Abstract
Glucocorticoids are extensively used to treat inflammatory diseases; however, their chronic intake increases the risk for mycobacterial infections. Meanwhile, the effects of glucocorticoids on innate host responses are incompletely understood. In this study, we investigated the direct effects of glucocorticoids on antimycobacterial host defense in primary human macrophages. We found that glucocorticoids triggered the expression of cathelicidin, an antimicrobial critical for antimycobacterial responses, independent of the intracellular vitamin D metabolism. Despite upregulating cathelicidin, glucocorticoids failed to promote macrophage antimycobacterial activity. Gene expression profiles of human macrophages treated with glucocorticoids and/or IFN-γ, which promotes induction of cathelicidin, as well as antimycobacterial activity, were investigated. Using weighted gene coexpression network analysis, we identified a module of highly connected genes that was strongly inversely correlated with glucocorticoid treatment and associated with IFN-γ stimulation. This module was linked to the biological functions autophagy, phagosome maturation, and lytic vacuole/lysosome, and contained the vacuolar H+-ATPase subunit a3, alias TCIRG1, a known antimycobacterial host defense gene, as a top hub gene. We next found that glucocorticoids, in contrast with IFN-γ, failed to trigger expression and phagolysosome recruitment of TCIRG1, as well as to promote lysosome acidification. Finally, we demonstrated that the tyrosine kinase inhibitor imatinib induces lysosome acidification and antimicrobial activity in glucocorticoid-treated macrophages without reversing the anti-inflammatory effects of glucocorticoids. Taken together, we provide evidence that the induction of cathelicidin by glucocorticoids is not sufficient for macrophage antimicrobial activity, and identify the vacuolar H+-ATPase as a potential target for host-directed therapy in the context of glucocorticoid therapy.
Glucocorticoids have been widely used for decades as potent immunosuppressive drugs to treat human inflammatory diseases (1). However, long-term glucocorticoid therapy, for instance, in patients suffering from rheumatoid arthritis, constitutes a risk factor for developing active tuberculosis (TB) (2) and other mycobacterial infections (3). Meanwhile, the effects of glucocorticoids on innate host responses are incompletely understood. Despite obvious anti-inflammatory effects, emerging evidence suggests that glucocorticoids also enhance innate immune pathways, for instance, the NLRP3 inflammasome (4). Consistent with this idea, recent microarray transcriptional profiling revealed that long-term exposure of human macrophages to glucocorticoids strengthened components of the innate response, albeit weakening an acquired immune signature (5).
Human macrophages play a central role in the host defense against mycobacteria, because they are the natural niches for these intracellular pathogens. We have contributed to the understanding of how human macrophages kill mycobacteria by identifying a vitamin D–dependent host defense pathway (6–8). The vitamin D antimicrobial pathway is induced by innate and acquired immune signals, including TLR2/1 ligand, CD40L, and IFN-γ (6–9). Central early events include the induction of IL-32 and IL-15 (10, 11), the CYP27b1-hydroxylase, which converts 25-OH vitamin D (25D) into the active form 1,25-di-OH vitamin D (1,25D) in an autocrine manner, as well as the upregulation of the vitamin D receptor (VDR) (6–9). Subsequently, 1,25D triggers VDR-mediated induction of cathelicidin antimicrobial peptide (12, 13), pivotal for killing mycobacteria in phagolysosomes (14). Furthermore, this pathway involves the vitamin D–dependent induction of autophagy, which is important to overcome the phagosome maturation block (7, 15, 16), an important mycobacterial virulence strategy (17–22). Given the importance of macrophages in combating mycobacteria, we investigated the impact of glucocorticoids on the antimycobacterial response in primary human macrophages.
Materials and Methods
Reagents
Recombinant human M-CSF was purchased from Miltenyi Biotec and recombinant human IFN-γ from Becton Dickinson. M-CSF was used at a concentration of 50 ng/ml and IFN-γ at a concentration of 10 ng/ml. Dexamethasone, hydrocortisone, imatinib mesylate, and mifepristone were from Sigma-Aldrich and used at the following concentrations: dexamethasone 10–1000 nM, hydrocortisone 300 nM, imatinib mesylate 5–10 µM, and mifepristone 1 µM. 25D3 and 1,25D3 were obtained from Biomol and used as indicated. Bafilomycin A1 was purchased from Sigma-Aldrich and Invivogen and used at 100 nM. The mAbs used were: anti–IL-15 PE (clone 34559) and IgG1 PE isotype control (clone 11711) from R&D Systems; anti-LAMP1 (clone D2D11) from Cell Signaling; anti-LAMP1 (clone eBioH4A3) from eBioscience; IgG1 pure isotype control (clone X40) and IgG2a isotype control (clone X39) from Becton Dickinson; anti-cathelicidin (clone OSX12) from Abcam or Novus Biologicals; and monoclonal anti-TCIRG1 from Abcam. Anti-mouse IgG1 PE (clone A85-1) and anti-mouse IgG2a FITC (clone R19-15) were obtained from Becton Dickinson. LysoSensor Green DND-189, DAPI, and secondary Abs labeled with Alexa Fluor 488 or Alexa Fluor 594 were purchased from Life Technologies. FCS and human AB serum were purchased from PAA Laboratories and PAN Biotech. RPMI 1640 and macrophage serum-free media (SFM) were obtained from Life Technologies.
Ethics statement
This study was conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the local Ethics Committee (Ethikkommission) of the University of Cologne, Germany. All donors provided written informed consent for the collection of peripheral blood and subsequent analyses.
Cell preparations and macrophage culture
Whole blood or buffy coats from healthy donors or patients on long-term glucocorticoid therapy were obtained with informed consent. Patients were receiving dosages of prednisolone between 7.5 and 45 mg/d for at least 4 wk for treatment of chronic inflammatory diseases. Their clinical diagnoses were pyoderma gangrenosum, polymyalgia rheumatica, psoriatic arthritis, and pyoderma gangrenosum, respectively. PBMCs were isolated by Ficoll-Paque (GE Healthcare). Monocytes were isolated via CD14+ MACS cell separation (Miltenyi Biotec) according to the manufacturer’s instructions. Monocyte-derived macrophages (MDMs) were prepared by culturing peripheral blood monocytes in RPMI 1640 media containing 10% FCS for 4–7 d in the presence of M-CSF (50 ng/ml). Afterward cells were cultured in fresh media with 10% vitamin D–sufficient human AB serum (supplemented with 25D3 to reach 25D serum levels between 100 and 180 nmol/l) unless indicated otherwise.
Real-time quantitative PCR
mRNA was isolated from MDMs or monocytes using the RNeasy mini kit (Qiagen) according to the manufacturer’s recommended protocol. cDNA was prepared using iScript cDNA Synthesis kit (BioRad), and mRNA levels were assessed by quantitative PCR using Power SYBR Green Master Mix (Life Technologies) as previously described (7) and GoTaq qPCR Master Mix (Promega) according to the manufacturer’s recommended protocol. Primer sequences for human CYP27B1, cathelicidin, CYP24A1, IL-15, VDR, TCIRG1 (vacuolar H+-ATPase [v-ATPase] V0 subunit a3) and h36B4 were previously reported (6, 7, 23). Primer sequences for human TNF-α were: TNF-α forward, 5′-GGAGAAGGGT-GACCGACTCA-3′, TNF-α reverse, 5′-CTGCCCAGACTCGGCAA-3′. Human IL-1β, IL-10, IL-32, and DRAM1 Quantitect Primer Assays were from Life Technologies.
Gene expression profiling
Total RNA of cells was isolated with TRIzol (Life Technologies), and RNA quality was confirmed using microcapillary electrophoresis (2100 Bioanalyzer; Agilent). A total of 100 ng RNA was labeled and hybridized to Sureprint G3 human GE 8x60K whole genome mRNA microarray according to the manufacturer’s specifications. The arrays were scanned (Agilent G2595C scanner), and data were extracted and processed using the Genespring XII software (Agilent). Macrophage gene expression files containing array data are available under accession no. GSE79077 in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/). Weighted gene coexpression network analysis (WGCNA) was performed on the microarray data set to identify networks of correlated genes using the R library ‘wgcna’ (22). Gene modules were generated using the function blockwiseModules(), with a soft thresholding power of 12. Module eigengenes (MEs), defined as the first principal component of each gene module, were correlated to experimental treatments of media, IFN-γ, dexamethasone, and IFN-γ/dexamethasone. Correlation was calculated using a binary matrix representation where a particular sample had a value of 1 for the treatment used and 0 for all other treatments. Correlations and corresponding p values were displayed in a heat map using the labeled-Heatmap() function. For each module, hub genes, or genes with high intermodular connectivity, were identified using the signedKME() function. Functional gene annotation enrichment analysis was done using Ingenuity Pathways Analysis (IPA) (http://www.ingenuity.com) or Cytoscape v3.2.0 software with ClueGO v2.1.5 plugin (PMID: 19237447). Functional annotation network was then generated with Gephi v0.8.2 β (24).
Flow cytometry
Surface staining with IL-15 PE or the appropriate isotype control was performed in PBS containing 20% FCS and 0.1% sodium azide. For intracellular measurement of cathelicidin and TCIRG1, cells were stained using Cytofix/Cytoperm kit (Becton Dickinson) according to the manufacturer’s recommended instructions. For acidification assays, MDMs or monocytes were incubated with LysoSensor Green (1–2 or 1 µM, respectively) for 30 min and subsequently washed with PBS. Flow cytometry was conducted on a FACSCalibur (BD Biosciences), and data were analyzed using FlowJo software (Tree Star). MDM and monocyte gates were set based on forward and sideward scatters.
