Abstract
Poorly organized tumour vasculature often results in areas of limited nutrient supply and hypoxia. Despite our understanding of solid tumour responses to hypoxia, how nutrient deprivation regionally affects tumour growth and therapeutic response is poorly understood. Here, we show the core region of solid tumours displayed glutamine deficiency compared to other amino acids. Low glutamine in tumour core regions led to dramatic histone hyper-methylation due to decreased α-ketoglutarate levels, a key cofactor for the Jumonji-domain containing (JmjC) histone demethylases (JHDMs). Using patient-derived V600EBRAF melanoma cells, we found that low glutamine-induced histone hyper-methylation resulted in cancer cell de-differentiation and resistance to BRAF inhibitor treatment, which was largely mediated by methylation on H3K27, as knockdown of the H3K27-specific demethylase KDM6B and methyltransferase EZH2 respectively reproduced and attenuated the low glutamine effects in vitro and in vivo. Thus, intra-tumoural regional variation in the nutritional microenvironment contributes to tumour heterogeneity and therapeutic response.
INTRODUCTION
Glutamine is one of the major carbon and nitrogen sources to support cancer cell survival and proliferation1. Glutamine catabolism is required to maintain pools of TCA cycle intermediates. For example, glutamine can be converted by glutaminase (GLS) to glutamate, which can be further converted to α-ketoglutarate in the TCA cycle2. Similar to glucose metabolism, increased glutamine uptake is controlled by oncogenes, such as c-MYC and KRAS3–5. On the other hand, as tumours grow, increased glutamine catabolism may deplete the local supply and lead to periods of glutamine deprivation. This is supported by in vivo studies wherein numerous tumours, including hepatomas and sarcomas, glutamine falls to almost undetectable levels relative to normal tissues6. A recent study using metabolomics analysis comparing paired pancreatic tumour patient samples with benign adjacent tissue specimens revealed that glutamine is one of the most strongly depleted metabolites in tumours7. However, how low glutamine levels in solid tumours affects tumour growth and therapeutic response remains largely unknown.
Reversible histone lysine methylation controlled by a variety of histone methyl-transferases (HMTs) and histone demethylases (HDMs) modulates chromatin structure and thereby contributes to a variety of cellular processes such as transcription, replication and repair8. A subset of HDMs named Jumonji-domain containing HDM (JHDM) utilizes α-ketoglutarate (αKG), oxygen and Fe (II) as cofactors and releases succinate and formaldehyde as by-products9. The requirement for αKG in mediating histone demethylase activity implied a potential interplay between metabolism and epigenetic modification. For example, a recent report demonstrated that glutamine deprivation in ES cells leads to increased methylation on H3K27me3 and H3K9me310. In addition, mutant forms of the metabolic enzymes isocitrate dehydrogenase 1 (IDH1) and IDH2, which display neomorphic functions by producing 2-hydroxyglutarate (2HG) from αKG11, can increase histone methylation and block tumour cell differentiation via inhibiting JHDM activity, as 2HG is a structural analog of αKG12. However, although αKG is a metabolite generated from glutamine, whether low glutamine levels in solid tumours affect the activity of these αKG-dependent JHDM and hence modulate histone methylation remains unclear.
In this study, we measured the extent of glutamine heterogeneity regionally within solid tumours, dissected the impact of low glutamine on tumour cell de-differentiation and drug sensitivity, and mechanistically linked specific histone demethylation to the impact of microenvironmental glutamine levels on tumour cell plasticity. Our study provides important evidence that regional glutamine deficiency leads to de-differentiation and drug resistance via inhibition of histone demethylation.
RESULTS
Tumour core regions display low glutamine levels and hyper-methylation of histone H3
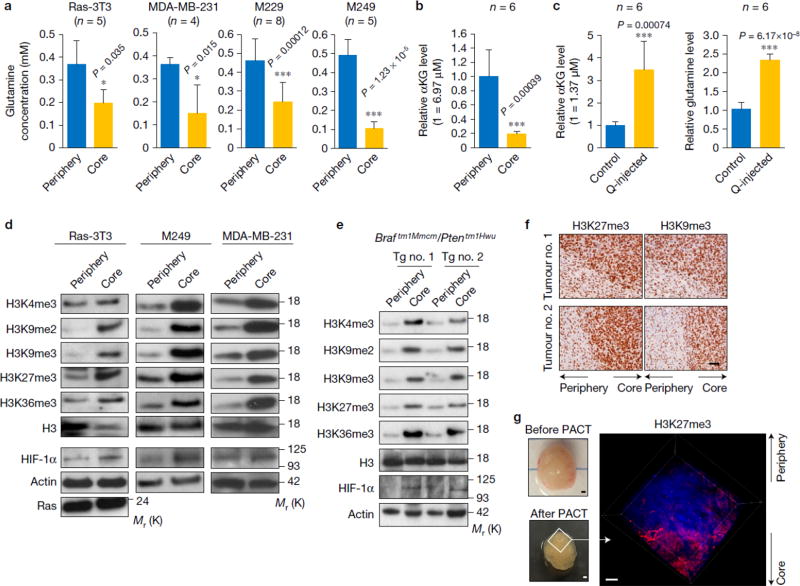
To assess potential differences in glutamine levels in the periphery vs. core region of tumours, we dissected different tumour xenografts and measured the glutamine concentrations in these distinct intra-tumoural regions (Supplementary Fig. 1a). We found that glutamine concentrations were consistently and significantly lower in the tumour core regions compared to the tumour periphery (Fig. 1a). We next measured the αKG level in the tumour tissues by liquid chromatography mass spectrometry (LC-MS) analysis. As with glutamine, compared to the periphery regions, αKG levels were significantly decreased in the tumour cores (Fig.1b). Moreover, αKG level was largely restored by injecting glutamine into the tumour cores, suggesting glutamine is sufficient to maintain tumour αKG levels (Fig. 1c). Since αKG is an essential co-factor for the JHDMs, we tested whether low glutamine levels in the tumour core regions correlated with differential histone methylation levels. At all the histone H3 methylation sites, which are the targets of JHDMs13–15, histone methylation levels dramatically increased in the core region (Fig. 1d). HIF-1α was used as a “marker” of the core tissues as it is induced in the core region of solid tumours due to hypoxia16. In addition, KRAS expression was used to distinguish tumour tissues from tumour adjacent normal tissues. We further examined histone methylation using melanoma tumours spontaneously developed from mice carrying a Braftm1Mmcm/Ptentm1Hwu allele17. Similarly, increased histone methylation was found in the core regions compared to the periphery in tumours from transgenic mice (Fig. 1e). In addition, histone hyper-methylation in the tumour core regions was also confirmed by immunohistochemistry staining using antibody against H3K27me3 or H3K9me3 (Fig. 1f). Moreover, we directly visualized H3K27me3 levels in intact tumours using the recently established technology for intact, slice-free, whole tissue imaging and phenotyping (PACT-defined as Passive CLARITY)18 (Supplementary Fig. 1b). In agreement with Western blot and IHC analysis, H3K27me3 staining significantly increased in the melanoma tumour core regions (Fig. 1g). Furthermore, HIF-1α and H3K27me3 staining strongly overlapped (Supplementary Fig. 1c). Thus, low glutamine and high histone methylation levels commonly co-exist in the core regions of tumours. Tumour cores often contain extensive dead cells. Despite the increased cell death (Supplementary Fig. 1d,e), there were still a certain amount of live cells in the tumour cores as cells in tumour cores could be stained by Ki67 antibody and displayed a negative staining of cleaved caspase-3 (Supplementary Fig. 1c,d).
