Abstract
Objective
Oesophageal squamous cell carcinoma (ESCC) is a heterogeneous disease with variable outcomes that are challenging to predict. A better understanding of the biology of ESCC recurrence is needed to improve patient care. Our goal was to identify small non-coding RNAs (sncRNAs) that could predict the likelihood of recurrence after surgical resection and to uncover potential molecular mechanisms that dictate clinical heterogeneity.
Design
We developed a robust prediction model for recurrence based on the analysis of the expression profile data of sncRNAs from 108 fresh frozen ESCC specimens as a discovery set and assessment of the associations between sncRNAs and recurrence-free survival (RFS). We also evaluated the mechanistic and therapeutic implications of sncRNA obtained through integrated analysis from multiple data sets.
Results
We developed a risk assessment score for recurrence (RAS) with three sncRNAs (miR-223, miR-1269a, and nc886) whose expression was significantly associated with RFS in the discovery cohort (N=108). RAS was validated in an independent cohort of 512 patients. In multivariable analysis, RAS was an independent predictor of recurrence (hazard ratio, 2.27; 95% confidence interval, 1.26 to 4.09; P=0.007). This signature implies the expression of ΔNp63 and multiple alterations of driver genes like PIK3CA. We suggested therapeutic potentials of immune checkpoint inhibitors in low-risk patients, and Polo-like kinase, mTOR, and HDAC inhibitors in high-risk patients.
Conclusion
We developed an easy-to-use prognostic model with three sncRNAs as robust prognostic markers for postoperative recurrence of ESCC. We anticipate that such a stratified and systematic, tumor-specific biologic approach will potentially contribute to significant improvement in ESCC treatment.
Keywords: non-coding RNA, oesophageal cancer, recurrence, prognostic biomarker, TP63, mass cytometry
INTRODUCTION
Oesophageal cancer (ESCA) is the eighth most common cancer worldwide. Of the 456,000 new cases estimated in 2012, 398,000 were squamous cell carcinoma (ESCC), 52,000 adenocarcinoma (EAC), and 6,000 other carcinomas. ESCA results in about 400,000 deaths, making the disease the sixth leading cause of cancer-related deaths.1,2 ESCC is particularly prevalent in Asia and Africa where it accounts of more than 90% of all ESCA cases.3 Although combination therapy with surgery, chemotherapy, and radiotherapy has improved survival of ESCC, the five-year overall survival (OS) rate is at best 20%.1
Reliable and reproducible prognostic markers identifying patients at high risk of ESCC recurrence after surgery have not been established. In recent decades, non-coding RNAs (ncRNAs), especially microRNAs (miRNAs), have been found to play important roles in carcinogenesis and tumor progression. For ESCC, several miRNAs4–7 and long ncRNAs8 have been investigated as biomarkers or therapeutic targets. The translation of these findings into clinical approaches should be supported by robust validation in large independent patient cohorts and a better understanding of the underlying biology associated with ncRNAs.
In this study, we applied systems biology approaches to evaluate the expression patterns of small ncRNAs (sncRNAs) and develop and validate a robust prognostic prediction model for ESCC. We propose molecular mechanisms and potential therapeutics dictating clinical outcomes that were well reflected in the prediction model.
METHODS
PATIENTS AND STUDY PROCEDURES
Eligibility and exclusion criteria
We enrolled patients who: (1) had histologically confirmed ESCC confined to the thorax, (2) had undergone only complete esophagectomy with adequate lymph node dissection, (3) had available paired tumor and adjacent normal tissue in the tumor bank (discovery set) or formalin-fixed, paraffin-embedded (FFPE) tissue block (validation set), and (4) were followed-up completely.
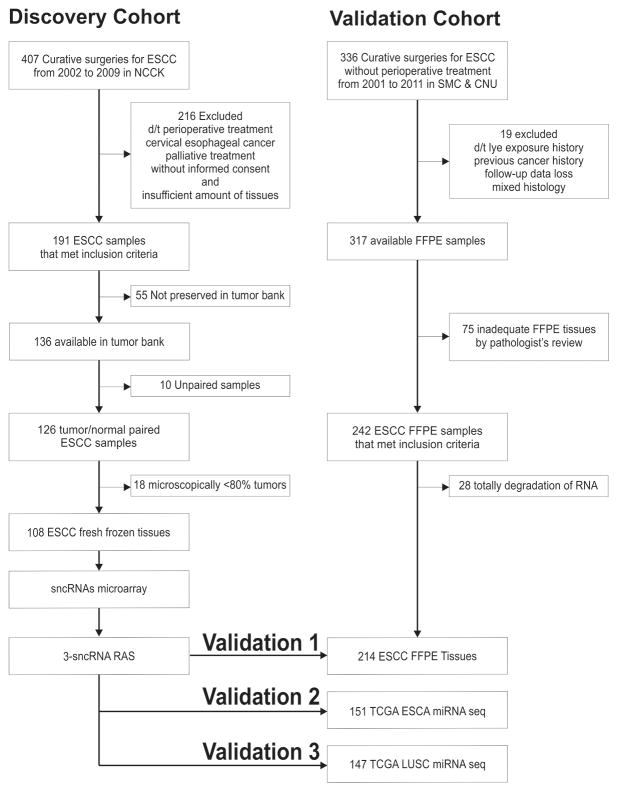
The patient enrollment process and study scheme are illustrated in figure 1 and supplementary figure S1. The clinical and pathologic characteristics of the discovery and validation sets appear in supplementary table S1. To generate the sncRNA expression profile (supplementary table S2), we obtained 108 pairs of fresh frozen ESCC and adjacent normal tissue samples from surgically resected specimens collected between 2002 and 2009 at the National Cancer Center Korea (discovery set). We included samples from patients with thoracic ESCC who underwent complete oesophagectomy with adequate lymph node dissection and did not receive any perioperative chemotherapy or radiotherapy. For external validation, we used 214 FFPE ESCC specimens collected at Samsung Medical Center and Chonnam National University between 2001 and 2011. For external validation in the Western cohorts, we utilized The Cancer Genome Atlas (TCGA) Database. All human samples were collected with informed patient consent, and the study protocol was approved by each institution’s ethics committee.