Infection of MDMs, treatment with stimuli, and quantification of mycobacterial growth
Mycobacterium bovis Bacille Calmette–Guérin strain Pasteur (BCG) was grown in suspension with constant, gentle rotation at 37°C in 7H9 media containing Difco Middlebrook 7H9 broth supplemented with 0.5% glycerol, 0.05% tyloxapol, and 10% BBL Middlebrook OADC Enrichment.
MDMs were infected for 3–4 h with single-cell suspensions of BCG at a multiplicity of infection of three to five in a 24-well plate. Afterward extracellular bacteria were removed by vigorous washing. For CFU assays, MDMs were treated with dexamethasone or IFN-γ and incubated for 3 d. To determine the number of viable intracellular bacilli, cells were lysed with 0.05% SDS (Serva Electrophoresis). Lysates of infected cells were resuspended vigorously and diluted in 0.05% tyloxapol (Sigma-Aldrich). Four to five dilutions of each sample were plated on 7H10 agar plates (Becton Dickinson and Merck Millipore) and incubated for 21 d at 37°C.
Immunostaining for TCIRG1 and LAMP1
For intracellular staining of TCIRG1 and LAMP1, MDMs were grown on eight-well chamber slides (Becton Dickinson) and stimulated with dexamethasone or IFN-γ, or left untreated for 20 h. Cells were fixed with methanol and permeabilized with saponin (0.01%) and Triton X-100 (0.1%). Next, cells were stained with mAbs against TCIRG1 and LAMP1, detected with anti-mouse Alexa Fluor 488 and anti-rabbit Alexa Fluor 594, and costained with DAPI. In some experiments, MDMs were infected for 3 h with single-cell suspensions of BCG. Cells were analyzed with a Keyence BZ-9000 microscope. For quantification of TCIRG1bright cells, counts from three independent investigators of ~50 cells (uninfected MDMs) or 200 cells (infected MDMs) per condition were averaged. To quantify lysosomes that recruited TCIRG1, three to five image sections of ~50 cells (uninfected MDMs) or 200 cells (infected cells) per condition were analyzed by calculating the Manders overlap coefficient (M2 = fraction of LAMP1 overlapping TCIRG1) using ImageJ with JACoP plugin. Identical threshold settings were applied to every condition.
Confocal microscopy
MDMs were grown on eight-well chamber slides (Becton Dickinson) and infected for 3 h with single-cell suspensions of M. bovis BCG Pasteur (pMV261::dsRed) expressing dsRed2 fluorescent protein under control of the constitutive Hsp60 promoter. Next, MDMs were stimulated with dexamethasone or IFN-γ, or left untreated for 20 h. In some experiments, cells were fixed with methanol and permeabilized with saponin (0.01%) and Triton X-100 (0.1%). Infected MDMs were stained with mouse mAb against TCIRG1, followed by anti-mouse Alexa Fluor 488 and DAPI. In some experiments, cells were fixed with paraformaldehyde (4%) and permeabilized with saponin (0.2%). Infected MDMs were stained with mouse mAb against cathelicidin, followed by anti-mouse Alexa Fluor 488 and DAPI. Cells were analyzed on an Olympus FV1000 confocal microscope. For quantification of TCIRG1 recruitment to intracellular BCG/clusters of BCG, counts from three independent investigators of ~150 infected cells per condition were averaged. In addition, ~20 image sections of ~150 cells were analyzed by calculating the Manders overlap coefficient (M2 = fraction of LAMP1 overlapping TCIRG1) using ImageJ with JACoP plugin. Identical threshold settings were applied to every condition.
Statistics
The p values were calculated by two-tailed Student t tests in Fig. 8C. Differences in all other quantitative PCR, FACS, and CFU data were analyzed by Friedman test for k dependent samples to compare more than two groups. If this test yielded significant results, we performed a Wilcoxon test as a post hoc test to compare two groups. n refers to the number of repeat experiments performed with cells from individual human donors.
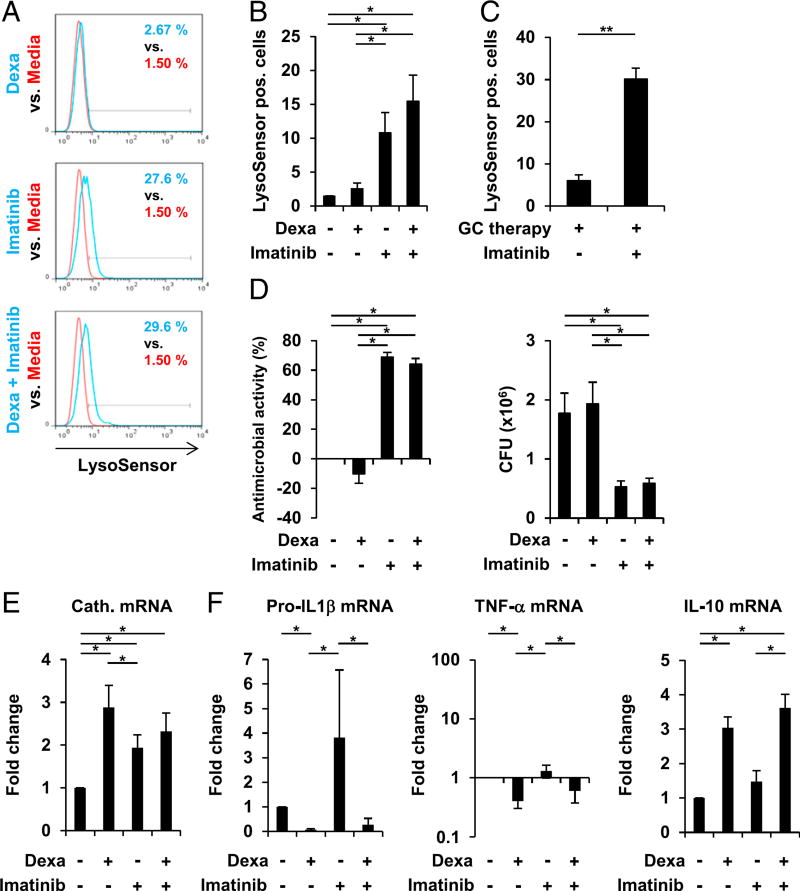
FIGURE 8.
Imatinib promotes lysosome acidification and antimycobacterial activity in dexamethasone (Dexa)-treated human macrophages. (A and B) MDMs were stimulated with Dexa (100 nM), Imatinib, Dexa and imatinib in combination, or media alone for 20 h. Cells were stained with LysoSensor Green. Acidification was determined by FACS analyses (% LysoSensor-positive cells). (A) Histogram plots from one representative donor out of seven. (B) Summary of seven donors (% LysoSensor-positive cells ± SEM, n = 7). (C) Monocytes from patients on long-term glucocorticoid therapy (GC therapy) were stimulated with imatinib or media alone for 20 h. Cells were stained with LysoSensor Green. Acidification was determined by FACS analyses (% LysoSensor-positive cells ± SEM, n = 4). (D) MDMs were infected with BCG for 3–4 h before stimulation with Dexa (100 nM), Imatinib, Dexa and imatinib in combination, or media alone for 3 d. Viable bacteria were quantified by CFU assay on day 3. Percentage antimicrobial activity relative to media (left panel) and CFU counts (right panel) are shown (± SEM, n = 5). (E) MDMs were stimulated as in (A). Cathelicidin mRNA expression was assessed by quantitative PCR (mean fold change ± SEM, n = 6). (F) MDMs were stimulated as in (A). Pro–IL1-β (n = 5), TNF-α (n = 5), and IL-10 (n = 5) mRNA expression were assessed by quantitative PCR (mean fold change ± SEM). *p < 0.05, **p < 0.01.
Results
Dexamethasone promotes an anti-inflammatory macrophage cytokine profile
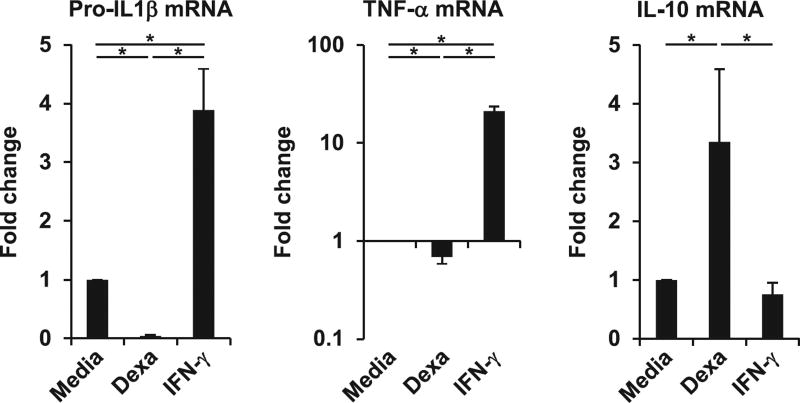
Because the therapeutic benefits of glucocorticoids are fundamentally linked to glucocorticoid-mediated anti-inflammatory effects, we tested whether glucocorticoid treatment of primary human macrophages results in a shift from a proinflammatory toward an anti-inflammatory profile. Therefore, we compared mRNA expression of key proinflammatory and anti-inflammatory cytokines, specifically pro–IL-1β, TNF-α, and IL-10, in MDMs cultured with dexamethasone, the inflammatory stimulus IFN-γ, or media alone for 20 h. We found that dexamethasone treatment significantly inhibited the mRNA expression of pro–IL-1β and TNF-α by 95 and 30%, respectively, when compared with the media control (both p <0.05; Fig. 1), whereas IFN-γ induced pro–IL-1β expression by 3.9-fold and TNF-α expression by 21-fold (both p <0.05; Fig. 1). Moreover, dexamethasone induced IL-10 by 3.4-fold (p < 0.05; Fig. 1). IFN-γ had no significant effect on IL-10 gene expression. Taken together, these data confirm that dexamethasone promotes an anti-inflammatory macro-phage cytokine profile.
FIGURE 1.