Figure 1. Tumour core regions display low glutamine levels and hyper-methylation of histone H3.
(a) Tissues from periphery and core regions of xenograft tumours were homogenized and used for amino acids extraction. Glutamine concentration was determined by glutamine assay. Data represent mean ± S.D. from independent xenografts (Ras-3T3, n=5; MDA-MB-231, n=4; M229, n=8; M249, n=5). *P<.05, ***P<.001 by unpaired two-tailed Student’s t-test. (b) M229 xenograft tumours were harvested and separated into periphery and core samples. Metabolites were extracted from each sample and αKG concentration was measured by LC-MS. (Y axis 1=6.97µM αKG in tumour tissue.) Data represent mean ± S.D., n=6 tumours. ***P<.001 by unpaired two-tailed Student’s t-test. (c) 4mM glutamine or PBS control injected tumours were harvested. Metabolites were extracted from the core tissues, then αKG (left, Y axis 1=1.37µM αKG in tumour tissue) and glutamine (right) concentrations were measured by LC-MS. Data represent mean ± S.D., n=6 tumours. ***P<.001 by unpaired two-tailed Student’s t-test. Source data for a, b and c are shown in supplementary Table 4. (d) Tissues from periphery and core regions of xenograft tumours were used for histone extraction or lysed to collect whole cell lysate. Histone methylation levels and other proteins were assessed by western blotting with specified antibodies. Total histone H3 and Actin were used as loading controls. (e) Melanoma tumour tissues were harvested from transgenic mice. Histone methylation levels in different tumours were assessed by western blotting. Total histone H3 and Actin were used as loading controls. (f) Immunohistochemistry staining in xenograft tumours. Whole tumours were sliced and stained with antibodies against H3K9me3 and H3K27me3. Scale bar = 50 µm. (g) Photograph of the same M229 xenograft tumour before and after clearing (scale bar = 1.5 mm) (left), and 3D imaging of M229 xenograft tumour depicting DAPI nuclear stain (blue) and H3K27me3 antibody staining (red) (right). Scale bar = 300 µm. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
Low glutamine level leads to increased histone methylation in tumour cells
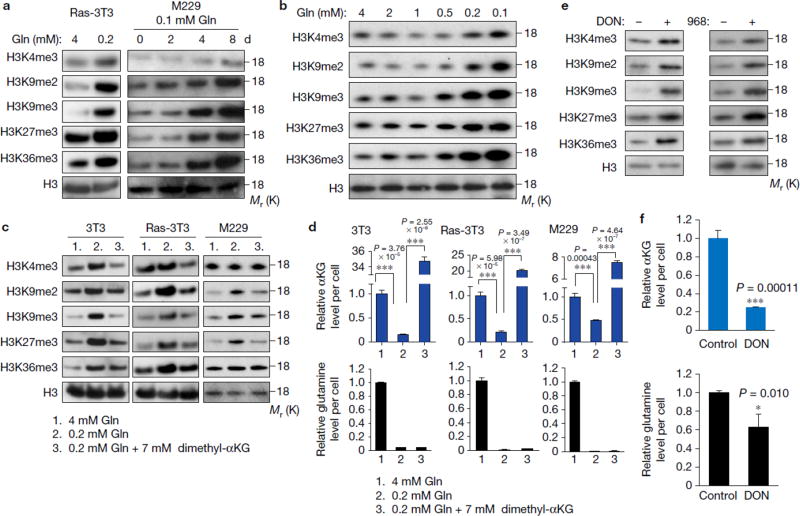
Next we determined if increased histone methylation in tumour core regions was induced by glutamine deficiency. We found that low glutamine was sufficient to induce histone hyper-methylation, particularly on K9, K27 and K36, in both a time and dose-dependent manner (Fig. 2a,b). Moreover, histone hyper-methylation required 0.2 mM for melanoma cells (Fig. 2b), which was similar to the glutamine concentration in tumour core regions (Fig. 1a). Furthermore, cell permeable dimethyl-αKG was sufficient to prevent low glutamine-induced histone methylation in all tumour cell types tested (Fig. 2c), suggesting that increased histone methylation induced by low glutamine levels resulted from low αKG. As a control, intracellular glutamine and αKG under these conditions were also measured (Fig. 2d). Furthermore, melanoma cells treated with glutaminase inhibitors L-DON and compound 96819, 20 also displayed increased histone methylation (Fig. 2e). As expected, DON treatment led to an 80% decreased αKG level (Fig. 2f). Interestingly, we found that the glutamine level decreased by 40% upon DON treatment. In addition, to understand whether cell proliferation rate had any effect on histone methylation, we examined H3 lysine methylation in confluent cells and observed no increase in histone methylation as compared to cells cultured in low glutamine (Supplementary Fig. 2a). Furthermore, cell number and cell survival were assayed under these conditions, but no correlation between cell proliferation and histone methylation was found (Supplementary Fig. 2b,c).
Figure 2. Low glutamine level leads to increased histone methylation in tumour cells.