Figure 1. Patient cohort enrollment.
Summary of the clinical study design. Left panel: the discovery cohort enrollment comprised 108 ESCC samples obtained from fresh frozen tissues in NCCK. Right panel: independent validation processes for other 214 ESCCs, 151 TCGA ESCAs, and 147 TCGA LUSCs.
ESCA denotes oesophageal cancer, ESCC oesophageal squamous cell carcinoma, LUSC lung squamous cell carcinoma, NCCK National Cancer Center Korea, sncRNAs small non-coding RNAs, RAS Risk Assessment Score for Recurrence, SMC Samsung Medical Center, CNU Chonnam National University, FFPE formalin-fixed, paraffin-embedded, and TCGA The Cancer Genome Atlas.
Determination of sample size in the discovery and validation sets
The discovery study was designed to detect sufficient difference in the five-year recurrence-free survival (RFS) rate between the low- and high-risk groups, as defined by the GeneChip miRNA 2.0 array (Affymetrix®, Santa Clara, CA, USA). The sample size and power calculations for the discovery study were determined in two ways considering the difference in survival and fold change. First, we determined that a total of 52 events were needed to predict survival using log2 sncRNA expression with a standard deviation of 1.67 according to our own experimental data and to detect a hazard ratio of 1.5 with 80% power. Since the internally validated five-year RFS rate was about 50%, a total of 104 samples were needed to observe 52 events. With this sample size, the study had 90% power to detect a five-year RFS rate difference of 30% from the 30% reference rate of the high-risk group defined by sncRNA expression at a two-sided type I error rate of 0.05. Second, the study was powered to detect significantly over- or under-expressed sncRNA in cancer cells compared with normal cells. To detect two-fold expression change with 90% power at a multiple testing corrected type I error rate of 0.05/900, a total of 79 samples were needed. Accounting for about 10% data loss to experimental failure and drop-outs, a total sample size of over 90 was needed. According to these two calculations, we sought to study a sample size of over 100 for the discovery study.
The validation study was designed to detect sufficient difference in the five-year RFS rate between the low- and high-risk groups, as defined by the three candidates of sncRNAs found in the discovery set. We assumed that the proportion of the low-risk group would be about 30%. Moreover, the internally validated five-year RFS rate of the high-risk group was about 30%, whereas the RFS rate of the low-risk group was over 70%. We therefore powered the study to detect a 30% five-year RFS rate difference from the 30% reference rate of the high-risk group, a conservative effect of size derived from the previous finding. To have 90% power at a two-sided 5% type I error rate, the study required 190 patients (57 in the low-risk group and 133 in the high-risk group) to observe 104 events. Accounting for data loss (about 10%) to experimental failure and dropouts, we set the sample size of the study to exceed 210.
Small non-coding RNA microarray hybridization and analysis
For sncRNA expression array analysis, we used a GeneChip miRNA 2.0 array. We labeled 1 μg of total RNA with the FlashTag Biotin HSR RNA Labeling Kit (Genisphere LLC, Hatfield, PA, USA) following the manufacturer’s recommendations. Then, labeled sncRNA was hybridized to the array and incubated as described in the manufacturer’s protocol (Affymetrix®). Unsupervised hierarchical clustering of sncRNA expression data from ESCC and adjacent normal tissues in the discovery set revealed marked distinctions between tumor tissues and normal tissues, suggesting that the expression of sncRNA well reflects the molecular characteristics of ESCC tumors (supplementary figure S2).
Selection of reference gene for normalization of quantitative real time-polymerase chain reaction
We adopted a normalization strategy with multiple reference genes, which was proposed in a previous study,9 to overcome the limitations of using a single reference gene for the normalization of miRNA expression from quantitative reverse transcription polymerase chain reaction (qRT-PCR) experiments.9–13 We first assessed the gene expression stability of putative normalizer genes with geNorm9 software in microarray data. Then, we selected two miRNAs (miR-132 and miR-652) with high expression stability (supplementary figure S3) and calculated the mean value of the two miRNAs as the normalizer of miRNAs.
To determine the reference gene for the normalization of nc886 expression, we searched previous studies of ncRNAs and selected three major reference genes: β-actin (ACTB), cyclophilin A (PPIA),14 and small nuclear RNA 6 (RNU6). We computed the expression stability with geNorm5 and selected endogenous gene, PPIA, because it showed a relatively stable expression in ESCC.
Integrative analysis from the TCGA dataset
To determine how the sncRNA signature is conserved in esophageal cancer from the Western cohort and the lung squamous cell carcinoma set, we obtained miRNA sequencing data of esophageal cancer (ESCA) and lung squamous cell carcinoma (LUSC) from The Cancer Genome Atlas (TCGA) portal (https://tcga-data.nci.nih.gov/tcga/). Normalized reads per kilobase per million values were transformed logarithmically and centralized by subtracting the mean of each channel, and further normalized to equalize the variance. To analyze TCGA and discovery set data together, we normalized the expression data from the discovery cohort, as previously described herein, and divided the data by the standard deviation of the channel to determine which cancer type was more similar to ESCC in terms of sncRNA expression.