Dexamethasone (Dexa) promotes an anti-inflammatory cytokine profile in human macrophages. MDMs were stimulated with Dexa (100 nM), IFN-γ, or media alone for 20 h. Gene expression of pro–IL1-β (n = 8), TNF-α (n = 5), and IL-10 (n = 8) was determined by quantitative PCR (mean fold change ± SEM). *p < 0.05.
Dexamethasone induces cathelicidin expression
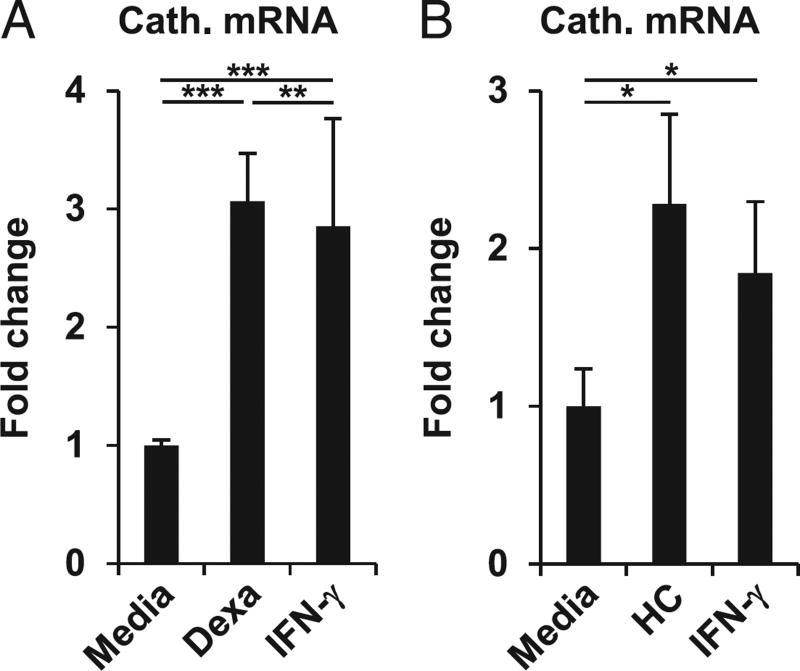
Anti-inflammatory macrophage phenotypes have a reduced capacity to control intracellular infections. For instance, in mouse macrophages, the expression of inducible NO synthase and the production of NO (25–27), a fundamental murine anti-mycobacterial molecule, are significantly reduced by glucocorticoids (28, 29). We and others previously reported that in humans, but not in mice, the vitamin D–mediated induction of cathelicidin antimicrobial peptide has a central role in the antimycobacterial activity in macrophages (6, 7, 14, 30, 31). Thus, we compared the ability of dexamethasone and IFN-γ to induce cathelicidin. MDMs were stimulated with dexamethasone, IFN-γ, or media alone for 20 h, and expression of cathelicidin mRNA was measured. Dexamethasone and IFN-γ each induced cathelicidin mRNA expression by 3.1- and 2.9-fold as compared with media (both p < 0.001; Fig. 2A). Furthermore, dexamethasone induction of cathelicidin gene expression was observed at 100 and 1000 nM, but not at 10 nM (data not shown). Next, we tested whether cathelicidin induction was specific for dexamethasone or whether a different member of the glucocorticoid family, hydrocortisone, also induces cathelicidin gene expression. We found that hydrocortisone induced cathelicidin expression by 2.3-fold compared with media (p < 0.05; Fig. 2B). Taken together, these data demonstrate that glucocorticoids induce cathelicidin expression.
FIGURE 2.
Dexamethasone (Dexa) induces cathelicidin gene expression in human macrophages. (A) MDMs were stimulated with Dexa (100 nM), IFN-γ, or media alone for 20 h. Cathelicidin gene expression was assessed by quantitative PCR (mean fold change ± SEM, n = 21). (B) MDMs were stimulated with hydrocortisone (HC), IFN-γ, or media alone for 20 h. Gene expression of cathelicidin was assessed by quantitative PCR (mean fold change ± SEM, n = 5). *p < 0.05, **p < 0.01, ***p < 0.001.
Dexamethasone induction of cathelicidin is independent of the macrophage vitamin D defense pathway
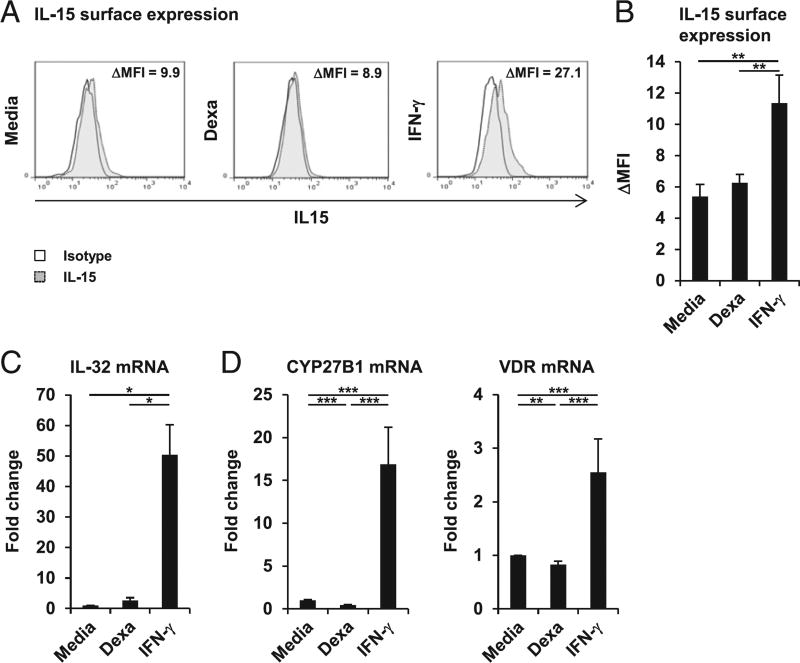
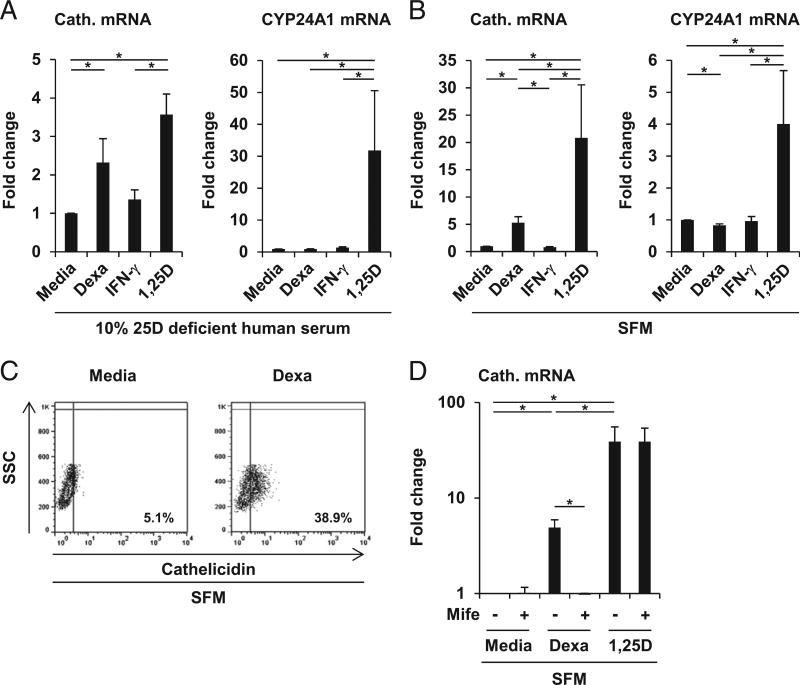
The induction of cathelicidin by TLR2/1 ligand, IFN-γ, or CD40L is dependent on the bioavailability of 25D in the sera added to the cultures, limiting the intracellular vitamin D metabolism in human macrophages (6–8). Thus, we compared the ability of dexamethasone and IFN-γ to regulate key components of the vitamin D defense pathway in macrophages. Dexamethasone had no effect on IL-15 mRNA (data not shown) and surface expression (Fig. 3A, 3B), nor on IL-32 mRNA expression (Fig. 3C), early events of the vitamin D defense pathway. In addition, dexamethasone significantly suppressed expression of the CYP27B1-hydroxylase by 56% and did not induce VDR mRNA expression (p < 0.001 and p < 0.01; Fig. 3D). As expected, IFN-γ induced IL-15 mRNA (data not shown) and surface expression when compared with media control (Δ mean fluorescence intensity [ΔMFI] = 11.4 versus 5.4, p < 0.01; Fig. 3A, 3B), as well as IL-32 (50-fold, p < 0.05; Fig. 3C), CYP27B1 (17.0-fold, p < 0.001; Fig. 3D), and VDR (2.5-fold, p < 0.001; Fig. 3D) mRNA expression. These findings indicate that the glucocorticoid-mediated induction of cathelicidin is not linked to the previously described induction of the intracellular vitamin D defense pathway in human macrophages. Thus, we hypothesized that, in contrast with the 25D-dependent IFN-γ induction of cathelicidin, dexamethasone triggers cathelicidin expression independently of serum 25D. Therefore, we stimulated MDMs with dexamethasone, IFN-γ, or media alone in vitamin D–deficient human serum (25D serum level = 31 nmol/l). The active form of vitamin D, 1,25D, was used as positive control. 1,25D and dexamethasone both induced cathelicidin (both p < 0.05; Fig. 4A) as compared with media control, yet, as expected, IFN-γ failed to induce cathelicidin mRNA expression in 25D-deficient culture conditions (Fig. 4A). CYP24, a classical genomic target of vitamin D used as a control readout, was induced by neither dexamethasone nor IFN-γ in 25D-deficient conditions (Fig. 4A). Given that the vitamin D-deficient serum used in these assays still contains 31 nmol/l 25D, we next analyzed dexamethasone induction of cathelicidin in SFM, which does not contain detectable amounts of 25D. Again, IFN-γ failed to induce, whereas dexamethasone and 1,25D were able to trigger, cathelicidin mRNA expression by 5.3- and 21-fold, respectively (both p < 0.05; Fig. 4B). CYP24 was induced by neither dexamethasone nor IFN-γ under these conditions.