(a) Cells (Ras-3T3 cultured in complete (4mM glutamine) or 0.2mM glutamine medium for 2 days, M229 cultured in complete (4mM glutamine) or 0.1mM glutamine medium for different days as indicated) were lysed for histone extraction and histone lysine methylation levels were assessed by western blotting. Total histone H3 was used as loading control. (b) M229 cells were cultured in medium with different concentrations of glutamine as indicated for 4 days, then cells were lysed for histone extraction and histone lysine methylation levels were assessed by western blotting. Total histone H3 was used as loading control. (c) Cells were cultured in complete medium, low glutamine medium, or low glutamine plus 7mM dimethyl-αKG for 4 days, then cells were lysed for histone extraction and histone lysine methylation levels were assessed by western blotting. Total histone H3 was used as loading control. (d) Cells were cultured in different conditions as indicated, then metabolites were extracted from the cells; αKG and glutamine concentrations were measured by LC-MS. Data represent mean ± S.D., n=3 independent experiments. ***P<.001 by unpaired two-tailed Student’s t-test. Source data are shown in supplementary Table 4. (e) In complete medium, 2mg/ml 968 or 100µM DON treated M229 cells were lysed for histone extraction, and histone lysine methylation levels were assessed by western blotting. Total histone H3 was used as loading control. (f) Metabolites were extracted from DON treated M229 cells, then αKG and glutamine concentrations were measured by LC-MS. Data represent mean ± S.D. n=3 independent experiments. *P<.05, ***P<.001 by unpaired two-tailed Student’s t-test. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
Glutamine deficiency drives histone hyper-methylation in tumour core regions
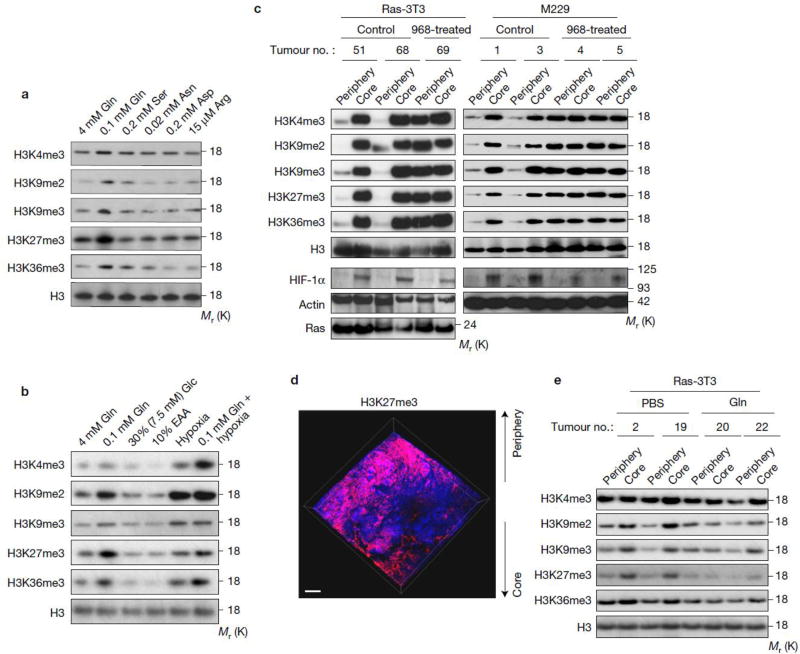
To assess the extent of selectivity of nutritional deficiency in the core regions of tumours, we measured the concentration of all amino acids in the core vs. peripheral regions by LC-MS. Interestingly, only the levels of five amino acids (including glutamine) were significantly lower in the core relative to the peripheral regions (Table 1), and this result was confirmed again by another independent metabolomics analysis (Supplementary Table 1). We further determined which relative amino acid deficiency was most responsible for increased histone methylation using medium that contained the corresponding concentration of each decreased amino acid (although the levels in cell culture may drop quickly due to consumption) and found only low glutamine levels resulted in increased histone methylation (Fig. 3a). In addition, we found histone methylation was only induced when glutamine was low, but not under a combinatorial deficiency of the other four amino acids (Arg, Ser, Asp, Asn) (Supplementary Fig. 3a). Moreover, other forms of metabolic stress had no significant effect on histone methylation levels (Fig. 3b and Supplementary Fig. 3b). Interestingly, the combination of low glutamine and hypoxia resulted in greater histone methylation than either alone, particularly on H3K4me3, suggesting that hypoxia is not the only mechanism contributing to this phenomenon in solid tumours (Fig. 3b).
Table 1.
Amino acid concentrations in tumour periphery and core regions. M229 xenograft tumours were harvested and separated into periphery and core samples (n=3 biologically independent xenografts). Metabolites were extracted from each sample and used for LC-MS analysis. Mean value of each amino acid concentration (µM) and core/periphery ratio were shown. p value was calculated by unpaired two-tailed Student’s t-test.
| Amino acid | Mean (uM)
|
Ratio (Core/periphery) |
p | |
|---|---|---|---|---|
| Periphery | Core | |||
|
| ||||
| Arg | 133.87 | 27.73 | 0.20 | 0.00030 |
| Asn | 121.41 | 28.99 | 0.29 | 0.022 |
| Gln | 487.66 | 229.15 | 0.46 | 0.0033 |
| Ser | 575.73 | 277.54 | 0.47 | 0.0060 |
| Asp | 666.86 | 387.92 | 0.55 | 0.0058 |
| Hyp | 18.46 | 14.40 | 0.69 | 0.040 |
| Trp | 22.11 | 20.21 | 0.71 | 0.24 |
| Phe | 82.47 | 65.75 | 0.72 | 0.047 |
| Tyr | 194.13 | 152.47 | 0.76 | 0.064 |
| Ala | 978.48 | 807.27 | 0.82 | 0.00045 |
| Gly | 1077.70 | 898.67 | 0.83 | 0.18 |
| Taurine | 378.89 | 350.62 | 0.85 | 0.25 |
| Met | 148.07 | 126.03 | 0.87 | 0.034 |
| Ile | 140.70 | 126.12 | 0.88 | 0.050 |
| Leu | 260.55 | 226.25 | 0.90 | 0.034 |
| Glu | 1870.06 | 1675.91 | 0.90 | 0.031 |
| Citrulline | 99.02 | 110.02 | 0.91 | 0.69 |
| Thr | 527.11 | 476.26 | 0.91 | 0.10 |
| Lys | 174.50 | 165.59 | 0.92 | 0.19 |
| His | 167.78 | 153.61 | 0.93 | 0.28 |
| Orn | 44.67 | 53.27 | 0.96 | 0.89 |
| Val | 284.05 | 271.62 | 0.97 | 0.48 |
| Pro | 301.11 | 330.89 | 1.16 | 0.048 |
| Cys-Cys | 12.15 | 8.69 | 50.82 | 0.23 |
Figure 3. Glutamine deficiency drives histone hyper-methylation in tumour core regions.
(a) M229 cells were cultured in complete medium and the medium with specified concentrations (which are similar as the concentration in tumour cores) of select amino acids for 4 days, then cells were lysed for histone extraction and histone lysine methylation levels were assessed by western blotting. Total histone H3 was used as loading control. (b) M229 cells were cultured under conditions indicated for 4 days, then cells were lysed for histone extraction and histone lysine methylation levels were assessed by western blotting. EAA, essential amino acid. Total histone H3 was used as loading control. (c) Control and 968-treated xenograft tumours were harvested. Tissues from the periphery and core regions were lysed for histone extraction and histone lysine methylation levels were assessed by western blotting. Total histone H3 and Actin were used as loading controls. (d) 3D imaging of M229 xenograft tumour treated with 968 depicting DAPI (blue) and H3K27me3 antibody (red). Scale bar = 300 µm. (e) 4mM glutamine or PBS control injected tumours were harvested. Tissues from the periphery and core regions were lysed for histone extraction and histone lysine methylation levels were assessed by western blotting. Total histone H3 was used as loading control. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
Moreover, we found that glutaminase inhibitor, compound 968, led to dramatic histone methylation in the tumour periphery, with levels similar to the core region of control treated tumours (Fig. 3c). Using PACT, we found that, in contrast to the untreated tumour in Fig. 1g, 968-treated tumour displayed higher levels of H3K27me3 methylation in both tumour periphery and core regions (Fig. 3d). In addition, increased histone methylation was largely attenuated by intra-tumour injections of glutamine, particularly on H3K9 and H3K27 (Fig. 3e). Taken together, these data demonstrate that low levels of glutamine are the major contributor to increased histone methylation in the tumour core regions.
