLRLSR – FITC, American Peptide); as all peptides were of similar length and N-terminus composition, we used GD6 as the model peptide for this experiment. After conjugation, the gels were washed 4 times for 5 minutes each to ensure complete removal of unattached peptide. Total peptide attached to each gel was determined using a plate reader to quantify fluorescence (Enspire MultiMode Plate Reader, Perkin Elmer). Each functionalized gel was detached from the glass microscope slide, finely diced, and suspended in PBS and placed in a clear 96-well plate. The fluorescence intensity of each well was read using a plate reader (490 nm excitation, 520 nm emission) and compared to a standard curve. All sample readings, except for the gels with no peptide attached, were above the lower limit of detection (LLOD = 3 μM) as determined by the linear portion of the standard curve. Peptide concentration for each sample was determined from the standard curve and total peptide per gel was calculated by dividing by the dilution factor.
To determine total gel height and depth of peptide conjugation, Z-stack images were obtained using confocal laser scanning microscopy (Zeiss LSM 510, Carl Zeiss) with 1μm thick optical slices (n≥3 gels). Peptide functionalization depth was calculated as the z-distance that the average pixel intensity for each planar image (each 1 μm thick) was over 25% of the image slice with maximum average pixel intensity. Total gel height was determined by subtracting the z-coordinate of initial FITC fluorescence from the z-coordinate of initial autofluorescence from the bottom glass microscope slide. To visualize cell interaction with peptide conjugated gels, human NP cells expressing a CMV-mCherry constitutive reporter construct were seeded onto the gels at 20,000 cells/cm2 and allowed to attach for 2 days. Representative z-stack images were taken on the confocal microscope to qualitatively identify if the cells penetrated the upper surface of the gel.
2.6 Analysis of NP cell attachment and clustering on peptide functionalized substrates
Cell attachment was determined on soft (0.3 kPa) and stiff (14 kPa) substrates functionalized with five different ligands (LM-111, AG73, P4, P678, or AG10). Human NP cells (n = 4 patients, ages 54 - 63) were seeded onto the gels at 60,000 cells/cm2 in culture media containing 10% FBS, 100 U/mL penicillin/100 μg/mL streptomycin and cultured for 4 days (cell doubling time ≥ 4d). To visualize the attached cells, the nuclei and filamentous actin (F-actin) were stained with propidium iodide (PI) and phalloidin, respectively. The cells on the gels were rinsed with PBS, fixed with 4% formaldehyde, permeabilized with 0.2% triton, and labeled with Alexa Fluor 488 phalloidin (Invitrogen) at a 1:200 dilution. Cell nuclei were counterstained with PI (Sigma) at 1 mg/mL. Stained slides were imaged using confocal laser scanning microscopy. For each gel, images from 3 fields of view were captured. Number of cells attached to each gel was determined by counting the nuclei in each field of view, averaging the three values for each gel, and extrapolating for the entire gel area. Percent cell attachment was determined by dividing the attached cells by the total cells seeded per well, averaged for all the gels, and differences between substrates was determined with a 2-way ANOVA (stiffness, ligand) and Tukey's HSD post-hoc test. Data normality was determined by the Shapiro-Wilk test (α = 0.05).
Cell clusters were manually outlined and counted using ImageJ software (NIH). Cell clusters were defined as having at least 3 cells, no gap between cells in a cluster, and at least 25% of each cell's perimeter was touching another cell in the cluster. The percent of attached cells in clusters was quantified for each field of view. These values were averaged for all gels of a given substrate, and differences between conditions was determined with a 2-way ANOVA (stiffness, ligand). Tukey's HSD post hoc test was used to test for differences between all conditions, and Dunnett's post hoc test was used to test for differences as compared to 14 kPa LM-111. Data normality was determined by the Shapiro-Wilk test (α = 0.05).
2.7 Sulfated glycosaminoglycan production
The production of sulfated glycosaminoglycan (sGAG) was determined on soft (0.3 kPa) and stiff (14 kPa) substrates functionalized with five different ligands (LM-111, P4, P678 or AG10 or AG73). Human NP cells (n = 4 patients, ages 32 - 63) were seeded onto the gels at 60,000 cells/cm2 in culture media. Cells were cultured on the gels at 5% CO2 and 37°C for 4 days before cell analysis. The production of sGAG by NP cells was quantified using the dimethylmethylene blue (DMMB) spectrophotometric assay. At day 4, media from each well was collected while the cells and substrates remaining in each well were digested in papain solution (125 μg/mL in PBS with 5 mM EDTA and 5 mM cysteine) for 4 hours at 55°C. As a control, media and papain digests were collected from 3 cell-free wells (PAAm only). sGAG was quantified from absorbance readings (535 nm), with chondroitin-4-sulfate (Sigma-Aldrich) as a standard (diluted in media or papain and corrected using reading from the cell-free control samples). The sGAG production was then normalized to DNA content (Quant-iT PicoGreen dsDNA kit, Invitrogen) for each sample. Differences in sGAG/DNA (μg/μg) amongst substrates were detected with a two-factor ANOVA (stiffness, ligand) and Dunnett's post hoc analysis. Data normality was determined by the Shapiro-Wilk test (α = 0.05).
2.8 mRNA extraction and quantification
Cells from patients (ages 36-67, n=5) were plated on gels prepared from each of the five different ligands (LM-111, AG73, P4, P678, or AG10) and cultured for 4 days in culture media. Cells were detached with 0.025% trypsin/EDTA, spun down, and lysed with RLT buffer (Qiagen) with 1% β-mercaptoethanol (Fisher Scientific). Total RNA was extracted with the RNAeasy Micro kit plus DNase I digestion (Qiagen). mRNA quality and concentration was determined (NanoDrop, Thermo Fisher Scientific) and reverse transcribed into cDNA using the iScript cDNA synthesis kit (Biorad). cDNA samples were diluted to a final concentration of 10 ng/μL using RNAse- and DNAse-free water. qRT-PCR was performed on cDNA samples to identify aggrecan (ACAN), type I collagen (COL1A1), type II collagen (COL2A1,), N-cadherin (CDH2), and glucose transporter (GLUT) 1 (SLC2A1) (Life Technologies). Duplicate reactions using cDNA (2 μL) were performed using the StepOnePlus real-time PCR system (Applied Biosystems, Waltham, MA) with each well containing 2× universal master mix (12.5 μl) (Applied Biosystems), Taqman primer probes (1.25 μL), and ddH2O (9.25 μL). Relative gene expression changes were quantified among the 10 substrate groups by comparing ΔCt values (Target Ct - GAPDH Ct). Data were normalized to LM-111. Fold change in expression was calculated for cDNA using the delta-delta Ct method (Fold change = 2-ΔΔCt) and Log2 transformed. The second Δ accounted for the fold change over cells cultured on LM-111 at 14 kPa. Differences in expression level for each gene was tested with a 2-way ANOVA (stiffness, ligand) with Dunnett's post hoc analysis. Additionally, differences between peptide and laminin substrates were tested for each gene (ANOVA). Data normality was determined by the Shapiro-Wilk test (α = 0.05).