We leveraged the ESCA (n=151) and LUSC data (n=147) including mRNA and miRNA sequencing, somatic mutation, DNA copy number alteration and clinical data from the TCGA portal and the cBioPortal for Cancer Genomics (http://www.cbioportal.org/public-portal/index.do).15 To visualize genomic alterations in multiple genes, we used OncoPrint generated by cBioPortal for Cancer Genomics.15 Then, we analyzed the frequency of genetic alteration in major genes. The experimental and analytical methods for generating sncRNA and mRNA expression profile data and the integrated analyses are in the Supplementary Appendix.
STATISTICAL ANALYSIS
Student t tests, chi-square tests, and Fisher exact tests were used to compare clinico-pathological data. Survival curves were generated with the Kaplan-Meier’s method and intergroup comparisons were performed with the log-rank test. Our primary endpoint was recurrence-free survival (RFS). Univariable Cox regression analysis was used to determine whether sncRNAs were associated with RFS. We performed multivariable Cox proportional hazards regression analysis to evaluate independent prognostic factors associated with survival, and we used gene signature, tumor-node-metastasis (TNM) stage, smoking, age, and sex as variables. We used data from the discovery cohort to develop a formula for a risk assessment score based on a linear combination of the expression level of sncRNAs weighed by regression coefficients derived from multivariable Cox regression analyses. We analyzed correlations between microarray data and quantitative real time-polymerase chain reaction (qRT-PCR) data with the Spearman correlation coefficient. To select genes with statistically significant alterations from integrated analysis of data from TCGA, we used chi-square or Fisher exact test. Statistical significance was accepted as P<0.05, and all tests were two-tailed. All statistical analyses were performed with SPSS 20.0 (SPSS, Inc., Chicago, Illinois) and R language and software environment (http://www.r-project.org).
RESULTS
Development of risk assessment score for ESCC recurrence with sncRNAs
Multi-step analyses were carried out to identify robust, prognostic sncRNAs from ESCC tumors. First, among 901 sncRNA with differential expression between tumors and adjacent normal tissues (supplementary figure S2), we selected sncRNAs whose expression showed at least a twofold difference from the median value of at least 30 samples, with a standard deviation > 1. This selection yielded 162 sncRNAs. Second, 8 of these 162 sncRNAs were significantly associated (P<0.05) with RFS in univariable Cox regression analysis (supplementary table S3). Finally, backward stepwise selection in multivariable Cox regression analysis revealed that only three sncRNAs (miR-223, miR-1269a, and miR-886-3p) were independent predictors of RFS (P<0.05). High expression of miR-886-3p and miR-223 was significantly associated with longer RFS (P=0.03 and P=0.004, respectively, by the log-rank test), but high expression of miR-1269a was significantly associated with shorter RFS (P=0.045) (supplementary figure S4).
We measured the size of miR-886 via Northern blot analysis of ESCC cell lines and confirmed that miR-886 is not a miRNA but rather a novel sncRNA 101 nucleotides long (supplementary figure S5) (ncRNA886 [hereafter, nc886]).16,17 We re-measured the expression of three sncRNAs (2 miRNAs and 1 sncRNAs) from the same tissues with qRT-PCR. The correlation between the two measurements was significantly high (r>0.6; P<0.001) (supplementary figure S6, and supplementary table S4).
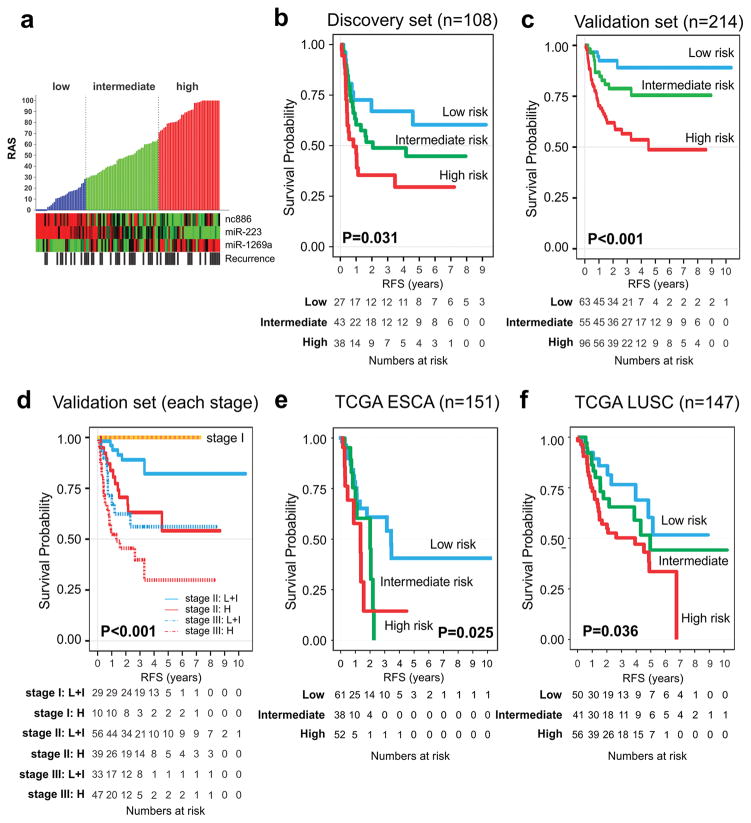
We next developed a recurrence risk assessment model with Cox regression coefficients and the expression level of each sncRNA measured with qRT-PCR: Raw Risk Assessment Score for Recurrence (RASraw)=(−0.084xexpression value of nc886)+(−0.227xexpression value of miR-223)+(0.137xexpression value of miR-1269a). To generate a dynamic range score from 0 to 100, we reformulated the RASraw: RAS=80xeRASraw–24.152. For results less than 0, the score was considered 0, and for results greater than 100, it was considered 100. Cutoff points were specified in advance to reflect prognostic differences observed in the training cohort: low (RAS≤30), intermediate (30<RAS≤65), and high risk (RAS>65) (figure 2A and supplementary figure S7). 18 In the discovery cohort, the five-year RFS rates were 60.8%, 44.8%, and 30.3% for the low-, intermediate-, and high-risk groups, respectively (P=0.031, log-rank test, figure 2B).