FIGURE 3.
Dexamethasone (Dexa) dissociates the vitamin D host defense pathway. (A and B) MDMs were stimulated with Dexa (100 nM), IFN-γ, or media alone for 20 h. IL-15 surface expression was determined by FACS analyses. (A) Histogram plots from 1 representative donor out of 12 (ΔMFI). (B) Summary of 12 donors (ΔMFI ± SEM). (C) Gene expression for IL-32 (n = 6), (D) CYP27B1 (n = 17), and VDR (n = 18) was determined by quantitative PCR (mean fold change ± SEM). *p < 0.05, **p < 0.01, ***p < 0.001.
FIGURE 4.
Dexamethasone (Dexa) induces cathelicidin independent of serum 25D in human macrophages. (A and B) MDMs were stimulated for 20 h either in (A) serum with 10% 25D-deficient human serum (25D serum level = 31 nmol/l) or (B) SFM with Dexa (100 nM), IFN-γ, 1,25D (10 nM), or media alone. Gene expression of cathelicidin and CYP24A1 was assessed by quantitative PCR [mean fold change ± SEM; (A), n = 5, (B), n = 6]. (C) MDMs were stimulated for 20 h in SFM with Dexa (1000 nM) or media alone. Intracellular cathelicidin protein expression was determined by FACS analyses (% cathelicidin-positive cells). (D) MDMs were stimulated for 20 h in SFM with either mifepristone (Mife; 1 µM), Dexa (1000 nM), 1,25D (10 nM), or media alone as indicated. Cathelicidin gene expression was determined by quantitative PCR (mean fold change ± SEM, n = 5). *p < 0.05.
Next, we analyzed whether dexamethasone induces expression of cathelicidin protein. MDMs were stimulated with dexamethasone in SFM for 20 h, and cathelicidin expression was measured by intracellular FACS. Dexamethasone triggered protein expression of cathelicidin in SFM (38.9 versus 5.1% positive cells in the media control; Fig. 4C). Finally, cathelicidin induction by glucocorticoids was dependent on the glucocorticoid receptor (GR), because mifepristone, a GR antagonist, completely blocked dexamethasone, but not 1,25D-induced cathelicidin gene expression (Fig. 4D). Taken together, these data showed that dexamethasone triggers cathelicidin expression by a mechanism that is different from the previously described IFN-γ–induced vitamin D–dependent pathway. Glucocorticoid induction of cathelicidin was observed at concentrations of dexamethasone (100 and 1000 nM), as well as of hydrocortisone (300 nM), that are within the range of the serum levels in individuals taking moderate-to-high therapeutic doses of glucocorticoids (32).
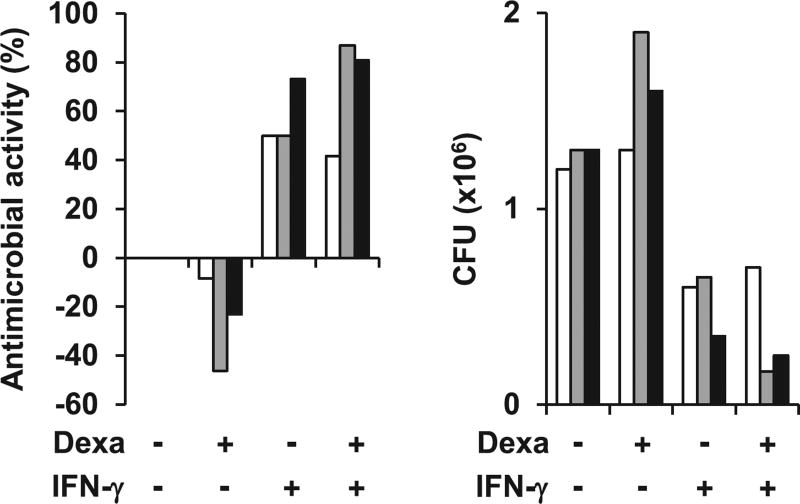
Dexamethasone fails to induce antimycobacterial activity
Cathelicidin is pivotal to the antimycobacterial response in human macrophages and kills bacteria by directly binding bacterial membranes (6, 7, 14–16). To investigate whether dexamethasone-induced cathelicidin targets intracellular mycobacteria, we infected MDMs with BCG (pMV261::dsRed) expressing dsRed2 fluorescent protein. Afterward, infected cells were stimulated with either dexamethasone or IFN-γ for 20 h and stained with anti-cathelicidin mAb. Confocal microscopy analysis showed colocalization of dexamethasone-induced and IFN-γ–induced cathelicidin with BCG (Supplemental Fig. 1A). Because dexamethasone induced cathelicidin expression and dexamethasone-induced cathelicidin colocalized with intracellular BCG, we asked whether dexamethasone promotes anti-mycobacterial activity in human macrophages. To test this, we infected human MDMs from three individual donors with BCG and treated the BCG-infected MDMs with dexamethasone and/or IFN-γ, or media alone. We measured between 50 and 73% bacterial growth restriction in the IFN-γ–treated MDMs as compared with media control (Fig. 5). In contrast, we observed no reduction in intracellular bacterial load in the dexamethasone-treated MDMs (Fig. 5). Moreover, we measured a similar bacterial growth pattern in the IFN-γ plus dexamethasone-treated cells as in the IFN-γ–treated cells (Fig. 5). IFN-γ plus dexamethasone also resulted in a comparable induction of cathelicidin gene expression as IFN-γ alone (Supplemental Fig. 1). Taken together, our data show that dexamethasone induces cathelicidin antimicrobial peptide expression but fails to promote antimicrobial activity against intracellular mycobacteria in human macrophages.
FIGURE 5.
Dexamethasone (Dexa) fails to induce antimycobacterial activity in human macrophages. MDMs were infected with BCG for 3–4 h before stimulation with Dexa (100 nM), IFN-γ, both in combination, or media alone for 3 d. Viable bacteria were quantified by CFU assay on day 3. Percentage antimicrobial activity relative to media (left panel) and CFU counts (right panel) are shown. Each colored row represents data obtained with MDMs from one individual donor (n = 3).
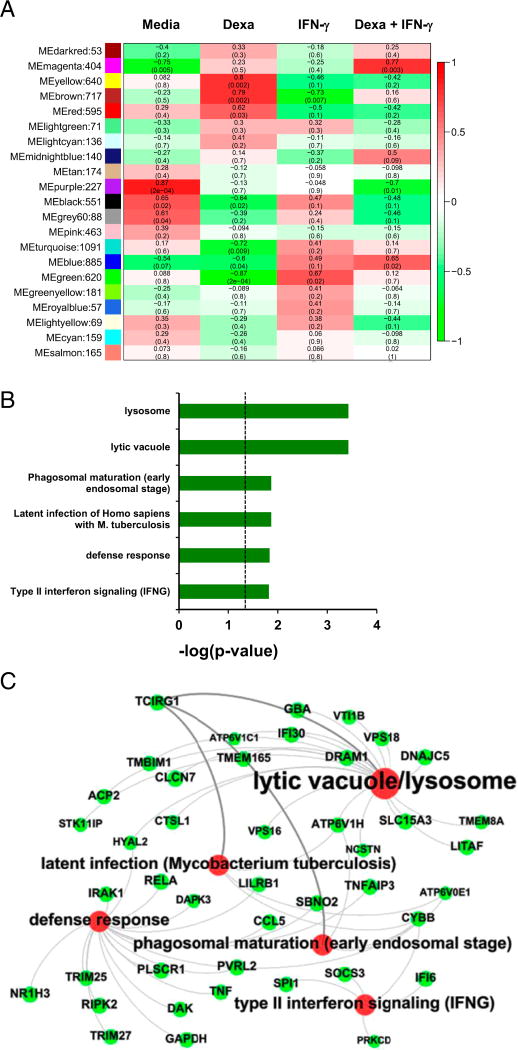
Identification of a macrophage host defense network associated with IFN-γ and inversely correlated with dexamethasone stimulation
The finding that dexamethasone triggers cathelicidin expression, yet fails to induce antimicrobial activity, suggested that dexamethasone does not induce or dexamethasone suppresses other critical factors involved in the macrophage antimycobacterial host response. To further investigate dexamethasone-regulated macrophage programs, we performed whole genome microarrays. Therefore, MDMs were cultured with dexamethasone and/or IFN-γ, or media alone. To uncover networks of highly interconnected genes, we subjected the microarray data to WGCNA, a systems biology approach. In brief, this analysis uses pairwise gene correlations, which are weighted to favor more robust coexpression values, to group genes into modules. MEs were calculated that represented a theoretical average gene for each module. Enrichment for particular modules in each treatment group was calculated by correlating MEs to experimental conditions (Fig. 6A). This approach identified a green module of 620 genes, which was significantly associated with IFN-γ stimulation and the most inversely correlated module to dexamethasone treatment. ClueGO functional enrichment analyses showed that the green module was significantly linked to key functional pathways in antimicrobial host defense, including lytic vacuole/lysosome, latent infection (Mycobacterium tuberculosis), phagosome maturation (early endosomal stage), and defense response (Fig. 6B, 6C). Among the genes in this module are several v-ATPase subunits, such as TCIRG1 alias v-ATPase V0 subunit a3, ATP6V1C1, ATP6V1H, and ATP6V0E1, which are involved in acidification of lysosomes; DRAM1, which promotes autophagic defense against mycobacteria (33); and VPS18, linked to autophagolysosome fusion. Several genes (CYBB, IFI6, PRKCD, SOCS3, SPI1) for IFN-γ signaling and TNF were also connected to this module. Taken together, our analyses revealed a network of host defense genes connected to phagosome maturation and the lysosome, fundamental aspects of the antimycobacterial host defense, which is associated with IFN-γ but not glucocorticoid treatment.