3. Results
3.1 NP cell attachment to peptide coated surfaces
On full length laminin LM-111 coated tissue culture plastic, primary human NP cells attach in an elongated morphology and form fewer clusters (Figure 1A). All peptides mediating cell attachment were found to be concentration dependent (Figure 1B) with a uniformly consistent monotonic increase in percent cells attached with increasing peptide concentration. At the highest peptide density, human NP cells attached to the LM-mimetic peptides IKVAV, GD6, P4, and AG10, and to the syndecan peptide AG73 at levels comparable to LM-111 (65% cell attachment at 20 μg/mL; Figure 1B); peptides AG73 and P678 increased NP cell attachment above BSA control levels at only 1 μg/ml. Of the integrin α3 binding peptides, P4 mediated the most attachment at a concentration of 200 μg/ml (59%), followed closely by GD6 (56%) and p678 (41%), while the cyclic peptide cNGQGEQ contributed to no significant increase in cell attachment compared to the BSA control (p > 0.05) (Figure 1C). Integrin α6 binding peptides AG10 and AG32 both mediated high levels of cell attachment (59% and 42%, respectively, at 200 μg/ml). RGD, unexpectedly, did not promote any increase in cell attachment over the BSA control at any concentration (p > 0.05). Based on these findings, peptides with maximal cell attachment through integrin α3 (P678 and P4), integrin α6 (AG10), and syndecans (AG73) were selected for further study via coupling to PAAm gels.
Figure 1.
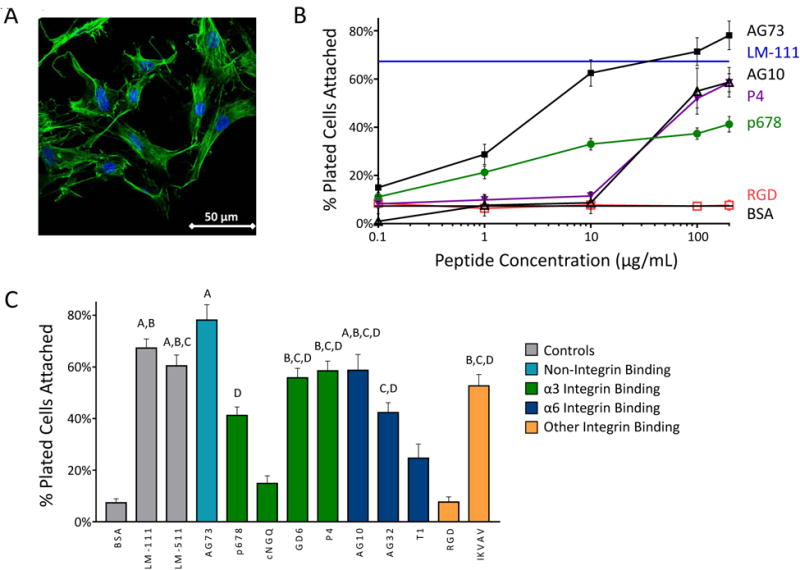
(A) Primary human NP cells display consistent morphology and higher attachment on LM-111-coated tissue culture plastic (scale bar = 50 μm). (B) NP cells attach to peptide-coated surfaces at levels equivalent to that for LM-111, but only at higher peptide densities and for select peptides as shown here. (C) Cell numbers attaching to peptide coated surfaces for all screened peptides at 200 μg/mL. Conditions labeled with the same letter (A, B, etc) are equivalent and significantly different than the BSA control (p<0.05, ANOVA with post-hoc tests). SEM are shown.
3.2 Characterization of peptide-PAAm conjugated substrates
Primary NP cells attached to the peptide coated substrates with varying morphologies from rounded and clustered, to more spread and elongated (Figure 2A). PAAm gels with NHS functionalization were formulated with stiffness of 0.3 kPa (soft) or 14 kPa (stiff), as confirmed by AFM (Figure 2B). Gels were functionalized with peptides P678, P4, AG10, and AG73, and with LM-111 via primary amine coupling. Total peptide attached to each gel was determined via fluorescence readings of GD6-FITC. Results show no variation in conjugated peptide with “soft” and “stiff” PAAm gels (p > 0.3, ANOVA) with an average surface density of 1,160 peptides/μm2 (Figure 2C, 2D) estimated from total conjugated peptide and gel area). The average depth of functionalization was found to be 11.3 ± 2.1 μm from the surface of the gel (Figure 2C), and total thickness of the PAAm gel was similar for both “soft” and “stiff” at 84.6 ± 6.9 μm.
Figure 2.