Low glutamine leads to de-differentiation in tumour cores
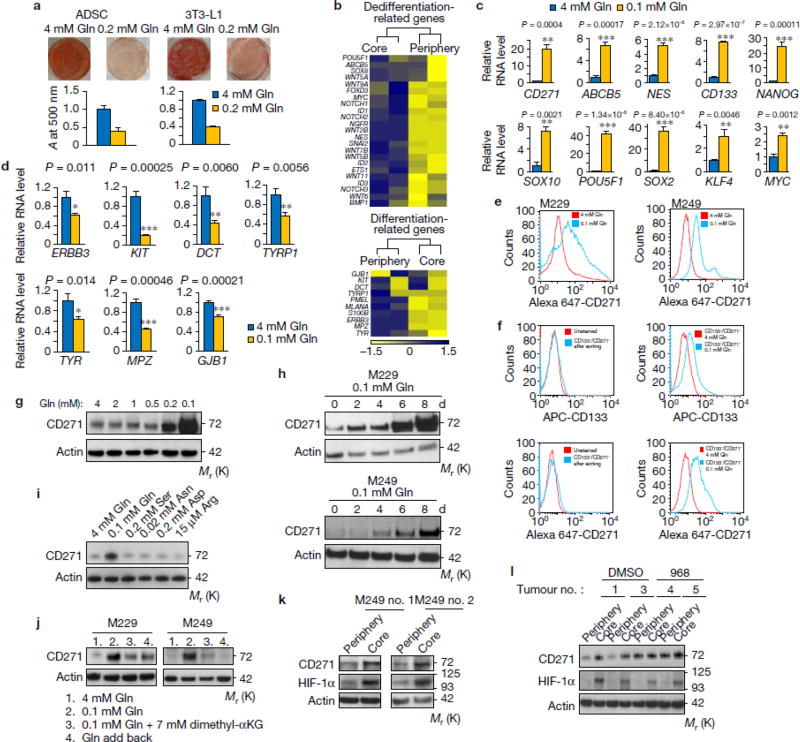
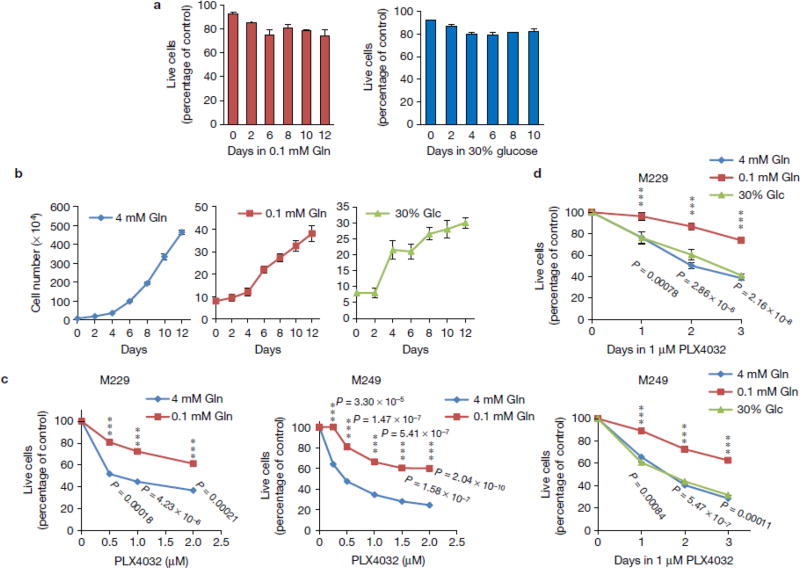
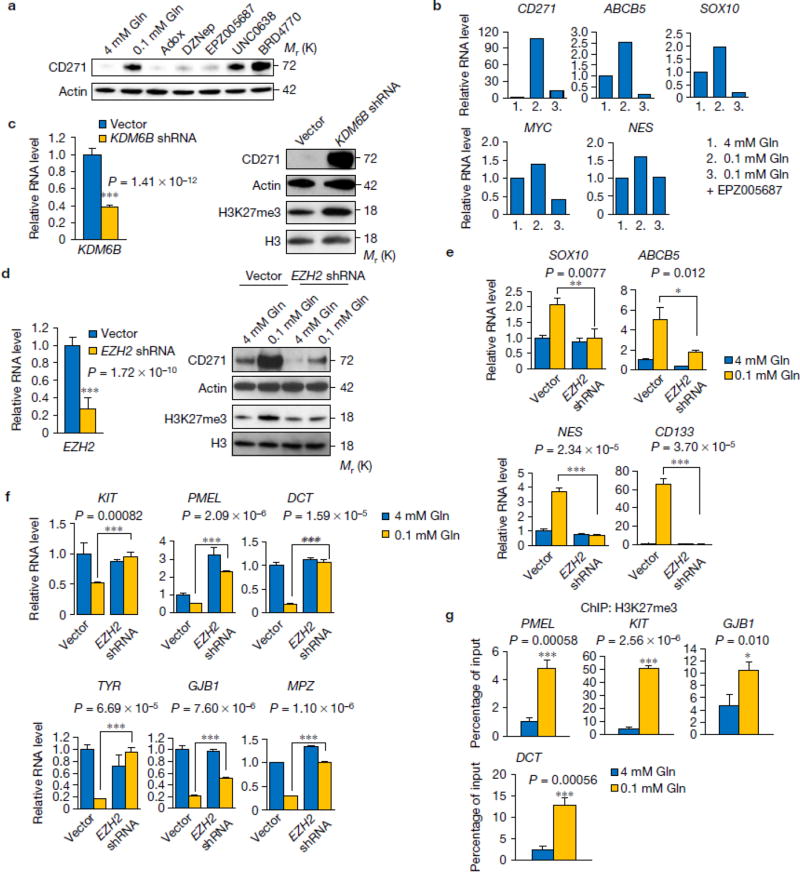
Previous reports indicate that histone hyper-methylation induced by IDH mutants prevents cell differentiation via inhibiting αKG-dependent histone demethylases12. Therefore, we examined whether decreased αKG in low glutamine conditions can affect cell differentiation using two adipocyte models, human adipocyte derived stem cells (ADSC) and 3T3-L1 cells. Interestingly, we found that cells cultured in low glutamine failed to differentiate compared to cells cultured in complete medium (Fig. 4a). In melanomas, markers for de-differentiation have been identified such as CD271, CD133, and ABCB521–24. Differentiation markers in melanomas have also been reported, such as ERBB3, KIT, PMEL, and GJB25–29. Next, we performed RNASeq and compared the genes related to melanoma de-differentiation and differentiation in core vs. periphery regions. Consistent with the result that low glutamine inhibited adipocyte differentiation, we found that de-differentiation related genes were up-regulated while differentiation genes were down-regulated in the tumour core regions compared with matched peripheral regions (Fig. 4b). This finding in xenograft tumours was further corroborated by the up-regulation of a panel of de-differentiation genes in melanoma cells cultured under low glutamine conditions (Fig. 4c,e), while the differentiation genes are simultaneously down-regulated (Fig. 4d & Supplementary Fig. 4a). In addition, low glutamine-induced temporal CD271 expression paralleled the dose response and time course of low glutamine-induced histone hyper-methylation (Fig. 4g vs. 2b, and Fig. 4h vs. 2a). To further distinguish whether low glutamine leads to de-differentiation or selectively expands a small subpopulation of cells that already carry the signatures, we sorted and collected CD133/CD271 double negative (CD133-/CD271-) cells (Supplementary Fig. 4b,c). We then cultured these cells in low glutamine followed by FACS analysis and found a large portion of the previously CD271-/CD133- cells now expressed CD271 and CD133 (Fig. 4f). Consistently, H3 lysine methylation and CD271 expression level increased in these cells after glutamine starvation (Supplementary Fig. 4d). In addition, CD271 expression was specifically induced by deficiency of glutamine but not other amino acids (Fig. 4i), which could be reversed by adding dimethyl-αKG or glutamine back in the medium (Fig. 4j). As expected, the expression of CD271 was increased in the core region of solid tumours, and compound 968 treatment induced CD271 expression also in the periphery of tumours (Fig. 4k,l).