FIGURE 6.
Identification of a macrophage host defense network inversely correlated with dexamethasone (Dexa) treatment and associated with IFN-γ stimulation. WGCNA analysis of MDMs stimulated with Dexa (100 nM) and/or IFN-γ or cultured in media alone for 20 h. (A) Heat map depicting correlation of ME to treatment condition with corresponding p values. Each row is labeled with a color corresponding to an individual module from the WGCNA analysis, and the number of probe sets per module is given after the color. Red indicates positive correlation; green indicates inverse correlation. (B and C) Association of the WGCNA green module to ClueGO functional groups lytic vacuole/lysosome, latent infection (M. tuberculosis), phagosome maturation (early endosomal stage), defense response, and IFN-γ signaling. (B) The p values of associations to ClueGO functional groups are displayed. (C) Functional annotation network of ClueGO analysis generated by Gephi. Red circle sizes increase with ClueGO analysis increasing p value; green circle sizes increase with WGCNA increasing eigengene-based connectivity (kME) values.
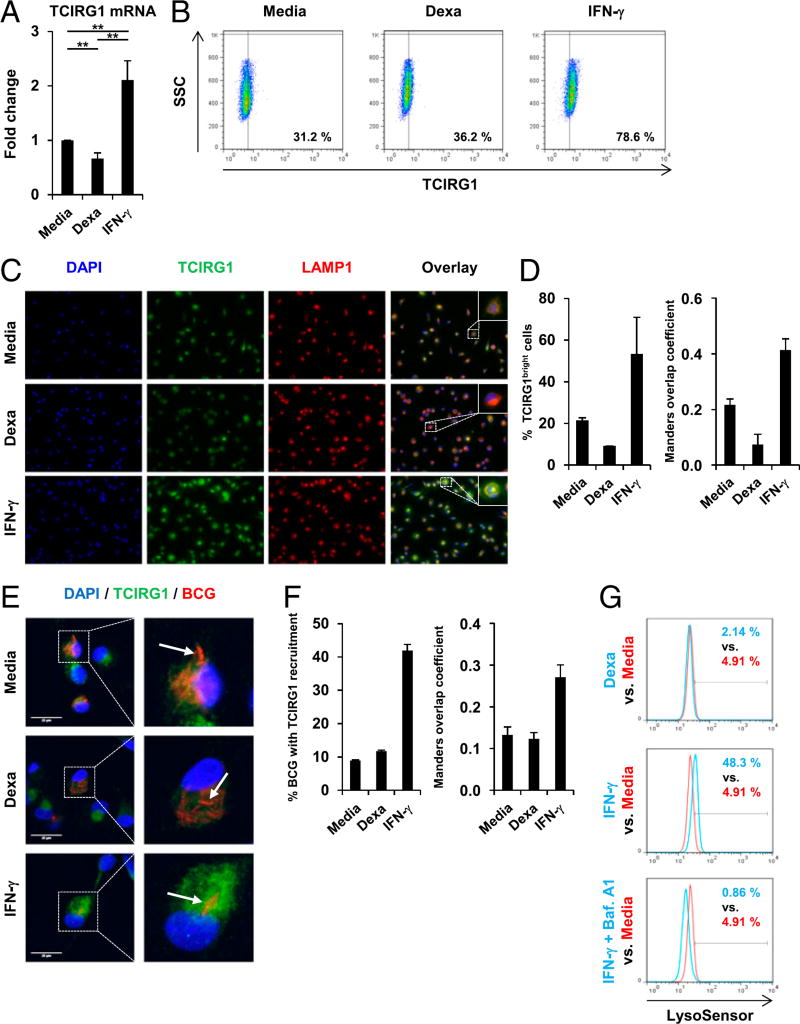
IFN-γ, yet not dexamethasone, induces TCIRG1 expression and phagolysosome recruitment, as well as lysosome acidification
The host defense genes in the green module identified by the ClueGO functional group analyses were next ranked according to their kME values (Fig. 6C), revealing TCIRG1 (kME = 0.98) as the top hub gene, which is the most connected gene. We and others previously showed that induction and lysosomal recruitment of TCIRG1 is linked to lysosome acidification, a hallmark of phagolysosome maturation, and subsequent antimycobacterial activity in human macrophages (23, 34), prompting us to study TCIRG1 in more detail. First, we measured TCIRG1 mRNA expression using quantitative PCR, demonstrating that dexamethasone suppressed TCIRG1 mRNA expression by 33% compared with media in MDMs after 20 h (p < 0.01; Fig. 7A). In contrast, IFN-γ triggered TCIRG1 mRNA expression by 2.1-fold when compared with media (p < 0.01; Fig. 7A). Similar results were observed in BCG-infected macrophages (Supplemental Fig. 2A). Next, we investigated the effect of dexamethasone and IFN-γ on TCIRG1 protein expression. MDMs were stimulated with dexamethasone, IFN-γ, or media alone for 20 h, and TCIRG1 expression was measured by intracellular FACS IFN-γ induced protein expression of TCIRG1 (78.6 versus 31.2% in the media control; Fig. 7B), whereas dexamethasone had no effect on TCIRG1 expression compared with media (Fig. 7B). Using immunofluorescence microscopy, we analyzed the expression of TCIRG1 and its lysosomal recruitment. IFN-γ upregulated TCIRG1 expression and induced TCIRG1 recruitment to lysosomes in both uninfected (Fig. 7C, 7D) and BCG-infected macrophages (Supplemental Fig. 2B, 2C). In contrast, dexamethasone failed to induce TCIRG1 expression and lysosome recruitment in uninfected (Fig. 7C, 7D) and BCG-infected macrophages (Supplemental Fig. 2B, Fig. 7C).
FIGURE 7.
IFN-γ triggers but dexamethasone (Dexa) fails to induce TCIRG1 expression, phagolysosome recruitment, and lysosome acidification in human macrophages. (A–C) MDMs were stimulated with Dexa (100 nM), IFN-γ, or media alone for 20 h. (A) Gene expression of TCIRG1 was assessed by quantitative PCR (mean fold change ± SEM, n = 12). (B) Intracellular TCIRG1 protein expression was determined by FACS analyses (% TCIRG1+ cells). (C) MDMs were fixed and immunostained with anti-TCIRG1 and anti-LAMP1. Original magnification ×40. (D) Quantification of (C). Left panel, Percentage of TCIRG1bright cells. Right panel, Manders overlap coefficient M2 (fraction of LAMP1 overlapping TCIRG1). (E) MDMs were infected with BCG (pMV261::dsRed) for 3 h and stimulated with Dexa (100 nM), IFN-γ, or media alone for 20 h, fixed, immunostained with anti-TCIRG1, and analyzed by confocal microscopy. Image brightness was enhanced equally across the entire image per journal policy. (F) Quantification of (E). Left panel, Percentage of BCG/BCG clusters with TCIRG1 recruitment. Right panel, Manders overlap coefficient M2 (fraction of BCG overlapping TCIRG1). (G) MDMs were stimulated with Dexa (100 nM), IFN-γ, IFN-γ and Bafilomycin A1 in combination, or media alone for 20 h and stained with LysoSensor Green. Acidification was determined by FACS analyses (% LysoSensor-positive cells). **p < 0.01.
To investigate whether TCIRG1 is recruited to the mycobacteria-containing vacuoles, we infected MDMs with BCG (pMV261:: dsRed). Afterward, infected cells were stimulated with dexamethasone, IFN-γ, or media alone for 20 h and stained with anti-TCIRG1 mAb. Confocal microscopy analysis showed enhanced recruitment of TCIRG1 to intracellular BCG (pMV261:: dsRed) in IFN-γ– but not in dexamethasone-treated cells (Fig. 7E, 7F).
Next, we tested the ability of dexamethasone and IFN-γ to regulate lysosome acidification. MDMs were stimulated with dexamethasone, IFN-γ, or media alone for 20 h, and acidification was measured by FACS using LysoSensor Green pH indicator. IFN-γ stimulation resulted in an increase of LysoSensor-positive cells (48.3 versus 4.91%; Fig. 7G), whereas dexamethasone had no effect (2.14 versus 4.91%; Fig. 7G). Furthermore, Bafilomycin A1, a specific inhibitor of v-ATPase, entirely reverted the IFN-γ– mediated induction of lysosome acidification (0.86 versus 48.3%; Fig. 7G), indicating that IFN-γ–mediated lysosome acidification was dependent on a functional v-ATPase. Together, our data showed that IFN-γ triggers but dexamethasone fails to induce TCIRG1 expression, recruitment to phagolysosomes, and induction of lysosome acidification in human macrophages.
We and others previously reported that the induction of autophagy is a central mechanism by which IFN-γ induced phagosome maturation and lysosome acidification (7, 8, 35, 36). Moreover, a recent study showed that glucocorticoids failed to induce autophagy in M. tuberculosis–infected macrophages (37). In line with these findings, IPA revealed a highly significant association of the green module with the biological function autophagy (p < 10−8; Supplemental Fig. 3A). Among the genes associated with the IPA biofunctional category autophagy were five genes also associated with the lytic vacuole/lysosome: APC2, DRAM1, CTSL, VPS18, and VTI1B, of which DRAM1 had the highest kME (DRAM1 kME = 0.95; Supplemental Fig. 3B). DRAM1 mRNA induction can be used to monitor induction of autophagy (38). We found that dexamethasone inhibited DRAM1 mRNA expression by 58% (p < 0.05; Supplemental Fig. 3C), whereas IFN-γ triggered DRAM1 mRNA expression by 4.3-fold (p < 0.05; Supplemental Fig. 3C) as measured by quantitative PCR. The induction of DRAM1 mRNA expression and the association of the green module to autophagy supported the known role of autophagy in the IFN-γ–mediated phagosome maturation and lysosome acidification (7, 8, 35, 36), which we further investigated. Macrophages were activated with IFN-γ in the absence or presence of two inhibitors of autophagy, specifically 3-MA and Wortmannin, both of which blocked induction of lysosome acidification by IFN-γ (Supplemental Fig. 3D), supporting the pivotal role of autophagy in promoting lysosome acidification (7, 8, 35, 36).