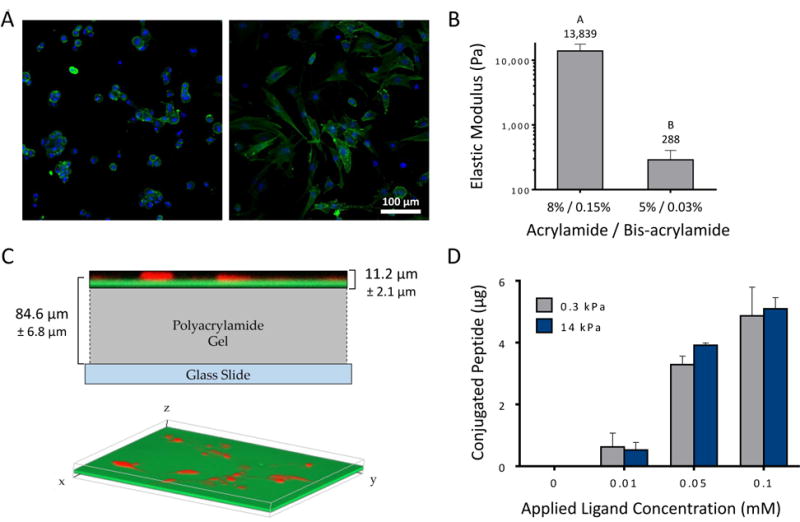
(A) Primary human NP cells attach to soft LM-111 coupled substrates (0.3 kPa polyacrylamide; left image) as rounded cells forming multi-cell clusters, while cells attaching to stiff (14 kPa; right image) substrates may elongate and form fewer cell clusters. (B) PAAm gels were formed with varying bisacrylamide ratios to achieve compressive moduli of 0.3 kPa (soft) and 14 kPa (stiff) as measured by atomic force microscopy. Conditions not connected by the same letters are significantly different from each other (p<0.0001, Mann-Whitney test). SD are shown. (C) Representative confocal Z-stack images of fluorescently labeled peptides conjugated to polyacrylamide gel. Peptides (green, FITC) and cells (red, mCherry) are shown in relation to each other, superimposed over a scaled diagram of the polyacrylamide gel (top). A three-dimensional isometric confocal image illustrates the even distribution of peptides throughout the gel (bottom). (D) Peptide functionalized substrates were found to have equivalent peptide densities on soft and stiff gels, with differences that vary with the amount of applied peptide. All studies were performed at the highest degree of peptide functionalization, 0.1 mM. SEM are shown.
3.3 NP cell attachment to peptide functionalized substrates
Primary human NP cells were analyzed for biosynthesis, morphology, and NP-specific marker expression after 4 days of culture on peptide-functionalized PAAm. NP cells had generally high levels of cell attachment to all peptide-conjugated substrates, soft or stiff, with the exception of P4 and AG10 upon stiff PAAm when compared against stiff LM-111 (p < 0.05, ANOVA, Dunnett's post-hoc). Less than 20% of NP cells were able to attach to the integrin α3 binding peptide, P4, and less than 50% of NP cells attached to AG10 under these circumstances. This result suggests that NP cells had an affinity for all peptide-conjugated surfaces, except these two, at levels nearly matched to that of stiff LM-111 conjugated substrates.
All peptide-conjugated substrates led to increases in the numbers of cells forming multi-cell clusters, as compared to stiff LM-111 conjugated substrates (Figure 3A, p < 0.05, Dunnett's post-hoc test). Cells generally attached as rounded and did not exhibit rapid spreading (i.e., spreading in first 24-48 hours) even when not in clusters (Figure 3B). Further work is needed to determine if this observation is due to substrate softness, peptide density, or duration of culture. NP cells on all peptide-conjugated substrates produced higher sGAG as compared to stiff surfaces functionalized with LM-111 (Figure 3C), a finding that was statistically significant for soft surfaces conjugated with the integrin-binding peptides p678 and AG10 only. While soft substrates were previously shown to promote increased cell clustering and sGAG synthesis when functionalized with full-length LM-111, there was no apparent correspondence between this clustering behavior and sGAG synthesis for the p678 and AG10 functionalized surfaces. In general, the correspondence between cell cluster formation and sGAG synthesis could not be confirmed for any of the studied peptides.
Figure 3.
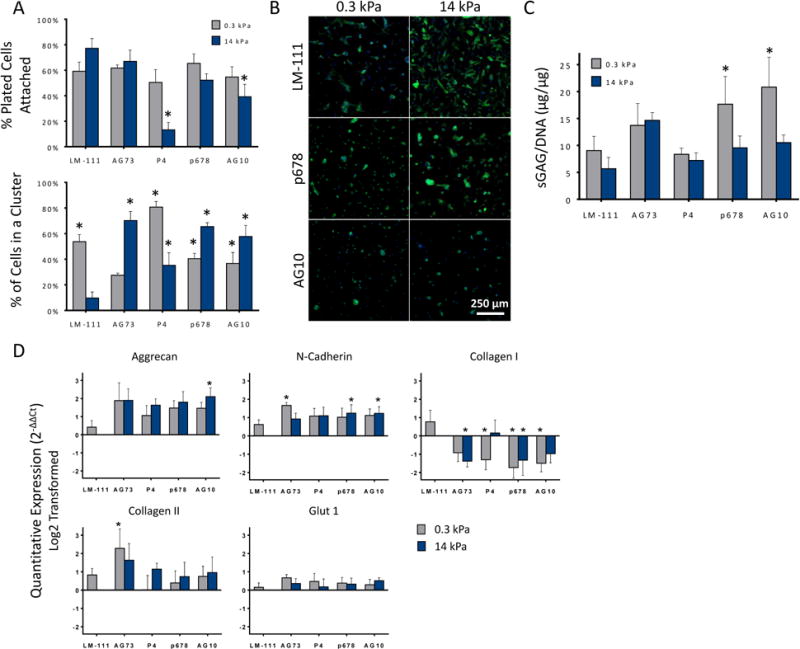
(A) Primary human NP cells attached to both soft and stiff peptide functionalized acrylamide gels. All peptide functionalized gels promoted greater cell clustering than for stiff LM-111 functionalized substrates (* p<0.05, significantly different from stiff LM-111 functionalized gels) SEM are shown. (B) Representative day 4 images showing the effect of substrate stiffness and ligand for NP cell morphologies. A majority of attached cells interact to form rounded, multi-cell clusters, in comparison to cells on the stiff (14 kPa) LM-111 control showing flattened morphology with stress fiber formation (scale bar = 250 μm). (C) NP cell production of sGAG per DNA varied across peptide-functionalized substrates. As found previously, sGAG/DNA for NP cells upon soft LM-111-functionalized substrates was greater (1.6-fold) than for stiff substrates. While similar elevations in sGAG/DNA were observed for many peptide-functionalized gels, only soft gels functionalized with P678 and AG10 were associated with higher sGAG/DNA (* p<0.05, significantly greater than stiff LM-111, 2-way ANOVA, Dunnett's post hoc). SEM are shown. (D) mRNA levels for aggrecan (AGC), N-cadherin (CDH2), type I collagen (COLA1), type II collagen (COL2A1), and GLUT-1 in primary human NP cells. (*p<0.05, significantly different from stiff LM-111 functionalized polyacrylamide gels, Dunnett's post hoc). SEM are shown.