Figure 4. Low glutamine leads to de-differentiation in tumour cores.
(a) Human adipocyte-derived stem cells (ADSC) and mouse pre-adipocytes, 3T3-L1, were induced to differentiate into mature adipocytes. The lipid droplets were stained by Oil-Red-O and quantified by measuring absorbance at 500nm. Representative wells from three independent experiments are shown. Data represent mean ± S.D. of three independent experiments. (b) Heatmap of de-differentiation gene (upper) and differentiation gene (lower) expression in periphery and core of M229 xenograft tumours in duplicate. (c, d) M229 cells were cultured in complete or 0.1mM glutamine medium for 4 to 12 days, de-differentiation (c) and differentiation (d) gene expression were assessed by qPCR. Data represent mean ± S.D., n=3 biologically independent RNA extracts. Two experiments were repeated independently with similar results. Source data are shown in supplementary Table 4. *P<.05, **P<.01, ***P<.001 by unpaired two-tailed Student’s t-test. (e) Complete or 0.1mM glutamine medium cultured M229 cells were stained with Alex647-CD271 antibody and analyzed by flow cytometry. Data is representative of three independent experiments. (f) CD271-/CD133- cells were cultured in complete (4mM Gln) or 0.1mM Gln medium for 8 days, then cells were stained with APC-CD133 or Alex647-CD271 antibody and analyzed by flow cytometry. Data is representative of three independent experiments. (g-h) M229 cells were cultured in medium with different concentrations of glutamine as indicated for 4 days (g), or in complete medium or 0.1mM glutamine medium for different days as indicated (h), then cell lysates were collected and CD271 expression was assessed by western blotting. (i-j) M229 or M249 cells were cultured in under different conditions as indicated for 4 days (i) or 8 days (j), whole cell lysates were collected and protein levels were assessed by western blotting. (k) Tissues from periphery and core regions of M249 xenograft tumours were lysed to collect whole cell lysate. Protein levels were assessed by western blotting with specified antibodies. (l) Control and 968 treated M229 xenograft tumours were harvested. Tissues from the periphery and core regions were lysed and protein levels were assessed by western blotting with specified antibodies. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
Low glutamine-induced de-differentiation results in resistance to BRAF inhibitor treatment
We then assessed whether glutamine deficiency-induced de-differentiation would impact responses to targeted therapies. First, we found that V600EBRAF melanoma cells maintained over 80% viability in low glutamine (0.1 mM) or low glucose (5–30 % of complete medium) (Fig. 5a & Supplementary Fig. 3c). Compared to melanoma cells in complete medium, cells proliferated at a slower rate in medium containing 0.1mM glutamine or in 30% glucose (Fig. 5b). Interestingly, we found that melanoma cells cultured in low glutamine prior to BRAF inhibitor, PLX4032, treatment were less sensitive to the drug compared to those cultured in complete medium (Fig. 5c). Although cells displayed a similar proliferation rate under low glutamine or low glucose conditions, we found only the low glutamine condition resulted in drug resistance, but not the low glucose condition (Fig. 5d). These data correlated with the previous observation that only low glutamine, but not low glucose conditions, induced hyper-methylation of histones (Fig. 3b).
Figure 5. Low glutamine-induced de-differentiation results in resistance to BRAF inhibitor treatment.
(a) M229 cells were cultured in the medium with 0.1mM glutamine or 30% (7.5mM) glucose for different days as indicated. Cell viability was assessed by Propidium Iodide exclusion. Data represent mean ± S.D., n=3 independent experiments. (b) M229 cells were cultured in complete, 0.1mM glutamine or 30% glucose (7.5mM) medium for different days as indicated. Cell number was counted every two days. Data represent mean ± S.D., n=4 independent experiments. (c) M229 and M249 cells were cultured in complete or 0.1mM glutamine medium for 4 days, and then treated with different doses of PLX4032 for 72 hours. MTS assay was performed to measure cell viability. Data represent mean ± S.D., n=3 independent cell culture for M229; n=4 for M249. ***P<.001 by unpaired two-tailed Student’s t-test. (d) M229 and M249 cells were cultured in complete, 0.1mM glutamine, or 30% glucose (7.5mM) medium for 4 days, then treated with 1µM PLX4032 for 24, 48, or 72 hours. MTS assay was performed to measure cell viability. Data represent mean ± S.D., n=4 independent cell culture. ***P<.001 by unpaired two-tailed Student’s t-test. Source data are shown in supplementary Table 4.
Low glutamine-induced de-differentiation is mediated by histone methylation on H3K27
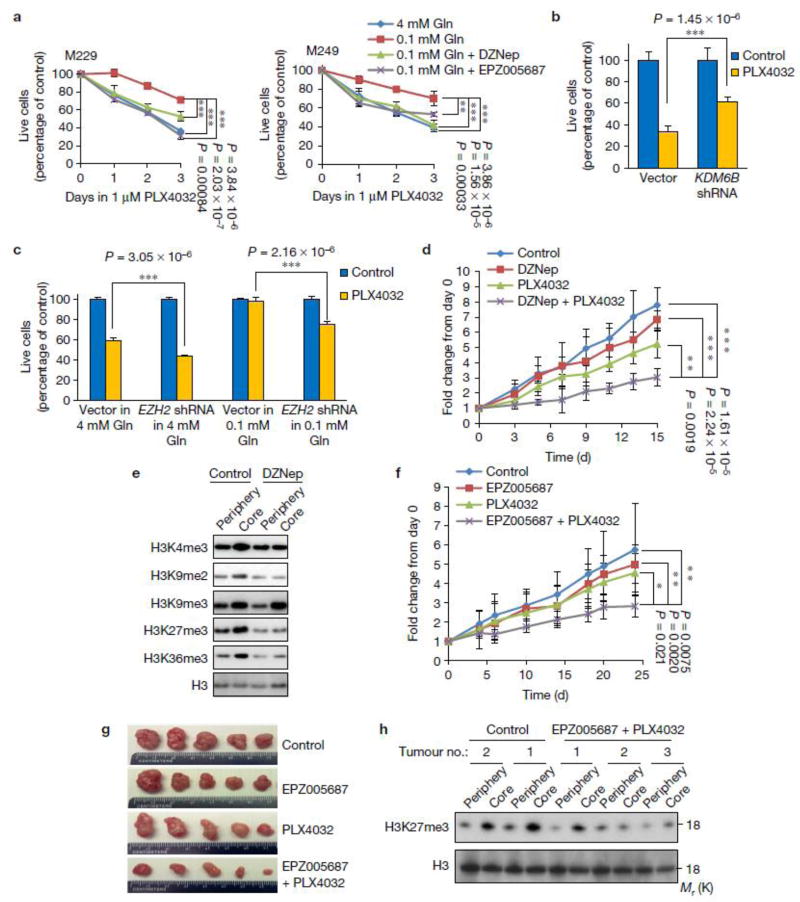
We next investigated whether low glutamine-induced cell de-differentiation is mediated by histone hyper-methylation. We found that global histone methylation inhibitors, including Adox and DZNep30, 31 reversed low glutamine-induced CD271 expression (Fig. 6a). Furthermore, an inhibitor targeting H3K27me3, EPZ00568732, suppressed the low glutamine-induced CD271 expression to a similar extent as the global inhibitors. In contrast, H3K9 inhibitors BRD477033 and UNC063834 had no effect on low glutamine-induced CD271 expression (Fig. 6a and Supplementary Fig. 5a,b). These data suggest that tri-methylation on H3K27 plays a major role in low glutamine-induced melanoma tumour cell de-differentiation. Consistently, the H3K27me3 methylation inhibitor EPZ005687 prevented low glutamine-induced methylation on H3K27 and CD271 expression in a dose-dependent manner (Supplementary Fig. 5c) as well as the expression of de-differentiation genes in general (Fig. 6b). To further confirm that changes in CD271 expression were dependent on H3K27 methylation, we knocked down KDM6B, a H3K27-specific