Figure 2. Development and validation of the prediction model with recurrence-associated small non-coding RNAs.
(A) Distribution of recurrence assessment score. We constructed a risk score formula with Cox regression coefficients of three sncRNAs. Patients were divided into three categories according to the modified risk prediction model on a scale from 0 to 100 (Panel A, upper). Each column represents an individual patient. The color in cells reflects relative expression levels. Kaplan-Meier curves in discovery (B) and validation sets (C,D) show discrete survival curves according to risk categories. (E,F) recurrence-free survival according to risk categories in TCGA ESCA, and TCGA LUSC, respectively.
ESCA denotes oesophageal cancer, LUSC lung squamous cell carcinoma, RAS Risk Assessment Score for Recurrence, RFS recurrence-free survival, and TCGA The Cancer Genome Atlas.
Validation of RAS in independent cohorts
We sought to validate RAS in an independent, blinded cohort of ESCC patients with RNAs from FFPE tissues and qRT-PCR methods. When patients in the validation cohort (N=214) were stratified according to their RAS with the same cutoff values used in the discovery set, the five-year RFS rate for the low-, intermediate-, and high-risk patient groups were 89.1%, 75.5%, and 48.7%, respectively (P<0.001, log-rank test) (figure 2C). The difference in the survival between discovery and validation cohorts is attributed to accessibility of FFPE specimens in the validation cohort. The validation cohort had earlier cancer staging and lower tumor location than those in the discovery cohort. In each stage, especially stages II and III, the high-risk group had worse prognosis in the validation set (figure 2D). Interestingly, the RFS rate of patients with high risk in stage II was similar to that of patients with low/intermediate-risk in stage III, suggesting RAS is an independent predictor of RFS over stages. In addition, to determine whether this signature is conserved in Western cohorts and in other surgically resected squamous cell carcinomas, we analyzed genomic data of ESCA (N=151) and lung squamous cell carcinoma (LUSC; N=147) from TCGA, because LUSC shares highly similar genetic alterations with ESCC.19–21 Patients with high-RAS levels consistently showed the worst prognosis (figure 2E, figure 2F, and supplementary figure S8). This result shows the robustness of RAS based on three sncRNAs, regardless of the technological platform (supplementary figure S9) or the source of RNAs.
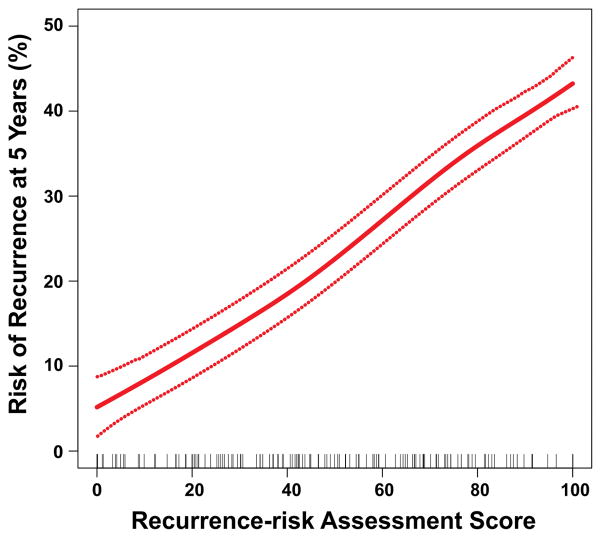
In the validation ESCC cohort, continuous RAS was significantly associated with risk of recurrence (hazard ratio [HR] for a 25-unit increase in score, 1.78; 95% confidence interval, 1.35 to 2.34; P=3.7×10−5; figure 3). In univariable analysis, TNM stage, age, smoking status, and RAS were significant predictors of RFS and OS (supplementary table S5). Multivariable regression analysis revealed that RAS (HR=2.27, P=0.007) and TNM stage (HR=4.13, P<0.001) were independent prognostic factors of RFS and OS in ESCC patients (supplementary table S6).
Figure 3. Relationship between the continuous risk assessment score for recurrence (RAS) and five-year recurrence risk with 95% confidence intervals.
Red dotted lines represent 95% confidence intervals. Rug plots represent the distribution of RAS in the validation cohort.