Substrate stiffness and peptide presentation regulated gene expression for key markers of the NP phenotype. Aggrecan, N-cadherin, types I and II collagen, but not GLUT-1, indicated notable differences on peptide-functionalized substrates compared to the full-length laminin (Figure 3D). The integrin-binding peptides, P4, P678 and AG10, yielded comparatively similar effects on mRNA for these targets independent of soft or stiff substrate, with values that were higher than LM-111 functionalized stiff substrates. Only the syndecan-mediated binding peptide, AG73, yielded a stiffness-dependent behavior for NP cells with higher levels of mRNA for N-cadherin and type II collagen on soft substrates. An interesting observation was the lower type I collagen mRNA levels for NP cells upon any peptide-functionalized substrate as compared to LM-111 upon stiff substrates (p<0.05, ANOVA). Type I collagen expression is commonly related to fibroblastic phenotypes that may be associated with the more spread NP cells upon the stiff LM-111 substrate.
4. Discussion
The findings of this study suggest NP cell interactions with ECM-mimetic peptides likely occur beyond the common binding sequence RGD. Of particular relevance is our finding illustrating that human NP cells will attach to many laminin-derived peptides as well as the syndecan binding partner, AG73, at levels nearly equivalent to that of the full-length laminins. Both LM-511 and LM-111 were included as controls based on previous studies showing NP cell attachment is promoted on these coatings compared to other ECM-mimetic ligands [13]. Peptides GD6, P4, p678, and cNGQGEQ were selected for their ability to engage integrin α3 [32, 35, 44, 45] and contribute to attachment and spreading of keratinocytes, melanoma cells, or carcinoma cells [32, 34-36, 40, 44, 45]. Peptides AG32, AG10, and T1 have been shown to engage integrin α6. AG10 and AG32 are from the LG1 and LG2 domains of the laminin α1 chain, respectively, and have been shown to promote cell attachment, spreading, tumor invasion, and to maintain the stem-ness of precursor cells [27, 29, 38]. The peptide T1 is derived from the full-length protein angiogenesis inducer T1, and has been shown to promote attachment of embryonic stem cells (human and mouse), human foreskin fibroblasts, and human umbilical vein endothelial cells [31, 46, 47]. Peptide T1 has additionally been shown to support early mesodermal differentiation of human embryonic stem cells when presented in conjunction with RGD [46], as well as maintain stem-ness of murine embryonic stem cells in conjunction with ligands for integrins α5β1, αVβ5, and α9β1 [31]. Peptide IKVAV is one of the most commonly studied laminin-derived peptides and has been shown to promote cell adhesion (melanoma cells, adipose derived stem cells, astrocytes, fibroblasts, endothelial cells), and neurite outgrowth [33, 39, 48, 49]. The peptide RGD was chosen for its demonstrated ability to mediate NP cell interactions and widespread use in biomedical engineering to increase cell attachment of multiple cell types [16, 18, 50-52]. Peptide AG73 (RKRLQVQLSIRT, derived from the α1 chain of mouse laminin) is the only non-integrin binding peptide that was studied here. AG73 has been shown to elicit a variety of cell behaviors by binding to syndecan cell-surface receptors [25, 27, 53-56]. While not widely studied in the NP [57, 58], syndecans have been shown to cooperatively bind with different integrin subunits on many cell types to differentially modulate cell behaviors, including cell spreading, focal adhesion formation and organization [37]. Furthermore, an interesting observation was that binding to RGD and the integrin α3-mediated ligand, cNGQ, were notably lower than binding to laminin-coated substrates or any other peptide coated substrate. It was not surprising that higher cell attachment was observed on laminin-coated substrates, as earlier studies indicated NP cells prefer laminins over other ECM proteins like fibronectin and collagen [13, 21]. Nevertheless, a prior study has suggested that cells of the intervertebral disc may rely upon RGD mediated interactions to sense mechanical stimuli so further studies of the ligand will be important in understanding cell-matrix interactions [7]. When conjugated to controlled PAAm hydrogels, cells attached as both single cells and in multi-cell clusters to P4, P678, AG10 or AG73 conjugated substrates, with a demonstrated ability to synthesize matrix at levels largely equivalent to that of full-length laminin. The healthy, biosynthetically active NP cell - typical of the non-degenerate intervertebral disc - may be characterized as having high sGAG synthesis, a higher ratio of type II to type I collagen synthesis, and elevated N-cadherin and GLUT-1 protein or mRNA levels [2]. Both the peptide sequence and the substrate stiffness have a significant influence on these phenotypic features of the human NP cells, as well as cell attachment numbers and cell cluster formation, as illustrated by heat maps in Figure 4. Importantly, we found that soft PAAm substrates conjugated with AG73 or the integrin binding peptides P678 and AG10 can reproduce many of the features associated with the healthy, biosynthetically active cell phenotype at the mRNA level. Additional work would be required to verify these differential effects on protein levels measured via ELISA or other methods.
Figure 4.
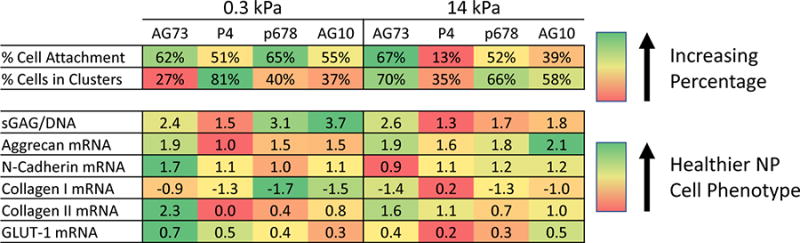
Heat map illustrating average values within each variable for each peptide functionalized substrates. Green denotes values closer to those recognized as healthy NP-specific. Each row demonstrates the best condition for each parameter, but is not compared to different items (i.e. other rows). As an example, NP cells with higher sGAG synthesis and lower COL1A1 mRNA may be produced by periods of culture upon soft substrates functionalized with P678 or AG10.