Imatinib induces antimicrobial activity but has no effect on inflammatory cytokine patterns in dexamethasone-treated macrophages
The tyrosine kinase inhibitor imatinib has emerged as a host-directed therapy for TB and other infections (23, 39, 40). In human macrophages imatinib was shown to induce the v-ATPase, leading to lysosome acidification and antimicrobial activity against M. tuberculosis (23). This prompted us to investigate whether imatinib can trigger lysosome acidification in dexamethasone-treated human macrophages. We found that imatinib alone and in combination with dexamethasone promoted lysosome acidification (10.8 and 15.5% LysoSensor-positive cells versus 1.4% in the media control, both p < 0.05; Fig. 8A, 8B). The ability to promote lysosome acidification was next tested in monocytic cells from patients on long-term glucocorticoid therapy. We decided to use monocytes directly isolated from blood, instead of MDMs, for this experiment to avoid decline of glucocorticoid effects during the in vitro differentiation period. Of relevance for this model we showed that IFN-γ induced, yet dexamethasone failed to trigger TCIRG1 and DRAM1 mRNA expression, as well as lysosome acidification in monocytes from healthy donors (Supplemental Fig. 4A, 4B). We cultured blood-derived monocytes of four patients receiving long-term glucocorticoid therapy for pyoderma gangrenosum, polymyalgia rheumatica, psoriatic arthritis, and pyoderma gangrenosum, respectively, with imatinib or media alone for 20 h. Imatinib stimulation increased the number of LysoSensor-positive cells when compared with media as measured in patient monocytes by FACS (30.1% LysoSensor-positive cells versus 6.0% in the media control, p < 0.01; Fig. 8C, Supplemental Fig. 4C). Next, we investigated whether imatinib promotes anti-mycobacterial activity in dexamethasone-treated MDMs and found that imatinib alone and in combination with dexamethasone induced mycobacterial growth restriction by 69 and 64%, respectively, as compared with media control (both p < 0.05; Fig. 8D), whereas dexamethasone alone failed to induce antimicrobial activity (Fig. 8D). Imatinib had no effect on the dexamethasone induction of cathelicidin (Fig. 8E).
Given that the therapeutic benefits of glucocorticoids are fundamentally linked to glucocorticoid-mediated anti-inflammatory effects, we tested whether imatinib modifies the dexamethasone-induced anti-inflammatory cytokine profile in human macrophages. We found that imatinib treatment did not reverse dexamethasone-mediated suppression of baseline pro–IL-1β and TNF-α expression, as well as induction of IL-10 expression as compared with the media control (Fig. 8F). Taken together, these data showed that imatinib promotes lysosome acidification and antimicrobial activity in glucocorticoid-treated macrophages, without reversing the anti-inflammatory properties of glucocorticoids.
Discussion
In this study, we provide evidence that glucocorticoids induce cathelicidin in human macrophages. However, glucocorticoid induction of cathelicidin occurs independently of autophagy, phagosome maturation, and lysosome acidification, known to be crucial for host defense (6–8, 35, 36). Thus, although critical (6, 7, 14–16), the induction of cathelicidin is not sufficient to promote antimycobacterial host defense. Nevertheless, we found that imatinib promotes lysosome acidification and antimycobacterial activity in glucocorticoid-stimulated macrophages. Strikingly, imatinib did not reverse the anti-inflammatory effects of glucocorticoids, thereby suggesting a preventive and therapeutic potential of imatinib in the context of glucocorticoid therapy.
A key finding of our study was the discovery of a glucocorticoid-repressed and IFN-γ–induced host defense network linked to phagosome maturation and lytic vacuole/lysosome, which we identified the v-ATPase subunit a3 TCIRG1 as a top hub gene. A central role for the v-ATPase V0 in the host response to mycobacterial infections was suggested by Russell and colleagues (41), who demonstrated that the v-ATPase V0 is excluded from the nascent mycobacteria phagosome. In this study we found that IFN-γ induces TCIRG1 expression (42), its recruitment to bacterial-loaded vacuoles, and promoted lysosome acidification (23, 43–45). We previously reported that TCIRG1, in concert with other subunits of the v-ATPase V0, was induced and recruited to the mycobacterial phagolysosome by pharmacological activation with imatinib, resulting in phagosome maturation and lysosome acidification, and subsequent antimicrobial activity against M. tuberculosis in human macrophages (23). The ability of imatinib to trigger lysosome acidification in glucocorticoid-treated macrophages identifies a strategy for augmenting antimicrobial responses in the context of glucocorticoid treatment.
We observed that in dexamethasone-treated macrophages cathelicidin colocalizes with intracellular mycobacteria. Therefore, the crucial role of phagolysosome maturation and acidification in the antimicrobial response could mean that cathelicidin-mediated killing is pH dependent. Another, yet not mutually exclusive explanation is that dexamethasone-induced cathelicidin is not efficiently processed from its inactive precursor hCAP18 into the active peptide LL-37. Despite the pivotal role of cathelicidin in the macrophage host response, only few mechanisms involved in regulating its expression are known. The human cathelicidin promoter contains three vitamin D response elements, and 1,25D is a direct inducer of cathelicidin (12, 13). Because physiological 1,25D serum levels are not sufficient to trigger the VDR-mediated induction of cathelicidin, macrophages rely on the intracellular conversion of 25D to 1,25D by the CYP27B1-hydroxylase. In fact, described immune mechanisms resulting in upregulation of cathelicidin gene expression to date, including activation by TLR2/1 ligand and IFN-γ, converge on the induction of CYP27B1 and the conversion of 25D to 1,25D (6, 7). However, glucocorticoid induction of cathelicidin was not linked to CYP27B1 induction, nor was it dependent on serum 25D levels. Thus, glucocorticoid induction of cathelicidin does not involve the previously described vitamin D pathway. The detailed molecular mechanisms by which glucocorticoids induce cathelicidin remain to be investigated. One mechanism, by which glucocorticoids induce transcription, is via GR binding to GR response elements on target genes. GR response elements are found within the promoter, introns, or exons of target genes (1). However, the human cathelicidin gene does not contain GR binding sites based on the DNA consensus sequences 5′∓2GGTACAnnnTGTTCT-3′ (46) and 5′-AGAA-CAnnnTGTTCT-3′ (1). Notably, besides the vitamin D–mediated induction, which is amplified by increased histone acetylation (47), little is known about pathways activating cathelicidin. In keratinocytes, but not myeloid cells, endoplasmic reticulum stress or resveratrol were shown to trigger induction of sphingosine-1-phosphate, which in turn activates NF-κB–mediated activation of C/EBP leading to cathelicidin production (48, 49). In human macrophages activated with dexamethasone, we observed repression of the NF-κB and NF-κB signaling pathways. This suggests that the induction of cathelicidin by dexamethasone is not mediated via this pathway in human macrophages. Nevertheless, induction of cathelicidin by glucocorticoids seems not to be macrophage specific, because microarray analyses showed that dexamethasone also triggered cathelicidin gene expression in the human lung A549 cell line (50).
An important mechanism of phagolysosome maturation and acidification is the induction of autophagy (7, 8, 35, 36). Consistently, our data showed that inhibition of autophagy by 3-MA and Wortmannin blunts IFN-γ–induced lysosome acidification. In addition, glucocorticoids reportedly did not trigger autophagy in M. tuberculosis–infected human monocytic cells (37). In line with these findings, the green module, correlated to IFN-γ stimulation and inversely correlated with glucocorticoid treatment of macrophages, was strongly associated with the IPA biofunctional category autophagy. Among autophagy-associated genes in our analysis of the green module, DRAM1, which has been used as a marker of autophagy (38), had the highest hierarchical position. Consistently, IFN-γ, but not glucocorticoids, promoted induction of DRAM1. Recently, DRAM1 was identified as a key host defense gene in antimycobacterial host defense in zebrafish and human macrophages (33). DRAM1 was found to trigger selective autophagy downstream of TLR signaling promoting mycobacterial phagosome maturation, phagolysosome fusion, and anti-mycobacterial activity (33). Knockdown of DRAM1 inhibits the v-ATPase and acidification of lysosomes (51). Thus, it is tempting to speculate that DRAM1 links IFN-γ induction of autophagy, TCIRG1, and acidification of lysosomes.
There is a great interest in the biomedical community regarding the development of host-directed therapies to treat infections. In TB, for instance, the induction of cathelicidin, autophagy, and phagosome maturation are potential targets (52). In our study the ability of imatinib to trigger an antimicrobial response was performed using an avirulent mycobacterium, such that future studies are required to examine the role of imatinib against virulent mycobacteria. Nevertheless, imatinib has been demonstrated effective as a host-directed therapy in a mouse model of TB infection (39, 40) and is currently being developed for a TB clinical trial (53). Future studies in animal models and clinical trials should investigate the effectiveness of imatinib in combination with glucocorticoids in the host-directed treatment of mycobacterial infections. This may be particularly useful in the management of inflammatory reactions (54–58), including immune reconstitution inflammatory syndrome, which occurs in HIV/TB-coinfected individuals at the initiation of chemotherapy and which is treated with glucocorticoids (59).