demethylase, and observed dramatic induction of H3K27 methylation and CD271 expression in cells cultured in complete medium (Fig. 6c). Next, we knocked down EZH2, the H3K27 methyltransferase in melanoma cells and found that cells cultured in 0.1mM glutamine medium with EZH2 knockdown failed to induce methylation on H3K27me3 and CD271 expression (Fig. 6d and Supplementary Fig. 6a,b). Importantly, the knockdown effect of EZH2 shRNAs could be rescued by over expression of an shRNA-resistant EZH2 cDNA in the melanoma cells (Supplementary Fig. 6a,b). Consistently, we found the induction of de-differentiation genes by low glutamine was dramatically blocked by EZH2 knockdown (Fig. 6e). Because H3K27me3 functions to suppress transcriptional activation, we next examined whether H2K27me3 plays a role in inhibiting expression of differentiation genes. First, suppression of differentiation markers in low glutamine medium was reversed by treating cells with H3K27me3 methylation inhibitor EPZ005687 (Supplementary Fig. 5d,e) or knocking down EZH2 (Fig. 6f). Chromatin Immunoprecipitation (ChIP) analysis using antibodies against H3K27me3 revealed that differentiation genes are directly repressed by this H3K27me3 methylation marker at the promoter regions (Fig. 6g).
Figure 6. Low glutamine-induced de-differentiation is mediated by histone methylation on H3K27.
(a) M229 cells were cultured in complete medium or 0.1mM glutamine medium with different histone methylation inhibitors for 4 days, whole cell lysates were collected and protein levels were assessed by western blotting. Actin was used as loading control. (b) M229 cells were cultured in complete medium or 0.1mM glutamine medium with or without H3K27 specific methylation inhibitor EPZ005687 for 4 days, then RNA was extracted and de-differentiation related gene expression was assessed by qPCR. Data represent the mean of 2 biologically independent RNA extracts. Two experiments were repeated independently with similar results. Source data are shown in supplementary Table 4. (c) M229 cells were transduced with lenti viral KDM6B shRNA. Knockdown efficiency was measured by qPCR (left). Data represent mean ± S.D., n=8 biologically independent RNA extracts. ***P<.001 by unpaired two-tailed Student’s t-test. H3K27me3 and CD271 level were measured by western blotting (right). (d) M229 cells were transduced with lenti viral EZH2 shRNA. Knockdown efficiency was assessed by qPCR (left). ). Data represent mean ± S.D., n=9 biologically independent RNA extracts. ***P<.001 by unpaired two-tailed Student’s t-test. CD271 and H3K27me3 level were measured by western blotting (right). (e, f) After transduction of lenti viral EZH2 shRNA, cells were cultured in complete or 0.1mM glutamine for at least 4 days, then RNA was extracted from bulk M229 cells, de-differentiation (e) and differentiation (f) related gene expression was checked by qPCR. Data represent mean ± S.D., n=3 biologically independent RNA extracts. *P<.05, **P<.01, ***P<.001 by unpaired two-tailed Student’s t-test. (g) Cells were cultured in complete or 0.1mM glutamine medium for 4 days, then cells were harvested for ChIP assay using antibody against H3K27me3. Immunoprecipitation of the neural differentiation-related gene promoters (GJB1 and DCT in M229, PMEL and KIT in M249) were analyzed by qPCR. Data represent mean ± S.D., n=3 independent experiments. *P<.05, ***P<.001 by unpaired two-tailed Student’s t-test. Source data are shown in supplementary Table 4. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
Besides JmjC family histone demethylases, there are other αKG dependent enzymes like HIF1-α prolyl-hydroxylase (HIF-PH) and TET enzymes that promote DNA demethylation. Knockdown of HIF-1α had no effect on histone hyper-methylation in low glutamine conditions (Supplementary Fig. 7a,b). Moreover, whole genome DNA methylation sequencing found no significant differences between complete and low glutamine cultured melanoma cells (Supplementary Fig. 7c), and DNA methylation inhibitors 5-Azacytidine and 5-Aza-2′-deoxycytidine failed to inhibit CD271 induction in 0.1mM glutamine medium (Supplementary Fig. 7d).
Low glutamine-induced drug resistance is mediated by histone methylation on H3K27
We next tested if low glutamine-induced resistance to BRAF inhibitor, PLX4032, was mediated by hyper-methylation of H3K27. Melanoma cells cultured in low glutamine displayed resistance to PLX4032, but this effect was prevented when global histone demethylase inhibitor DZNep or H3K27 methylation specific inhibitor, EPZ005687, were supplemented in the low glutamine condition (Fig. 7a). In addition, knockdown of the H3K27 specific demethylase, KDM6B, which resulted in increased CD271 expression (Fig. 6c), led to resistance to PLX4032 treatment compared to the control cells (Fig. 7b). On the other hand, the protective effect against PLX4032 treatment in low glutamine conditions was reduced after knockdown of the H3K27 methyltransferase EZH2 (Fig. 7c). Finally, we performed a mouse xenograft experiment using melanoma cells and treated the mice with PBS control, DZNep, PLX4032, or the combination of DZNep and PLX4032. We found that treatment with PLX4032 alone inhibited tumour growth, especially at early time points; however, when combined with DZNep, PLX4032 treatment led to dramatic decreases in tumour size (Fig. 7d). As expected, histone methylation was attenuated in the DZNep and PLX4032 combination treated tumour core regions (Fig. 7e). Interestingly, no impairment in tumour growth was found upon DZNep treatment, suggesting it may take a long time (more than 10 weeks) to see the significant effects on tumour growth35. We also observed similar results on tumour growth and core histone methylation when we replaced DZNep with H3K27me3 specific methylation inhibitor EPZ005687 (Fig. 7f-h).
Figure 7. Low glutamine-induced drug resistance is mediated by histone methylation on H3K27.