NP cell cluster formation is believed to be key to maintenance of the healthy, juvenile phenotype and has been previously shown to relate to the stiffness of the gel substrate [20, 21]. It is important that the peptide-functionalized surfaces promoted many behaviors for NP cells, independent of their ability to drive cell cluster formation. But it must be noted that the observed differences in cell clusters is not necessarily, nor likely, due to differences in proliferation. Based on earlier unpublished data and one published study [59], the duration of these experiments were not long enough to allow for proliferation of primary NP cells. With the exception of P4, peptides show increased cell cluster formation on stiff versus soft when compared to the full length laminin, which may be due to peptide density or spacing where a more controlled ligand presentation is able to overcome the stiffness-mediated effects on cell cluster formation. Previously, NP cells upon laminin-functionalized PEG crosslinked hydrogels [60] or laminin-functionalized PAAm exhibited elevated sGAG biosynthesis and markers of the healthy NP cell phenotype only when associated with cadherin-mediated cell cluster formation. Here, a majority of NP cells did attach as cell clusters, most notably to surfaces conjugated with P4, P678 or AG73. However, their ability to express healthy NP markers was apparently independent of cell cluster formation. There are several possibilities for this observation. Ligand density is certainly a critical parameter for modulating cell attachment [18, 61-64] with prior studies showing that 6 peptides/μm2 and 60 peptides/μm2 are required for cell spreading and formation of focal adhesions in fibroblasts, respectively [65]. Additional work has shown that co-presentation of syndecans may be necessary to promote focal adhesion formation in select cell types [66, 67]. For peptides conjugated to non-fouling polymers [68], cell attachment was promoted with increasing RGD density for endothelial cells up to 9,000 peptides/μm2 with evidence of ERK phosphorylation at 1,200 peptides/μm2. While NP cells are neither fibroblasts nor endothelial cells, our use of peptide densities exceeding 1,100 peptides/μm2 are consistent with values that may not optimize cell spreading (for RGD), but still promote cell signaling events upon attachment. This is consistent with the attachment of cells not as single, spread cells upon these substrates, and also with demonstrated effects on promoting cellular biosynthesis and gene expression. Given the absence of a broad variation in peptide density, which we sought to control with our PAAm gel system, and consequent absence of a broad variation in cell morphology, it is likely that phenotypic differences were obscured between spread cells upon stiff substrates and rounded, clustered cells upon soft substrates. Additional work would be required to verify an ability for increasing peptide density to regulate cell spreading for NP cells upon any substrate, soft or stiff.
In summary, many of the ECM-mimetic peptide conjugated substrates were able to reproduce behaviors in NP cells observed when attached to soft, full-length laminin substrates. Because the ECM peptides used in this study are derived from integrin binding domains, we expect cells are binding directly to the peptides and not through any secondary mechanisms to ECM molecules. These findings show that cell-matrix interactions with specific ECM protein sequences may play a critical role in the influencing the NP phenotype during development, maturation, and degeneration. In this regard, controlled presentation of specific ECM-mimetic peptides may provide a means of inducing a regenerative response or phenotype in NP cells. It is important to note that here, only a subset of individual ECM peptide sequences were screened, and only those derived from laminin or syndecan. Some peptide ligands can act synergistically when co-presented at a surface, as has been repeatedly shown for AG73 that will enhance and promote cell spreading in multiple cell types when also coupled with an integrin-binding peptide [69, 70]. With a clear idea of target values for the healthy, biosynthetically active NP cell, this work suggests that surfaces can be designed to exploit both integrin and non-integrin binding mechanisms for NP cells, in order to modulate the phenotype, morphology and biosynthesis towards targeted and healthy values.
Significance.
NP cells reside in a laminin-rich environment that deteriorates with age, including a loss of water content and changes in the extracellular matrix (ECM) structure that may lead to the development of a degenerated IVD. There is great interest in methods to re-express healthy, biosynthetically active NP cells using laminin-derived biomimetic peptides toward the goal of using autologous cell sources for tissue regeneration. Here, we describe a novel study utilizing several laminin mimetic peptides conjugated to polyacrylamide gels that are able to support an immature, healthy NP phenotype after culture on “soft” peptide gels. These findings can support future studies in tissue regeneration where cells may be directed to a desired regenerative phenotype using niche-specific ECM peptides.
Acknowledgments
Work was supported by the NIH (AR047442, EB002263, AR069588, AG015768, AR057410) and Arthritis Foundation.
Footnotes