Although we demonstrate the role of glucocorticoids in modulating the innate macrophage antimicrobial response, there is evidence for immunosuppressive effects on innate instruction of the adaptive T cell response as well as a direct effect on T cells (60). In this context, glucocorticoid therapy increases the likelihood of false-negative or indeterminate results of the IFN-γ release assay used to diagnose TB in humans (61, 62). Although our data indicate that imatinib can overcome the deleterious effects of glucocorticoids on innate antimicrobial responses, imatinib is also known to suppress adaptive T cell responses (23). Nevertheless, there are only sporadic reports of infections in imatinib-treated patients (23). In this regard, our study, by showing that imatinib promotes lysosome acidification and antimicrobial activity in glucocorticoid-stimulated macrophages without reversing glucocorticoid-mediated anti-inflammatory effects, suggests one promising strategy to maintain innate antimicrobial activity in patients receiving glucocorticoids for treatment of chronic inflammatory diseases.
Supplementary Material
Acknowledgments
This work was supported by the Ministry of Innovation, Science, Research and Technology of the German State of North Rhine-Westphalia, the Center for Molecular Medicine (B4), the Deutsche Forschungsgemeinschaft (Grant SFB829), and the Juergen Manchot Foundation (to R.K.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Anja Niehoff for excellent support regarding the statistical analyses, Julian Nüchel for help with automated image analyses, Birgit Gathof for providing buffy coats, and all blood donors who participated in this study.
Abbreviations used in this article
- BCG
Mycobacterium bovis Bacille Calmette-Guérin
- 1,25D
1,25-di-OH vitamin D
- 25D
25-OH vitamin D
- GR
glucocorticoid receptor
- IPA
Ingenuity Pathway Analysis
- MDM
monocyte-derived macrophage
- ME
module eigengene
- ΔMFI
Δ mean fluorescence intensity
- SFM
serum-free media
- TB
tuberculosis
- v-ATPase
vacuolar H+-ATPase
- VDR
vitamin D receptor
- WGCNA
weighted gene coexpression network analysis
Footnotes
ORCIDs: 0000-0002-1772-510X (S.R.); 0000-0002-3378-2067 (R.K.); 0000-0003-4174-3268 (P.H.); 0000-0001-5471-0807 (S.S.); 0000-0003-4720-031X (R.L.M.).
The array data presented in this article have been submitted to the Gene Expression Omnibus database (http://www.mcbi.nlm.nih.gov/geo/) under accession number GSE79077.
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Busillo JM, Cidlowski JA. The five Rs of glucocorticoid action during inflammation: ready, reinforce, repress, resolve, and restore. Trends Endocrinol. Metab. 2013;24:109–119. doi: 10.1016/j.tem.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horsburgh CR, Jr, Rubin EJ. Clinical practice. Latent tuberculosis infection in the United States. N. Engl. J. Med. 2011;364:1441–1448. doi: 10.1056/NEJMcp1005750. [DOI] [PubMed] [Google Scholar]
- 3.Kothavade RJ, Dhurat RS, Mishra SN, Kothavade UR. Clinical and laboratory aspects of the diagnosis and management of cutaneous and subcutaneous infections caused by rapidly growing mycobacteria. Eur. J. Clin. Microbiol. Infect. Dis. 2013;32:161–188. doi: 10.1007/s10096-012-1766-8. [DOI] [PubMed] [Google Scholar]
- 4.Busillo JM, Azzam KM, Cidlowski JA. Glucocorticoids sensitize the innate immune system through regulation of the NLRP3 inflammasome. J. Biol. Chem. 2011;286:38703–38713. doi: 10.1074/jbc.M111.275370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van de Garde MD, Martinez FO, Melgert BN, Hylkema MN, Jonkers RE, Hamann J. Chronic exposure to glucocorticoids shapes gene expression and modulates innate and adaptive activation pathways in macrophages with distinct changes in leukocyte attraction. J. Immunol. 2014;192:1196–1208. doi: 10.4049/jimmunol.1302138. [DOI] [PubMed] [Google Scholar]
- 6.Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, Ochoa MT, Schauber J, Wu K, Meinken C, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311:1770–1773. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 7.Fabri M, Stenger S, Shin DM, Yuk JM, Liu PT, Realegeno S, Lee HM, Krutzik SR, Schenk M, Sieling PA, et al. Vitamin D is required for IFN-gamma-mediated antimicrobial activity of human macrophages. Sci. Transl. Med. 2011;3:104ra102. doi: 10.1126/scitranslmed.3003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klug-Micu GM, Stenger S, Sommer A, Liu PT, Krutzik SR, Modlin RL, Fabri M. CD40 ligand and interferon-γ induce an antimicrobial response against Mycobacterium tuberculosis in human monocytes. Immunology. 2013;139:121–128. doi: 10.1111/imm.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edfeldt K, Liu PT, Chun R, Fabri M, Schenk M, Wheelwright M, Keegan C, Krutzik SR, Adams JS, Hewison M, Modlin RL. T-cell cytokines differentially control human monocyte antimicrobial responses by regulating vitamin D metabolism. Proc. Natl. Acad. Sci. USA. 2010;107:22593–22598. doi: 10.1073/pnas.1011624108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montoya D, Inkeles MS, Liu PT, Realegeno S, Teles RM, Vaidya P, Munoz MA, Schenk M, Swindell WR, Chun R, et al. IL-32 is a molecular marker of a host defense network in human tuberculosis. Sci. Transl. Med. 2014;6:250ra114. doi: 10.1126/scitranslmed.3009546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krutzik SR, Hewison M, Liu PT, Robles JA, Stenger S, Adams JS, Modlin RL. IL-15 links TLR2/1-induced macrophage differentiation to the vitamin D-dependent antimicrobial pathway. J. Immunol. 2008;181:7115–7120. doi: 10.4049/jimmunol.181.10.7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang TT, Nestel FP, Bourdeau V, Nagai Y, Wang Q, Liao J, Tavera-Mendoza L, Lin R, Hanrahan JW, Mader S, White JH. Cutting edge: 1,25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J. Immunol. 2004;173:2909–2912. doi: 10.4049/jimmunol.173.5.2909. [DOI] [PubMed] [Google Scholar]
- 13.Gombart AF, Borregaard N, Koeffler HP. Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up-regulated in myeloid cells by 1,25-dihydroxyvitamin D3. FASEB J. 2005;19:1067–1077. doi: 10.1096/fj.04-3284com. [DOI] [PubMed] [Google Scholar]
- 14.Liu PT, Stenger S, Tang DH, Modlin RL. Cutting edge: vitamin D-mediated human antimicrobial activity against Mycobacterium tuberculosis is dependent on the induction of cathelicidin. J. Immunol. 2007;179:2060–2063. doi: 10.4049/jimmunol.179.4.2060. [DOI] [PubMed] [Google Scholar]
- 15.Yuk JM, Shin DM, Lee HM, Yang CS, Jin HS, Kim KK, Lee ZW, Lee SH, Kim JM, Jo EK. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe. 2009;6:231–243. doi: 10.1016/j.chom.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Campbell GR, Spector SA. Vitamin D inhibits human immunodeficiency virus type 1 and Mycobacterium tuberculosis infection in macrophages through the induction of autophagy. PLoS Pathog. 2012;8:e1002689. doi: 10.1371/journal.ppat.1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armstrong JA, Hart PD. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J. Exp. Med. 1971;134:713–740. doi: 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deghmane AE, Soualhine H, Bach H, Sendide K, Itoh S, Tam A, Noubir S, Talal A, Lo R, Toyoshima S, et al. Lipoamide dehydrogenase mediates retention of coronin-1 on BCG vacuoles, leading to arrest in phagosome maturation. J. Cell Sci. 2007;120:2796–2806. doi: 10.1242/jcs.006221. [DOI] [PubMed] [Google Scholar]
- 19.Sun J, Deghmane AE, Soualhine H, Hong T, Bucci C, Solodkin A, Hmama Z. Mycobacterium bovis BCG disrupts the interaction of Rab7 with RILP contributing to inhibition of phagosome maturation. J. Leukoc. Biol. 2007;82:1437–1445. doi: 10.1189/jlb.0507289. [DOI] [PubMed] [Google Scholar]
- 20.Via LE, Deretic D, Ulmer RJ, Hibler NS, Huber LA, Deretic V. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J. Biol. Chem. 1997;272:13326–13331. doi: 10.1074/jbc.272.20.13326. [DOI] [PubMed] [Google Scholar]
- 21.Weiss G, Schaible UE. Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 2015;264:182–203. doi: 10.1111/imr.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell DG. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2001;2:569–577. doi: 10.1038/35085034. [DOI] [PubMed] [Google Scholar]
- 23.Bruns H, Stegelmann F, Fabri M, Döhner K, van Zandbergen G, Wagner M, Skinner M, Modlin RL, Stenger S. Abelson tyrosine kinase controls phagosomal acidification required for killing of Mycobacterium tuberculosis in human macrophages. J. Immunol. 2012;189:4069–4078. doi: 10.4049/jimmunol.1201538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bastian M, Heymann S, Jacomy M. Gephi: An Open Source Software for Exploring and Manipulating Networks. International AAAI Conference on Weblogs and Social Media; 2009. [Accessed: May 14, 2016]. Available at: https://gephi.org. [Google Scholar]