(a) M229 or M249 cells were cultured in complete medium or 0.1mM glutamine medium with histone methylation inhibitors DZNep or EPZ005687 for 4 days, and then cells were treated with 1µM PLX4032 for 3 days. MTS assay was performed to measure viability. Data represent mean ± S.D., n=4 (M229), or n=6 (M249) independent cell culture. ***P<.001 by unpaired two-tailed Student’s t-test. (b) Control vector and KDM6B shRNA lenti viral transduced M229 bulk cells were treated with DMSO control or 1µM PLX4032 for 72 hours. MTS assay was performed to measure viability. Data represent mean ± S.D., n=6 independent cell culture. ***P<.001 by unpaired two-tailed Student’s t-test. (c) Control vector and EZH2 shRNA lenti viral transduced M229 bulk cells were treated with DMSO control or 1µM PLX4032 for 48 hours. MTS assay was performed after treatment. Data represent mean ± S.D., n=5 independent cell culture. ***P<.001 by unpaired two-tailed Student’s t-test. Source data for a, b and c are shown in supplementary Table 4. (d) NSG mice were injected subcutaneously with 5×106 M229 cells. When the tumour size reached an average of 100mm3, mice were treated with vehicle control (n=6 mice), DZNep (I.P. injection 1.5mg/kg, n=4 mice), PLX4032 (Oral gavage, 10mg/kg, n=5 mice), or both DZNep and PLX4032 (n=5 mice) daily for 15 days. Tumour size was measured over time. Graph represents mean ± S.D., **P<.01, ***P<.001 by unpaired two-tailed Student’s t-test. (e) Samples from peripheral or core regions of the individual tumour as indicated were lysed and immunoblotting was performed with indicated antibodies. Total H3 was used as loading control. (f) Nude mice were injected subcutaneously with 5×106 M229 cells. When the tumour size reached an average of 100mm3, mice were treated with vehicle control (n=8 mice), EPZ005687 (I.P. injection, 2.5mg/kg, n=7 mice), PLX4032 (Oral gavage, 10mg/kg, n=6 mice), or both DZNep and PLX4032 (n=7 mice) every other day for 25 days. Tumour size was measured over time. Graph represents mean ± S.D. *P<.05, **P<.01 by unpaired two-tailed Student’s t-test. (g) Representative tumours in each group. (h) Samples from peripheral or core regions of the individual tumour as indicated were lysed and immunoblotting was performed with indicated antibodies. Total H3 was used as loading control. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
DISCUSSION
It is well established that the core of solid tumours is highly hypoxic and hypoxia is considered to be a major contributor to drug resistance36. However, whether nutrients are also depleted in regions of solid tumours similar to oxygen is not clear. Here, we found that glutamine is dramatically decreased in the tumour core regions compared to the periphery (Fig. 1a and Table 1). Interestingly, a few other amino acids, including serine, asparagine, arginine, and aspartate, are also significantly decreased in tumour core regions. A recent report demonstrated that glutamine, serine, and asparagine are of the most strongly depleted metabolites in human pancreatic tumors7. This result is highly consistent with our data that those amino acids are significantly decreased in the tumour core probably due to the heavy usages and poor tumour blood supply (Table 1, Supplementary Table 1). It supports the hypothesis that amino acids that are consumed by multiple anabolic processes (such as glutamine and serine) become depleted in the tumour core regions relative to those used mainly for protein synthesis.
It is worth mentioning that, different from cultured cells, whether glutamine is a major source for αKG in vivo is likely to depend on both the tumour genotype and tissue of origin, similar to glucose metabolism37. For example, 13C-glutamine tracing resulted in increased levels of TCA cycle metabolites in Myc-driven liver tumours38. In addition, analysis of NSCLC tumour metabolism in patients has indicated both glucose and glutamine can be important for TCA anaplerosis39, 40. Nonetheless, a recent report demonstrated that glutamine is not a major source for αKG in KRas-driven lung tumours41. Here, we demonstrated that in V600EBRAF melanoma tumours, that glutamine is sufficient to provide αKG in vivo (Fig. 1). Interestingly, we found that the decrease in αKG levels in melanoma tumour core regions are more extensive than the decrease in glutamine levels (Supplementary Table 1), suggesting that other sources beside glutamine could also contribute to αKG levels in vivo, such as low glucose or hypoxia in tumour core regions.
How does methylation on H3K27me3 promote de-differentiation? Although H3K27me3 is associated with repressed transcription, accumulating evidence support that H3K27me3 is essential for maintaining the self-renewal capability of embryonic and adult stem cells8, 42. In fact, it has been reported that critical genes involved in differentiation, such as HOX genes, are targeted for repression by H3K27 methylation43, supporting the hypothesis that H3K27me3 contributes to de-differentiation by inhibiting transcription of differentiation-related genes. Consistent with this model, we found that neural crest differentiation genes are directly repressed by this H3K27me3 methylation marker at promoter regions in melanoma cells (Fig. 6g). This is consistent with the previous study showing adipocyte differentiation genes such as Adipoq and Cebpa are suppressed by the binding of H3K9me3 and H3K27me3 at promoter regions12. In addition, several reports demonstrated the H3K27me3 specific methyl-transferase, EZH2 promotes cancer stem-like cell expansion44, 45. In agreement, our data demonstrate that EZH2 is required for low glutamine-induced expression of de-differentiation related genes (Fig. 6e).
Distinct tumour microenvironments may influence therapeutic responses mediated by inducing cancer cell de-differentiation46. For example, depletion of oxygen in tumours contributes to cancer stem-like cells and promotes drug resistance, where it is usually located at tumour ‘core’ or regions with a lack of blood vessel47–51 . Consistently, our data showed another example that regional glutamine deficiency can also induce cancer cell de-differentiation and lead to drug resistance (Fig. 5). It is worth noting that other studies suggested that cancer stem cells that promote metastasis are located at the tumour fronts or vascular niche52–54. It is not clear whether low glutamine-induced dedifferentiation enhances metastatic potential. However, it is possible that low glutamine/hypoxic regions will eventually result in VEGF expression and vascular formation over time, which will further provide other critical factors for cancer stem cell maintenance and metastasis, such as β-catenin, VEGF, Nrp1, and TGF52–54. In support of this, in the invasive front of pancreatic tumours, a distinct subpopulation of cancer stem cells was identified that determines the metastatic phenotype, but not the tumourigenic potential55.
In the past few years, targeting EZH2 as a potential cancer therapy has been explored. However, understanding where and how EZH2 inhibitors will be useful in cancer therapy is critical for the use of these drugs56. Our results provide the molecular basis for an important strategy to combine cancer drugs, such as BRAF inhibitor, with EZH2 inhibitor to achieve a more promising treatment efficacy.
Supplementary Material
Acknowledgments
We thank members of the Kong laboratory for helpful comments on the manuscript. This work was supported by National Institutes of Health (NIH)/National Cancer Institute (NCI) grants R01CA183989 (to M.K.), American Cancer Society Research Scholar RSG-16-085-01-TBE (to M.K.) and Stand up to Cancer Philip A. Sharp Innovation in Collaboration Award. M.K. is the Pew Scholar in the Biomedical Sciences and the V scholar in Cancer Research. X.H.L. is supported by DNA Damage Response and Oncogenic Signaling (DDROS) Training Program at City of Hope. Research reported here includes work carried out in Core Facilities supported by the NIH/NCI under grant number P30CA33572.