Disclosures: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sakai D, Nakamura Y, Nakai T, Mishima T, Kato S, Grad S, Alini M, Risbud MV, Chan D, Cheah KS, Yamamura K, Masuda K, Okano H, Ando K, Mochida J. Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nature communications. 2012;3:1264. doi: 10.1038/ncomms2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Risbud MV, Schoepflin ZR, Mwale F, Kandel RA, Grad S, Iatridis JC, Sakai D, Hoyland JA. Defining the phenotype of young healthy nucleus pulposus cells: recommendations of the Spine Research Interest Group at the 2014 annual ORS meeting. J Orthop Res. 2015;33(3):283–93. doi: 10.1002/jor.22789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi KS, Harfe BD. Hedgehog signaling is required for formation of the notochord sheath and patterning of nuclei pulposi within the intervertebral discs. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(23):9484–9. doi: 10.1073/pnas.1007566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tran CM, Schoepflin ZR, Markova DZ, Kepler CK, Anderson DG, Shapiro IM, Risbud MV. CCN2 suppresses catabolic effects of interleukin-1beta through alpha5beta1 and alphaVbeta3 integrins in nucleus pulposus cells: implications in intervertebral disc degeneration. The Journal of biological chemistry. 2014;289(11):7374–87. doi: 10.1074/jbc.M113.526111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Risbud MV, Guttapalli A, Albert TJ, Shapiro IM. Hypoxia activates MAPK activity in rat nucleus pulposus cells: regulation of integrin expression and cell survival. Spine (Phila Pa 1976) 2005;30(22):2503–9. doi: 10.1097/01.brs.0000186326.82747.13. [DOI] [PubMed] [Google Scholar]
- 6.Nettles DL, Richardson WJ, Setton LA. Integrin expression in cells of the intervertebral disc. J Anat. 2004;204(6):515–20. doi: 10.1111/j.0021-8782.2004.00306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Maitre CL, Frain J, Millward-Sadler J, Fotheringham AP, Freemont AJ, Hoyland JA. Altered integrin mechanotransduction in human nucleus pulposus cells derived from degenerated discs. Arthritis and rheumatism. 2009;60(2):460–9. doi: 10.1002/art.24248. [DOI] [PubMed] [Google Scholar]
- 8.Gilchrist CL, Chen J, Richardson WJ, Loeser RF, Setton LA. Functional integrin subunits regulating cell-matrix interactions in the intervertebral disc. J Orthop Res. 2007;25(6):829–40. doi: 10.1002/jor.20343. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Jing L, Gilchrist CL, Richardson WJ, Fitch RD, Setton LA. Expression of laminin isoforms, receptors, and binding proteins unique to nucleus pulposus cells of immature intervertebral disc. Connect Tissue Res. 2009;50(5):294–306. [PMC free article] [PubMed] [Google Scholar]
- 10.Bridgen DT, Gilchrist CL, Richardson WJ, Isaacs RE, Brown CR, Yang KL, Chen J, Setton LA. Integrin-mediated interactions with extracellular matrix proteins for nucleus pulposus cells of the human intervertebral disc. J Orthop Res. 2013;31(10):1661–7. doi: 10.1002/jor.22395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia M, Zhu Y. Fibronectin fragment activation of ERK increasing integrin alpha(5) and beta(1) subunit expression to degenerate nucleus pulposus cells. J Orthop Res. 2011;29(4):556–61. doi: 10.1002/jor.21273. [DOI] [PubMed] [Google Scholar]
- 12.Trout JJ, Buckwalter JA, Moore KC, Landas SK. Ultrastructure of the human intervertebral disc. I Changes in notochordal cells with age. Tissue & cell. 1982;14(2):359–69. doi: 10.1016/0040-8166(82)90033-7. [DOI] [PubMed] [Google Scholar]
- 13.Gilchrist CL, Francisco AT, Plopper GE, Chen J, Setton LA. Nucleus pulposus cell-matrix interactions with laminins. Eur Cell Mater. 2011;21:523–32. doi: 10.22203/ecm.v021a39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nature biotechnology. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 15.Pierschbacher MD, Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309(5963):30–3. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 16.Ruoslahti E. RGD and other recognition sequences for integrins. Annual review of cell and developmental biology. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 17.Mardilovich A, Kokkoli E. Biomimetic peptide-amphiphiles for functional biomaterials: the role of GRGDSP and PHSRN. Biomacromolecules. 2004;5(3):950–7. doi: 10.1021/bm0344351. [DOI] [PubMed] [Google Scholar]
- 18.Hersel U, Dahmen C, Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24(24):4385–415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 19.Hwang PY, Jing L, Michael KW, Richardson WJ, Chen J, Setton LA. N-Cadherin-Mediated Signaling Regulates Cell Phenotype for Nucleus Pulposus Cells of the Intervertebral Disc. Cell Mol Bioeng. 2015;8(1):51–62. doi: 10.1007/s12195-014-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang PY, Chen J, Jing L, Hoffman BD, Setton LA. The role of extracellular matrix elasticity and composition in regulating the nucleus pulposus cell phenotype in the intervertebral disc: a narrative review. Journal of biomechanical engineering. 2014;136(2):021010. doi: 10.1115/1.4026360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilchrist CL, Darling EM, Chen J, Setton LA. Extracellular matrix ligand and stiffness modulate immature nucleus pulposus cell-cell interactions. PloS one. 2011;6(11):e27170. doi: 10.1371/journal.pone.0027170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francisco AT, Mancino RJ, Bowles RD, Brunger JM, Tainter DM, Chen YT, Richardson WJ, Guilak F, Setton LA. Injectable laminin-functionalized hydrogel for nucleus pulposus regeneration. Biomaterials. 2013;34(30):7381–8. doi: 10.1016/j.biomaterials.2013.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francisco AT, Hwang PY, Jeong CG, Jing L, Chen J, Setton LA. Photocrosslinkable laminin-functionalized polyethylene glycol hydrogel for intervertebral disc regeneration. Acta biomaterialia. 2014;10(3):1102–11. doi: 10.1016/j.actbio.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki N, Yokoyama F, Nomizu M. Functional sites in the laminin alpha chains. Connect Tissue Res. 2005;46(3):142–52. doi: 10.1080/03008200591008527. [DOI] [PubMed] [Google Scholar]
- 25.Richard BL, Nomizu M, Yamada Y, Kleinman HK. Identification of synthetic peptides derived from laminin alpha 1 and alpha 2 chains with cell type specificity for neurite outgrowth. Experimental cell research. 1996;228(1):98–105. doi: 10.1006/excr.1996.0304. [DOI] [PubMed] [Google Scholar]
- 26.Nomizu M, Kuratomi Y, Song SY, Ponce ML, Hoffman MP, Powell SK, Miyoshi K, Otaka A, Kleinman HK, Yamada Y. Identification of cell binding sequences in mouse laminin gamma1 chain by systematic peptide screening. The Journal of biological chemistry. 1997;272(51):32198–205. doi: 10.1074/jbc.272.51.32198. [DOI] [PubMed] [Google Scholar]
- 27.Nomizu M, Kim WH, Yamamura K, Utani A, Song SY, Otaka A, Roller PP, Kleinman HK, Yamada Y. Identification of cell binding sites in the laminin alpha 1 chain carboxyl-terminal globular domain by systematic screening of synthetic peptides. The Journal of biological chemistry. 1995;270(35):20583–90. doi: 10.1074/jbc.270.35.20583. [DOI] [PubMed] [Google Scholar]