Integrated analysis based on sncRNA signature
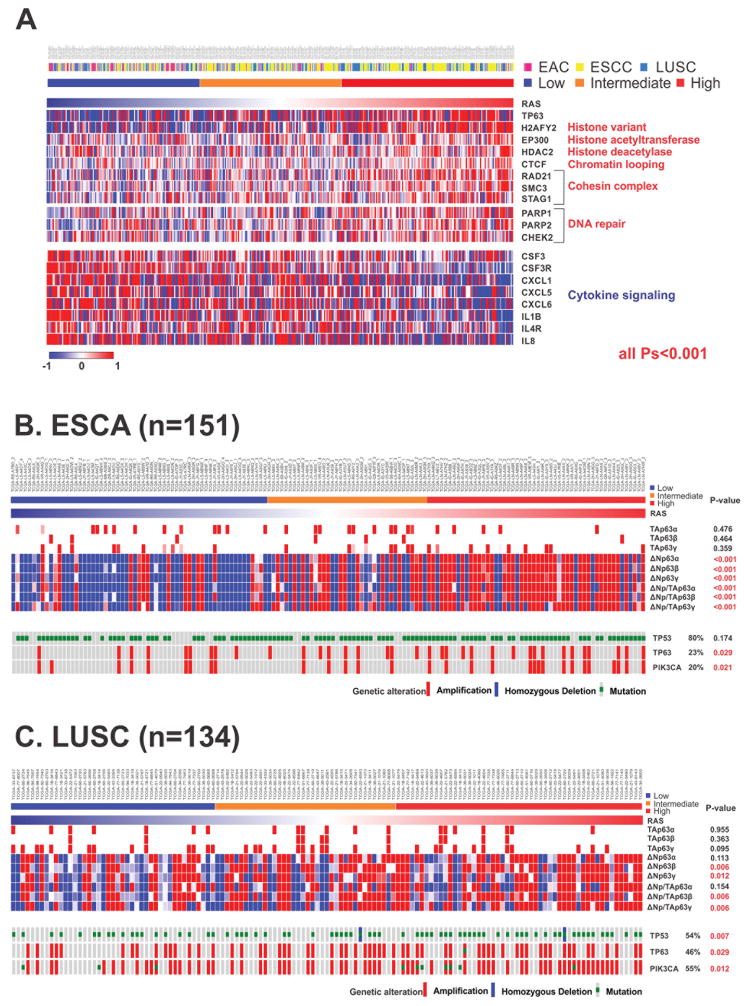
To gain insight into the biology of sncRNAs associated with prognosis, we performed mRNA microarray assays in the discovery set and analyzed mRNA sequencing data of ESCA and LUSC from the TCGA project. Common 613 genes were significantly correlated with RAS in all three cohorts (supplementary table S7 and supplementary figure S10). Enrichment analysis of these genes through gene ontology revealed that genes related to cell mitosis, chromatin remodeling, histone modification, and DNA repair were significantly activated in the high-RAS group, but genes related to cell-to-cell cytokine signaling were dominant in the low-RAS group. Among them, TP63, a marker of squamous histology and a member of the TP53 family, was overexpressed in the high-RAS group. In addition, the expression of genes related to histone modification, such as histone acetyltransferase (HAT), EP300, and histone deacetylase 2 (HDAC2), and the expression of chromatin remodeling genes, such as CTCF, were simultaneously increased. In contrast, the low-RAS group showed up-regulation of immunoregulatory genes with cell-to-cell cytokine signaling functions, such as CSF3, IL8, and IL4R (figure 4A).
Figure 4. Patterns of genetic alteration in oesophageal cancer and lung squamous cell carcinoma according to RAS.
(A) The expression of common 613 genes is correlated with the risk assessment score for recurrence (RAS). (B) Expression of the isoforms of TP63 (TAp63 [tumor suppressive] and ΔNp63 [oncogenic]), TP53 mutation, and copy number alteration of TP63 and PIK3CA in TCGA ESCA. (C) Expression of the isoforms of TP63, TP53 mutation, and copy number alteration of TP63 and PIK3CA in TCGA LUSC.
EAC denotes oesophageal adenocarcinoma, ESCA oesophageal carcinoma, ESCC oesophageal squamous cell carcinoma, LUSC lung squamous cell carcinoma, and RAS Risk Assessment Score for Recurrence.
To elucidate the biological linkage between sncRNA signature and TP63, we analyzed the expression patterns of TP63 isoforms in ESCA and LUSC (n=134) from the TCGA project. We investigated six isoforms of TP63 that have different biological activities. TAp63α, β, and γ with a transcription activation (TA) domain have tumor suppressive activity, but ΔNp63 α, β, and γ without a TA domain have oncogenic activity, as they serve as competitive inhibitors of TAp63α, β, and γ.22 As a result, the expression of oncogenic isoforms, ΔNp63β and γ were significantly higher in the high-RAS group of both ESCA and LUSC cohorts (figure 4B–C), suggesting that the tumor suppressive activity of TP63 is blocked by its oncogenic isoforms in this group. Interestingly, higher expression of ΔNp63β and γ correlated with amplification of TP63, further suggesting that the activation of alternative promoters and alternative splicing of TP63 is a major mechanism in the regulation of TP63 in squamous cell carcinoma. Additionally, TP53 mutation and PIK3CA amplification are significantly higher in the high-RAS group (figure 4C). Taken together, these data strongly support mechanistic roles of the sncRNA signature as a prognostic marker, as this signature is associated with the activation of ΔNp63 and multiple alterations of driving genes, such as TP53, TP63, and PIK3CA.