Footnotes
AUTHOR CONTRIBUTIONS
M.P. designed and performed most of the experiments, analyzed and interpreted the data and wrote the manuscript. M.K. conceived and supervised this study, designed experiments and wrote the paper. M.A.R. and X.H.L. helped to measure metabolites and assisted with mouse experiments. R.P.K. and V.G. performed PACT experiments. T.Q.T. assisted with flow cytometry experiments. Y.Y. assisted with qPCR experiments. J.E.H and K.K.R. helped set up melanoma cell culture. W.H., C.S. and R.S.L. provided patient-derived melanoma cells and conceptual advice on melanoma de-differentiation. X.X. assisted with IHC experiments. D.E.S assisted with ChIP experiments and H.L. performed the bioinformatics analyses. D.K.A provided conceptual advice on hypoxia and metabolism experiments. X.L. and J.W.L performed and helped to analyze the metabolomics experiments.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Le A, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell metabolism. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts E, Caldwell AL, et al. Amino acids in epidermal carcinogenesis in mice. Cancer research. 1949;9:350–353. [PubMed] [Google Scholar]
- 7.Kamphorst JJ, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer research. 2015;75:544–553. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annual review of biochemistry. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 9.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nature reviews. Molecular cell biology. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 10.Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Santa F, et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 14.Seward DJ, et al. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nature structural & molecular biology. 2007;14:240–242. doi: 10.1038/nsmb1200. [DOI] [PubMed] [Google Scholar]
- 15.Li KK, Luo C, Wang D, Jiang H, Zheng YG. Chemical and biochemical approaches in the study of histone methylation and demethylation. Medicinal research reviews. 2012;32:815–867. doi: 10.1002/mrr.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dewhirst MW, Cao Y, Moeller B. Cycling hypoxia and free radicals regulate angiogenesis and radiotherapy response. Nature reviews. Cancer. 2008;8:425–437. doi: 10.1038/nrc2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dankort D, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nature genetics. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang B, et al. Single-Cell Phenotyping within Transparent Intact Tissue through Whole-Body Clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conti F, Minelli A. Glutamate immunoreactivity in rat cerebral cortex is reversibly abolished by 6-diazo-5-oxo-L-norleucine (DON), an inhibitor of phosphate-activated glutaminase. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1994;42:717–726. doi: 10.1177/42.6.7910617. [DOI] [PubMed] [Google Scholar]
- 20.Wang JB, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boiko AD, et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 2010;466:133–137. doi: 10.1038/nature09161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Civenni G, et al. Human CD271-positive melanoma stem cells associated with metastasis establish tumor heterogeneity and long-term growth. Cancer research. 2011;71:3098–3109. doi: 10.1158/0008-5472.CAN-10-3997. [DOI] [PubMed] [Google Scholar]
- 23.Monzani E, et al. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur J Cancer. 2007;43:935–946. doi: 10.1016/j.ejca.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 24.Schatton T, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erickson SL, et al. ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2-and heregulin-deficient mice. Development. 1997;124:4999–5011. doi: 10.1242/dev.124.24.4999. [DOI] [PubMed] [Google Scholar]
- 26.Oetting WS. The tyrosinase gene and oculocutaneous albinism type 1 (OCA1): A model for understanding the molecular biology of melanin formation. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2000;13:320–325. doi: 10.1034/j.1600-0749.2000.130503.x. [DOI] [PubMed] [Google Scholar]
- 27.Aoki H, Tomita H, Hara A, Kunisada T. Conditional Deletion of Kit in Melanocytes: White Spotting Phenotype Is Cell Autonomous. The Journal of investigative dermatology. 2015;135:1829–1838. doi: 10.1038/jid.2015.83. [DOI] [PubMed] [Google Scholar]
- 28.Kwon BS, et al. A melanocyte-specific gene, Pmel 17, maps near the silver coat color locus on mouse chromosome 10 and is in a syntenic region on human chromosome 12. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:9228–9232. doi: 10.1073/pnas.88.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sargiannidou I, et al. Intraneural GJB1 gene delivery improves nerve pathology in a model of X-linked Charcot-Marie-Tooth disease. Annals of neurology. 2015;78:303–316. doi: 10.1002/ana.24441. [DOI] [PubMed] [Google Scholar]
- 30.Mortl M, Busse D, Bartel H, Pohl B. Partial purification and characterization of rabbit-kidney brush-border (Ca2+ or Mg2+)-dependent adenosine triphosphatase. Biochimica et biophysica acta. 1984;776:237–246. doi: 10.1016/0005-2736(84)90213-x. [DOI] [PubMed] [Google Scholar]
- 31.Miranda TB, et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Molecular cancer therapeutics. 2009;8:1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knutson SK, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nature chemical biology. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 33.Yuan Y, et al. A small-molecule probe of the histone methyltransferase G9a induces cellular senescence in pancreatic adenocarcinoma. ACS chemical biology. 2012;7:1152–1157. doi: 10.1021/cb300139y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vedadi M, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nature chemical biology. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang P, et al. Antitumor effects of pharmacological EZH2 inhibition on malignant peripheral nerve sheath tumor through the miR-30a and KPNB1 pathway. Molecular cancer. 2015;14:55. doi: 10.1186/s12943-015-0325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nature reviews. Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 37.Hensley CT, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuneva MO, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell metabolism. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sellers K, et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. The Journal of clinical investigation. 2015;125:687–698. doi: 10.1172/JCI72873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng T, et al. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davidson SM, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell metabolism. 2016;23:517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nature reviews. Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 43.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 44.Chang CJ, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonzalez ME, et al. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3098–3103. doi: 10.1073/pnas.1308953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 47.Conley SJ, et al. Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2784–2789. doi: 10.1073/pnas.1018866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Liu Y, Malek SN, Zheng P, Liu Y. Targeting HIF1alpha eliminates cancer stem cells in hematological malignancies. Cell stem cell. 2011;8:399–411. doi: 10.1016/j.stem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muz B, de la Puente P, Azab F, Luderer M, Azab AK. Hypoxia promotes stem cell-like phenotype in multiple myeloma cells. Blood cancer journal. 2014;4:e262. doi: 10.1038/bcj.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samanta D, Gilkes DM, Chaturvedi P, Xiang L, Semenza GL. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E5429–5438. doi: 10.1073/pnas.1421438111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Mao Q, et al. A tumor hypoxic niche protects human colon cancer stem cells from chemotherapy. Journal of cancer research and clinical oncology. 2013;139:211–222. doi: 10.1007/s00432-012-1310-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beck B, et al. A vascular niche and a VEGF-Nrp1 loop regulate the initiation and stemness of skin tumours. Nature. 2011;478:399–403. doi: 10.1038/nature10525. [DOI] [PubMed] [Google Scholar]
- 53.Zucchi I, et al. Distinct populations of tumor-initiating cells derived from a tumor generated by rat mammary cancer stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16940–16945. doi: 10.1073/pnas.0808978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malanchi I, et al. Cutaneous cancer stem cell maintenance is dependent on beta-catenin signalling. Nature. 2008;452:650–653. doi: 10.1038/nature06835. [DOI] [PubMed] [Google Scholar]
- 55.Hermann PC, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell stem cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 56.McCabe MT, Creasy CL. EZH2 as a potential target in cancer therapy. Epigenomics. 2014;6:341–351. doi: 10.2217/epi.14.23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.