- 28.Tibbitt MW, Anseth KS. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng. 2009;103(4):655–63. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng Y, Eshghi S, Li YJ, Schmidt R, Schaffer DV, Healy KE. Characterization of integrin engagement during defined human embryonic stem cell culture. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24(4):1056–65. doi: 10.1096/fj.08-126821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mann BK, Tsai AT, Scott-Burden T, West JL. Modification of surfaces with cell adhesion peptides alters extracellular matrix deposition. Biomaterials. 1999;20(23-24):2281–6. doi: 10.1016/s0142-9612(99)00158-1. [DOI] [PubMed] [Google Scholar]
- 31.Lee ST, Yun JI, Jo YS, Mochizuki M, van der Vlies AJ, Kontos S, Ihm JE, Lim JM, Hubbell JA. Engineering integrin signaling for promoting embryonic stem cell self-renewal in a precisely defined niche. Biomaterials. 2010;31(6):1219–26. doi: 10.1016/j.biomaterials.2009.10.054. [DOI] [PubMed] [Google Scholar]
- 32.Calzada MJ, Roberts DD. Novel integrin antagonists derived from thrombospondins. Current pharmaceutical design. 2005;11(7):849–66. doi: 10.2174/1381612053381792. [DOI] [PubMed] [Google Scholar]
- 33.Grant DS, Kinsella JL, Fridman R, Auerbach R, Piasecki BA, Yamada Y, Zain M, Kleinman HK. Interaction of endothelial cells with a laminin A chain peptide (SIKVAV) in vitro and induction of angiogenic behavior in vivo. Journal of cellular physiology. 1992;153(3):614–25. doi: 10.1002/jcp.1041530324. [DOI] [PubMed] [Google Scholar]
- 34.Guo N, Templeton NS, Al-Barazi H, Cashel JA, Sipes JM, Krutzsch HC, Roberts DD. Thrombospondin-1 promotes alpha3beta1 integrin-mediated adhesion and neurite-like outgrowth and inhibits proliferation of small cell lung carcinoma cells. Cancer research. 2000;60(2):457–66. [PubMed] [Google Scholar]
- 35.Kim JM, Park WH, Min BM. The PPFLMLLKGSTR motif in globular domain 3 of the human laminin-5 alpha3 chain is crucial for integrin alpha3beta1 binding and cell adhesion. Experimental cell research. 2005;304(1):317–27. doi: 10.1016/j.yexcr.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Krutzsch HC, Choe BJ, Sipes JM, Guo N, Roberts DD. Identification of an alpha(3)beta(1) integrin recognition sequence in thrombospondin-1. The Journal of biological chemistry. 1999;274(34):24080–6. doi: 10.1074/jbc.274.34.24080. [DOI] [PubMed] [Google Scholar]
- 37.Morgan MR, Humphries MJ, Bass MD. Synergistic control of cell adhesion by integrins and syndecans, Nature reviews. Molecular cell biology. 2007;8(12):957–69. doi: 10.1038/nrm2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakahara H, Nomizu M, Akiyama SK, Yamada Y, Yeh Y, Chen WT. A mechanism for regulation of melanoma invasion. Ligation of alpha6beta1 integrin by laminin G peptides. The Journal of biological chemistry. 1996;271(44):27221–4. doi: 10.1074/jbc.271.44.27221. [DOI] [PubMed] [Google Scholar]
- 39.Tashiro K, Sephel GC, Weeks B, Sasaki M, Martin GR, Kleinman HK, Yamada Y. A synthetic peptide containing the IKVAV sequence from the A chain of laminin mediates cell attachment, migration, and neurite outgrowth. The Journal of biological chemistry. 1989;264(27):16174–82. [PubMed] [Google Scholar]
- 40.Wilke MS, Skubitz AP. Human keratinocytes adhere to multiple distinct peptide sequences of laminin. The Journal of investigative dermatology. 1991;97(1):141–6. doi: 10.1111/1523-1747.ep12479311. [DOI] [PubMed] [Google Scholar]
- 41.Reinhart-King CA, Dembo M, Hammer DA. Endothelial cell traction forces on RGD-derivatized polyacrylamide substrata. Journal of cell science. 2003;113(10):1677–1686. [Google Scholar]
- 42.Darling EM, Wilusz RE, Bolognesi MP, Zauscher S, Guilak F. Spatial mapping of the biomechanical properties of the pericellular matrix of articular cartilage measured in situ via atomic force microscopy. Biophysical journal. 2010;98(12):2848–56. doi: 10.1016/j.bpj.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Substrate compliance versus ligand density in cell on gel responses. Biophysical journal. 2004;86(1 Pt 1):617–28. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lau D, Guo L, Liu R, Marik J, Lam K. Peptide ligands targeting integrin alpha3beta1 in non-small cell lung cancer. Lung Cancer. 2006;52(3):291–297. doi: 10.1016/j.lungcan.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 45.Gehlsen KR, Sriramarao P, Furcht LT, Skubitz AP. A synthetic peptide derived from the carboxy terminus of the laminin A chain represents a binding site for the alpha 3 beta 1 integrin. The Journal of cell biology. 1992;117(2):449–59. doi: 10.1083/jcb.117.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Wang X, Kaufman DS, Shen W. A synthetic substrate to support early mesodermal differentiation of human embryonic stem cells. Biomaterials. 2011;32(32):8058–66. doi: 10.1016/j.biomaterials.2011.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leu SJ, Liu Y, Chen N, Chen CC, Lam SC, Lau LF. Identification of a novel integrin alpha 6 beta 1 binding site in the angiogenic inducer CCN1 (CYR61) The Journal of biological chemistry. 2003;278(36):33801–8. doi: 10.1074/jbc.M305862200. [DOI] [PubMed] [Google Scholar]
- 48.Santiago LY, Nowak RW, Peter Rubin J, Marra KG. Peptide-surface modification of poly(caprolactone) with laminin-derived sequences for adipose-derived stem cell applications. Biomaterials. 2006;27(15):2962–9. doi: 10.1016/j.biomaterials.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 49.Kam L, Shain W, Turner JN, Bizios R. Selective adhesion of astrocytes to surfaces modified with immobilized peptides. Biomaterials. 2002;23(2):511–5. doi: 10.1016/s0142-9612(01)00133-8. [DOI] [PubMed] [Google Scholar]
- 50.Matsuda T, Kondo A, Makino K, Akutsu T. Development of a novel artificial matrix with cell adhesion peptides for cell culture and artificial and hybrid organs. ASAIO Trans. 1989;35(3):677–9. doi: 10.1097/00002480-198907000-00165. [DOI] [PubMed] [Google Scholar]