Therapeutic implication for risk-stratified ESCC
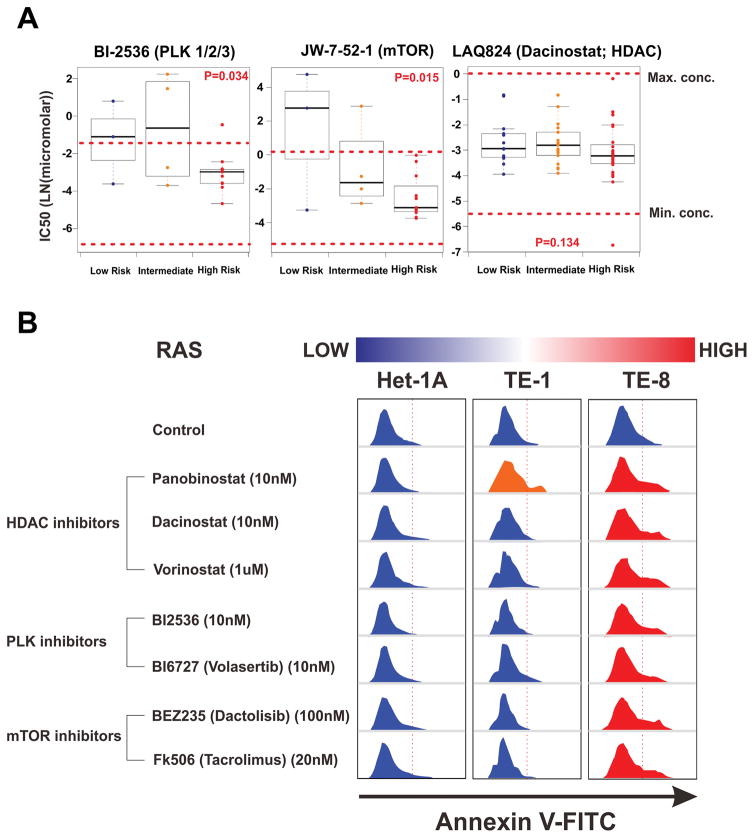
To detect suitable targeted drugs for low- and high-risk patients, we utilized the Genomics of Drug Sensitivity in Cancer Database (www.cancerRxgene.org), the largest public resource for information on the sensitivity of almost 700 cancer cells to 140 drugs.23,24 55 cell lines of ESCA, lung squamous cell, and head and neck squamous cell carcinoma were enrolled and classified into three risk groups by using a prediction model with proof of its reproducibility (supplementary figure S11 and supplementary figure S12).
The drug sensitivity test revealed that cell lines with a high-risk signature were sensitive to Polo-like kinase (PLK) inhibitor - which blocks cell mitosis and regulates chromosomal remodeling - and mTOR inhibitor - which inhibits down stream of PIK3CA directly. Likewise, all cell lines were sensitive to HDAC inhibitor - which disturbs the reset of chromatin by removing acetyl groups (figure 5A and supplementary figure S13). These results were consistent with our previous integrated genome analysis. Furthermore, the apoptosis assay of risk-stratified ESCC cell lines for three HDAC inhibitors (Panobisnotat, Dacinostat (LAQ824), and Vorinostat), two PLK inhibitors (BI2536 and BI6727 [Volasertib]), and two mTOR inhibitors (BEZ235 [Dactolisib] and Tacrolimus [FK506]) demonstrated that TE-8 high-risk cell line had dominant signals of the apoptosis compared with Het-1a and TE-1 cell lines with lower risk (figure 5B).
Figure 5. Therapeutic implications for high-risk oesophageal cancer.
(A) A prediction model derived from a RAS-correlated 613 mRNAs signature revealed that TE-8 ESCC cell lines are a high-risk group and TE-1 cell line is a lower-risk group. Drug rearrangement in high-risk group with the Genomics of Drug Sensitivity in Cancer Database. The IC50 values of each drug within the therapeutic range of concentration are compared between groups. (B) Apoptosis assay of risk-stratified esophageal cell lines in the candidate targeted drugs. The IC50 of each drug for high-risk cell line was administrated, and then all cells were captured 24-hours later.
HDAC denotes histone deacetylase inhibitor, IC50 half-maximal inhibitory concentration, mTOR mammalian target of rapamycin inhibitor, and PLK Polo-like kinase inhibitor.
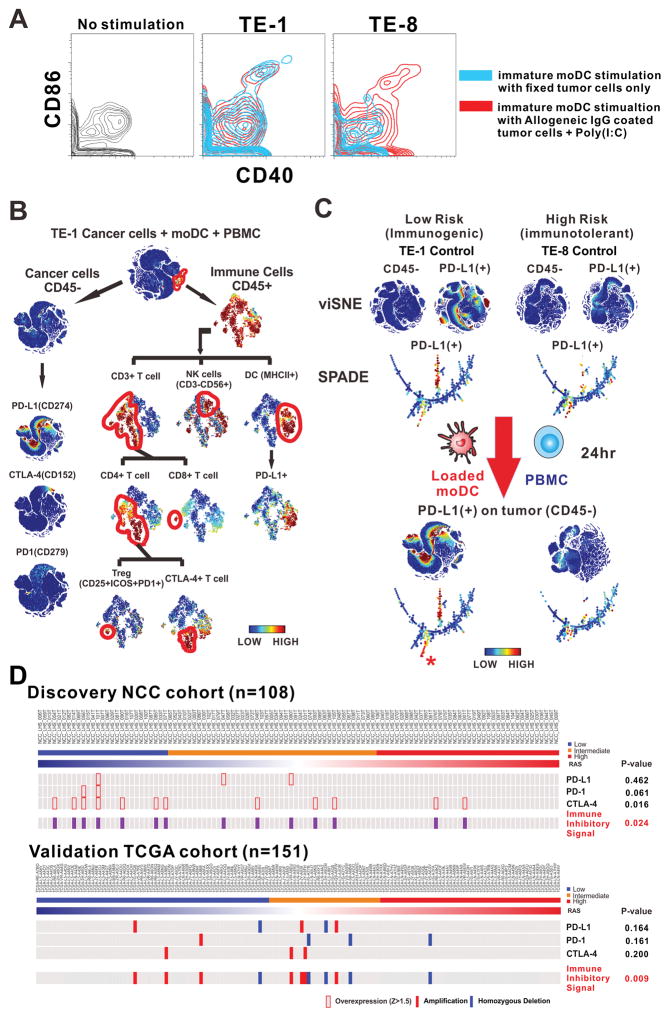
Given the upregulation of immunoregulatory genes in the low-RAS group, we hypothesized that such gene expression profile in the ESCC cell would stimulate an immunogenic tumor microenvironment.25 To investigate immunogenicity26 on ESCC cells and dendritic cells, we used a model system in which either the low-risk ESCC cell line TE-1 or the high-risk cell line TE-8 was cultured in the presence of immature dendritic cells (imoDCs) or matured monocyte-derived dendritic cells (moDCs) and peripheral blood mononuclear cells (PBMCs) obtained from healthy human donors. Flow cytometry showed that CD40 and CD86, markers of mature dendritic cells, were activated by pooled IgG-coated TE-1 or TE-8 tumor cells with poly(I:C) for 24 hours. Interestingly, imoDCs were also stimulated by TE-1 cells fixed with 2% paraformaldehyde only, not by fixed TE-8 cells, implying that TE-1 cells are more immunogenic to dendritic cells (figure 6A). We then performed single cell analysis with time-of-flight mass cytometry (supplementary table S8) and applied viSNE27 and SPADE28,29 software algorithms to evaluate the immune landscape. Immunophenotyping with 17 metal-conjugated antibodies (figure 6B) revealed that the expression of programmed cell death 1 ligand 1 (PD-L1) (whose expression on tumor or antigen-presenting cells suppresses antitumor CD8+ T cells by binding its receptor, programmed cell death 1 (PD-1), found on activated T cells30) was up-regulated in only TE-1 cancer cells, but not altered in TE-8 cells after incubation with MoDCs and PBMCs. These results suggest that low-risk ESCC cells have immunogenicity that leads to the expression of PD-L1 and coordinates a tumor-favorable milieu (figure 6C). Furthermore, PD-L1 was markedly expressed in dendritic cells in a tumor microenvironment with TE-1 but not with TE-8 (figure 6B and supplementary figure S14). Also, the expression of immune inhibitory PD-1, PD-L1, and CTLA-4 from discovery and validation cohorts was significantly up-regulated in the low-risk group compared with other risk groups (figure 6D).