- 25.Chan J, Xing Y, Magliozzo RS, Bloom BR. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J. Exp. Med. 1992;175:1111–1122. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan J, Tanaka K, Carroll D, Flynn J, Bloom BR. Effects of nitric oxide synthase inhibitors on murine infection with Mycobacterium tuberculosis. Infect. Immun. 1995;63:736–740. doi: 10.1128/iai.63.2.736-740.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thoma-Uszynski S, Stenger S, Takeuchi O, Ochoa MT, Engele M, Sieling PA, Barnes PF, Rollinghoff M, Bolcskei PL, Wagner M, et al. Induction of direct antimicrobial activity through mammalian toll-like receptors. Science. 2001;291:1544–1547. doi: 10.1126/science.291.5508.1544. [DOI] [PubMed] [Google Scholar]
- 28.Kunz D, Walker G, Eberhardt W, Pfeilschifter J. Molecular mechanisms of dexamethasone inhibition of nitric oxide synthase expression in interleukin 1 beta-stimulated mesangial cells: evidence for the involvement of transcriptional and posttranscriptional regulation. Proc. Natl. Acad. Sci. USA. 1996;93:255–259. doi: 10.1073/pnas.93.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc. Natl. Acad. Sci. USA. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu PT, Schenk M, Walker VP, Dempsey PW, Kanchanapoomi M, Wheelwright M, Vazirnia A, Zhang X, Steinmeyer A, Züugel U, et al. Convergence of IL-1beta and VDR activation pathways in human TLR2/1-induced antimicrobial responses. PLoS One. 2009;4:e5810. doi: 10.1371/journal.pone.0005810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dimitrov V, White JH. Species-specific regulation of innate immunity by vitamin D signaling. J. Steroid Biochem. Mol. Biol. 2015 doi: 10.1016/j.jsbmb.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 32.Wilson CG, May CS, Paterson JW. Plasma prednisolone levels in man following administration in plain and enteric-coated forms. Br. J. Clin. Pharmacol. 1977;4:351–355. doi: 10.1111/j.1365-2125.1977.tb00723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Vaart M, Korbee CJ, Lamers GE, Tengeler AC, Hosseini R, Haks MC, Ottenhoff TH, Spaink HP, Meijer AH. The DNA damage-regulated autophagy modulator DRAM1 links mycobacterial recognition via TLR-MYD88 to autophagic defense [corrected] Cell Host Microbe. 2014;15:753–767. doi: 10.1016/j.chom.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Sun-Wada GH, Tabata H, Kawamura N, Aoyama M, Wada Y. Direct recruitment of H+-ATPase from lysosomes for phagosomal acidification. J. Cell Sci. 2009;122:2504–2513. doi: 10.1242/jcs.050443. [DOI] [PubMed] [Google Scholar]
- 35.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 36.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 37.Bongiovanni B, Mata-Espinosa D, D’Attilio L, Leon-Contreras JC, Marquez-Velasco R, Bottasso O, Hernandez-Pando R, Bay ML. Effect of cortisol and/or DHEA on THP1-derived macrophages infected with Mycobacterium tuberculosis. Tuberculosis (Edinb.) 2015;95:562–569. doi: 10.1016/j.tube.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 38.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Napier RJ, Rafi W, Cheruvu M, Powell KR, Zaunbrecher MA, Bornmann W, Salgame P, Shinnick TM, Kalman D. Imatinib-sensitive tyrosine kinases regulate mycobacterial pathogenesis and represent therapeutic targets against tuberculosis. Cell Host Microbe. 2011;10:475–485. doi: 10.1016/j.chom.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Napier RJ, Norris BA, Swimm A, Giver CR, Harris WA, Laval J, Napier BA, Patel G, Crump R, Peng Z, et al. Low doses of imatinib induce myelopoiesis and enhance host anti-microbial immunity. PLoS Pathog. 2015;11:e1004770. doi: 10.1371/journal.ppat.1004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, Allen RD, Gluck SL, Heuser J, Russell DG. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- 42.Jutras I, Houde M, Currier N, Boulais J, Duclos S, LaBoissière S, Bonneil E, Kearney P, Thibault P, Paramithiotis E, et al. Modulation of the phagosome proteome by interferon-gamma. Mol. Cell. Proteomics. 2008;7:697–715. doi: 10.1074/mcp.M700267-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Jung JY, Robinson CM. IL-12 and IL-27 regulate the phagolysosomal pathway in mycobacteria-infected human macrophages. Cell Commun. Signal. 2014;12:16. doi: 10.1186/1478-811X-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montaner LJ, da Silva RP, Sun J, Sutterwala S, Hollinshead M, Vaux D, Gordon S. Type 1 and type 2 cytokine regulation of macrophage endocytosis: differential activation by IL-4/IL-13 as opposed to IFN-gamma or IL-10. J. Immunol. 1999;162:4606–4613. [PubMed] [Google Scholar]
- 45.Via LE, Fratti RA, McFalone M, Pagan-Ramos E, Deretic D, Deretic V. Effects of cytokines on mycobacterial phagosome maturation. J. Cell Sci. 1998;111:897–905. doi: 10.1242/jcs.111.7.897. [DOI] [PubMed] [Google Scholar]
- 46.Beato M, Chalepakis G, Schauer M, Slater EP. DNA regulatory elements for steroid hormones. J. Steroid Biochem. 1989;32:737–747. doi: 10.1016/0022-4731(89)90521-9. [DOI] [PubMed] [Google Scholar]
- 47.Schauber J, Oda Y, Büuchau AS, Yun QC, Steinmeyer A, Züugel U, Bikle DD, Gallo RL. Histone acetylation in keratinocytes enables control of the expression of cathelicidin and CD14 by 1,25-dihydroxyvitamin D3. J. Invest. Dermatol. 2008;128:816–824. doi: 10.1038/sj.jid.5701102. [DOI] [PubMed] [Google Scholar]
- 48.Park K, Elias PM, Oda Y, Mackenzie D, Mauro T, Holleran WM, Uchida Y. Regulation of cathelicidin antimicrobial peptide expression by an endoplasmic reticulum (ER) stress signaling, vitamin D receptor-independent pathway. J. Biol. Chem. 2011;286:34121–34130. doi: 10.1074/jbc.M111.250431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park K, Elias PM, Hupe M, Borkowski AW, Gallo RL, Shin KO, Lee YM, Holleran WM, Uchida Y. Resveratrol stimulates sphingosine-1-phosphate signaling of cathelicidin production. J. Invest. Dermatol. 2013;133:1942–1949. doi: 10.1038/jid.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lannan EA, Galliher-Beckley AJ, Scoltock AB, Cidlowski JA. Proinflammatory actions of glucocorticoids: glucocorticoids and TNFα coregulate gene expression in vitro and in vivo. Endocrinology. 2012;153:3701–3712. doi: 10.1210/en.2012-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang XD, Qi L, Wu JC, Qin ZH. DRAM1 regulates autophagy flux through lysosomes. PLoS One. 2013;8:e63245. doi: 10.1371/journal.pone.0063245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallis RS, Hafner R. Advancing host-directed therapy for tuberculosis. Nat. Rev. Immunol. 2015;15:255–263. doi: 10.1038/nri3813. [DOI] [PubMed] [Google Scholar]
- 53. [Accessed: May 14, 2016];Development of Gleevec for TB and TB/HIV. Available at: https://projectreporter.nih.gov//project_info_description.cfm?aid=9040684&icde=0.
- 54.Critchley JA, Young F, Orton L, Garner P. Corticosteroids for prevention of mortality in people with tuberculosis: a systematic review and meta-analysis. Lancet Infect. Dis. 2013;13:223–237. doi: 10.1016/S1473-3099(12)70321-3. [DOI] [PubMed] [Google Scholar]
- 55.Strang JI, Kakaza HH, Gibson DG, Girling DJ, Nunn AJ, Fox W. Controlled trial of prednisolone as adjuvant in treatment of tuberculous constrictive pericarditis in Transkei. Lancet. 1987;2:1418–1422. doi: 10.1016/s0140-6736(87)91127-5. [DOI] [PubMed] [Google Scholar]
- 56.Thwaites GE, Nguyen DB, Nguyen HD, Hoang TQ, Do TT, Nguyen TC, Nguyen QH, Nguyen TT, Nguyen NH, Nguyen TN, et al. Dexamethasone for the treatment of tuberculous meningitis in adolescents and adults. N. Engl. J. Med. 2004;351:1741–1751. doi: 10.1056/NEJMoa040573. [DOI] [PubMed] [Google Scholar]
- 57.Van Veen NH, Lockwood DN, Van Brakel WH, Ramirez J, Jr, Richardus JH. Interventions for erythema nodosum leprosum. A Cochrane review. Lepr. Rev. 2009;80:355–372. [PubMed] [Google Scholar]
- 58.Critchley JA, Orton LC, Pearson F. Adjunctive steroid therapy for managing pulmonary tuberculosis. Cochrane Database Syst. Rev. 2014;11:CD011370. doi: 10.1002/14651858.CD011370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meintjes G, Skolimowska KH, Wilkinson KA, Matthews K, Tadokera R, Conesa-Botella A, Seldon R, Rangaka MX, Rebe K, Pepper DJ, et al. Corticosteroid-modulated immune activation in the tuberculosis immune reconstitution inflammatory syndrome. Am. J. Respir. Crit. Care Med. 2012;186:369–377. doi: 10.1164/rccm.201201-0094OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coutinho AE, Chapman KE. The anti-inflammatory and immunosuppressive effects of glucocorticoids, recent developments and mechanistic insights. Mol. Cell. Endocrinol. 2011;335:2–13. doi: 10.1016/j.mce.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Calabrese C, Overman RA, Dusetzina SB, Hajj-Ali RA. Evaluating indeterminate interferon-γ-release assay results in patients with chronic inflammatory diseases receiving immunosuppressive therapy. Arthritis Care Res. (Hoboken) 2015;67:1063–1069. doi: 10.1002/acr.22454. [DOI] [PubMed] [Google Scholar]
- 62.Clifford V, Zufferey C, Germano S, Ryan N, Leslie D, Street A, Denholm J, Tebruegge M, Curtis N. The impact of anti-tuberculous antibiotics and corticosteroids on cytokine production in QuantiFERON-TB Gold In Tube assays. Tuberculosis (Edinb.) 2015;95:343–349. doi: 10.1016/j.tube.2015.02.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.