- 51.Massia SP, Hubbell JA. Covalent surface immobilization of Arg-Gly-Asp- and Tyr-Ile-Gly-Ser-Arg-containing peptides to obtain well-defined cell-adhesive substrates. Analytical biochemistry. 1990;187(2):292–301. doi: 10.1016/0003-2697(90)90459-m. [DOI] [PubMed] [Google Scholar]
- 52.Hubbell JA, Massia SP, Drumheller PD. Surface-grafted cell-binding peptides in tissue engineering of the vascular graft. Annals of the New York Academy of Sciences. 1992;665:253–8. doi: 10.1111/j.1749-6632.1992.tb42589.x. [DOI] [PubMed] [Google Scholar]
- 53.Weeks BS, Nomizu M, Ramchandran RS, Yamada Y, Kleinman HK. Laminin-1 and the RKRLQVQLSIRT laminin-1 alpha1 globular domain peptide stimulate matrix metalloproteinase secretion by PC12 cells. Experimental cell research. 1998;243(2):375–82. doi: 10.1006/excr.1998.4157. [DOI] [PubMed] [Google Scholar]
- 54.Mochizuki M, Philp D, Hozumi K, Suzuki N, Yamada Y, Kleinman HK, Nomizu M. Angiogenic activity of syndecan-binding laminin peptide AG73 (RKRLQVQLSIRT) Archives of biochemistry and biophysics. 2007;459(2):249–55. doi: 10.1016/j.abb.2006.12.026. [DOI] [PubMed] [Google Scholar]
- 55.Hoffman MP, Nomizu M, Roque E, Lee S, Jung DW, Yamada Y, Kleinman HK. Laminin-1 and laminin-2 G-domain synthetic peptides bind syndecan-1 and are involved in acinar formation of a human submandibular gland cell line. The Journal of biological chemistry. 1998;273(44):28633–41. doi: 10.1074/jbc.273.44.28633. [DOI] [PubMed] [Google Scholar]
- 56.Hoffman MP, Engbring JA, Nielsen PK, Vargas J, Steinberg Z, Karmand AJ, Nomizu M, Yamada Y, Kleinman HK. Cell type-specific differences in glycosaminoglycans modulate the biological activity of a heparin-binding peptide (RKRLQVQLSIRT) from the G domain of the laminin alpha1 chain. The Journal of biological chemistry. 2001;276(25):22077–85. doi: 10.1074/jbc.M100774200. [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Markova D, Anderson DG, Zheng Z, Shapiro IM, Risbud MV. TNF-alpha and IL-1beta promote a disintegrin-like and metalloprotease with thrombospondin type I motif-5-mediated aggrecan degradation through syndecan-4 in intervertebral disc. The Journal of biological chemistry. 2011;286(46):39738–49. doi: 10.1074/jbc.M111.264549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beckett MC, Ralphs JR, Caterson B, Hayes AJ. The transmembrane heparan sulphate proteoglycan syndecan-4 is involved in establishment of the lamellar structure of the annulus fibrosus of the intervertebral disc. Eur Cell Mater. 2015;30:69–88. doi: 10.22203/ecm.v030a06. discussion 88. [DOI] [PubMed] [Google Scholar]
- 59.Liu MC, Chen WH, Wu LC, Hsu WC, Lo WC, Yeh SD, Wang MF, Zeng R, Deng WP. Establishment of a promising human nucleus pulposus cell line for intervertebral disc tissue engineering. Tissue Eng Part C Methods. 2014;20(1):1–10. doi: 10.1089/ten.TEC.2013.0048. [DOI] [PubMed] [Google Scholar]
- 60.Hwang PY, Jing L, Chen J, Lim FL, Tang R, Choi H, Cheung KM, Risbud MV, Gersbach CA, Guilak F, Leung VY, Setton LA. N-cadherin is Key to Expression of the Nucleus Pulposus Cell Phenotype under Selective Substrate Culture Conditions. Sci Rep. 2016;6:28038. doi: 10.1038/srep28038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ward MD, Hammer DA. Focal contact assembly through cytoskeletal polymerization: steady state analysis. J Math Biol. 1994;32(7):677–704. doi: 10.1007/BF00163022. [DOI] [PubMed] [Google Scholar]
- 62.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. Journal of cell science. 2000;113(Pt 10):1677–86. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 63.Comisar WA, Kazmers NH, Mooney DJ, Linderman JJ. Engineering RGD nanopatterned hydrogels to control preosteoblast behavior: a combined computational and experimental approach. Biomaterials. 2007;28(30):4409–17. doi: 10.1016/j.biomaterials.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 64.Cavalcanti-Adam EA, Volberg T, Micoulet A, Kessler H, Geiger B, Spatz JP. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophysical journal. 2007;92(8):2964–74. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Massia SP, Hubbell JA. An RGD spacing of 440 nm is sufficient for integrin alpha V beta 3-mediated fibroblast spreading and 140 nm for focal contact and stress fiber formation. The Journal of cell biology. 1991;114(5):1089–100. doi: 10.1083/jcb.114.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woods A, Couchman JR. Couchman, Syndecan 4 heparan sulfate proteoglycan is a selectively enriched and widespread focal adhesion component. Molecular biology of the cell. 1994;5(2):183–92. doi: 10.1091/mbc.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woods A, McCarthy JB, Furcht LT, Couchman JR. A synthetic peptide from the COOH-terminal heparin-binding domain of fibronectin promotes focal adhesion formation. Molecular biology of the cell. 1993;4(6):605–13. doi: 10.1091/mbc.4.6.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patel S, Tsang J, Harbers GM, Healy KE, Li S. Regulation of endothelial cell function by GRGDSP peptide grafted on interpenetrating polymers. Journal of biomedical materials research Part A. 2007;83(2):423–33. doi: 10.1002/jbm.a.31320. [DOI] [PubMed] [Google Scholar]
- 69.Otagiri D, Yamada Y, Hozumi K, Katagiri F, Kikkawa Y, Nomizu M. Cell attachment and spreading activity of mixed laminin peptide-chitosan membranes. Biopolymers. 2013;100(6):751–9. doi: 10.1002/bip.22303. [DOI] [PubMed] [Google Scholar]
- 70.Hozumi K, Kobayashi K, Katagiri F, Kikkawa Y, Kadoya Y, Nomizu M. Syndecan- and integrin-binding peptides synergistically accelerate cell adhesion. FEBS Lett. 2010;584(15):3381–5. doi: 10.1016/j.febslet.2010.06.032. [DOI] [PubMed] [Google Scholar]
- 71.Min SK, Lee SC, Hong SD, Chung CP, Park WH, Min BM. The effect of a laminin-5-derived peptide coated onto chitin microfibers on re-epithelialization in early-stage wound healing. Biomaterials. 2010;31(17):4725–30. doi: 10.1016/j.biomaterials.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 72.Yamada Y, Hozumi K, Aso A, Hotta A, Toma K, Katagiri F, Kikkawa Y, Nomizu M. Laminin active peptide/agarose matrices as multifunctional biomaterials for tissue engineering. Biomaterials. 2012;33(16):4118–25. doi: 10.1016/j.biomaterials.2012.02.044. [DOI] [PubMed] [Google Scholar]