Figure 6. Therapeutic implications for low-risk oesophageal cancer.
(A) CD40 and CD86 expression on immature moDCs (imoDCs) from TE-1 and TE-8 ESCC cells. The imoDCs incubated with pooled IgG-coated tumor cells and poly(I:C) are used as positive control of the stimulation in dendritic cell. (B) Immunophenotyping of TE-1 ESCC cell lines after administration of mature moDCs and PBMCs. (C) CyTOF single cell analysis of TE-1 and TE-8 cells before and after incubation with mature moDCs and PBMCs. The alteration of PD-L1 expression on the surface of tumor cells (CD45-) responsive to immune stimulation was analyzed with viSNE and SPADE software. (*) = new expression of PD-L1 on tumor cells. (D) Expression of PD-1, PD-L1, and CTLA-4 in discovery and validation cohorts. Immune inhibitor signal represents any alteration of PD-1, PD-L1, and CTLA-4.
CTLA-4 denotes Cytotoxic T-Lymphocyte-Associated Protein 4, CyTOF Time-Of-Flight Mass Cytometry, imoDCs immature monocyte-derived dendritic cells, moDCs monocyte-derived dendritic cells, PBMCs peripheral blood mononuclear cells, PD-1 programmed cell death 1, PD-L1 Programmed Cell Death 1 Ligand 1 (=CD274), SPADE cyto spanning tree progression of density normalized events, and viSNE visualization of t-distributed stochastic neighboring embedding algorithm.
DISCUSSION
We developed and validated a RAS model based on the expression of three sncRNAs (miR-223, miR-1269a, and nc886). This model can easily and reproducibly quantify the likelihood of recurrence in patients with ESCC who have received surgical resection as primary treatment. This sncRNA signature reflects multiple alterations of driving genes such as TP53, ΔNp63, and PIK3CA. Furthermore, we provided evidence based on cancer genomics for clinical trials for immune checkpoint inhibitors in low-risk ESCC patients, and targeted agents, such PLK inhibitors, mTOR inhibitors, and HDAC inhibitors, in high-risk ESCC patients. These findings show the potential of sncRNA signatures as robust prognostic and predictive markers for precision medicine based on risk assessment of recurrence.
Previously reported risk prediction models in ESCA have had limited clinical use because, first, signatures derived from heterogeneous patient populations have not been thoroughly validated in independent cohorts.6,7,8 Second, many models have been based solely on correlative associations between dysregulated markers and prognostic endpoints without identifying the underlying molecular mechanisms and the biological and clinical implications of dysregulation.4,5,31–33 Lastly, most models were generated with data obtained from fresh frozen tissues, which are not readily available in general clinical practice.5–8,31,33 Our RAS model can overcome these limitations by developing an intuitive risk prediction scoring system with FFPE tissue data. This system has been validated in independent Eastern and Western ESCA cohorts and LUSC cohort comprising 512 patients, and uncovers genomic implications that may dictate clinical outcomes associated with the identified markers.
Among three sncRNAs, the role of miR-223 has been extensively analyzed since PARP1 was identified as a direct target gene of miR-223 in ESCA. Increased sensitivity to chemotherapy was observed in cells with increased miR-223 and reduced PARP1 expression.34 As for mir-1269a, its high expression was associated with increased risk of relapse and metastasis in colorectal cancer.35 Recently, we reported that nc886 knockdown induced oncogenes MYC and FOS.17,36 Combined alterations in these sncRNAs can reflect significant modifications in the tumor microenvironment. Most interestingly, our RAS signature was highly associated with the transcription of TP63, which is critical in the development of normal oesophageal and tracheobronchial epithelia and controls the commitment of early stem cells into basal cell progeny and the maintenance of basal cells. Networking between TP63 and miRNAs is also known in multiple cancers.37,38 Taken together, our findings suggest that combined alterations in these sncRNAs may control the differentiation of ΔNp63 and regulate driver genes.
Recently, among patients with previously treated advanced LUSC, survival and response rate were significantly better with nivolumab, a PD-1 inhibitor, than with docetaxel, but the rate of objective response to nivolumab was only 20%.39 In our study, the low-risk group presented a tumor microenvironment in which immune cells coexisted with tumor cells expressing PD-L1, which suppressed cytotoxic T cell functions. Thus, we can speculate that there will be a subgroup that responds well to immune checkpoint inhibitors, and the low RAS group, having overexpression of many immune regulators, can be a candidate for immunotherapy. In contrast, the alteration of the tumor microenvironment did not affect tumor cells in the high-risk group. This finding highlights that sncRNA signatures have predictive potential for immune checkpoint inhibitors.
Several comprehensive analyses of squamous cell carcinoma have revealed significant alterations in histone-modifying and chromatin-remodeling genes.40–46 Histone deacetylases (HDACs) are recruited to active genes to reset chromatin modification states and maintain an adequate level of histone acetylation, after the activation of RNA polymerase II and HATs. Indeed, this strategy of balancing between euchromatin and heterochromatin in active genes could in part account for the mechanism of drug resistance by preventing DNA damage.47 Moreover, HDAC-inhibitor treatment resulted in rapid upregulation of histone acetylation of the PD-L1 gene leading to enhanced and durable gene expression in melanoma.48 In addition, in vivo experiments revealed that the combination therapy of HDAC inhibitor with PD-1 blockade resulted in slower tumor progression and increased survival, when compared with that of control and single agent experiments. Our results suggest that the combination of immune checkpoint blockade or other targeted drugs with HDAC inhibitors can boost the therapeutic effect on ESCC with less concentrated doses.
In conclusion, our risk prediction model with three sncRNAs from easily accessible tissues can be potentially useful for identifying patients at high risk of recurrence after surgical resection. This study represents a comprehensive characterization of genomic alterations in squamous cell carcinoma, and provides insights into the genetic mechanism of ESCC oncogenesis and supporting evidence for therapeutics. Our findings can facilitate the rational design of future clinical studies that may ultimately lead to the development of effective biomarker-based therapeutic approaches for ESCC.
Supplementary Material
Summary box.
What is already known about this subject?
Oesophageal cancer results in about 400,000 deaths, making the disease the sixth leading cause of cancer-related deaths. Oesophageal squamous cell carcinoma (ESCC) is particularly prevalent in Asia and Africa where it accounts of more than 90% of all oesophageal cancer cases.
Although combination therapy with surgery, chemotherapy, and radiotherapy has improved survival of ESCC, the five-year overall survival (OS) rate is at best 20%.
Reliable and reproducible prognostic markers identifying patients at high risk of ESCC recurrence after surgery have not been established.
What are the new findings?
For easy translation of our findings to the clinic, we developed a recurrence risk assessment score (RAS) using three small non-coding RNAs (sncRNAs) from fresh frozen tissue or formalin-fixed, paraffin-embedded ESCC specimens as robust prognostic markers for predicting postoperative recurrence of oesophageal squamous cell carcinoma (ESCC) and validated this score in independent Eastern, Western, and lung squamous cell carcinoma (LUSC) cohorts comprising over 500 patients.
The expression of oncogenic isoforms ΔNp63 was significantly higher in the high-RAS group, and, interestingly, this higher expression correlated with amplification of TP63, further suggesting that the activation of alternative promoters and alternative splicing of TP63 are major mechanisms for the regulation of TP63 in squamous carcinoma.
For the low-risk group, we tested the immunogenicity of the tumor with Time-Of-Flight Mass Cytometry (CyTOF) under the hypothesis that the low-risk group has immunogenic tumors because a plethora of immune genes are up-regulated. The up-regulation of PD-L1 in low-risk ESCC and no alteration of PD-L1 in high-risk ESCC, in response to loaded dendritic cells, suggests the therapeutic potential of immune checkpoint inhibitors.
We found that the high-risk group was more sensitive to Polo-like kinase, mTOR, and HDAC inhibitors by utilizing the Genomics of Drug Sensitivity in Cancer Database, the largest public resource of information on the sensitivity of almost 700 cancer cells to 140 drugs.
How might it impact on clinical practice in the foreseeable future?
This study provides suggestive evidence, based on cancer genomics, for clinical trials for immune checkpoint inhibitors in low-risk ESCC patients and for targeted agents, such PLK inhibitors, mTOR inhibitors, and HDAC inhibitors, in high-risk ESCC patients. Our analyses of risk assessment score for recurrence demonstrate the potential of sncRNA signatures as robust prognostic and predictive markers for precision medicine.
Acknowledgments
This work was supported by National Cancer Center Korea Research Grant No. 1110260 to H-SL; National Institutes of Health grants CA150229 to J-SL; Research Scholar Grant, RSG-12-187-01 – RMC from the American Cancer Society to YSL; the R&D Program for the Society of the National Research Foundation (NRF) funded by the Ministry of Science, ICT & Future Planning (Grant number: 2013M3C8A1078501) to YLC; NIH/NCI award number P30CA016672. This project was supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the NIH (NIAID P30AI036211, NCI P30CA125123, and NCRR S10RR024574) and the assistance of Joel M. This research was performed in the Flow Cytometry & Cellular Imaging Facility, which is supported in part by the National Institutes of Health through M.D. Anderson’s Cancer Center Support Grant CA016672. The authors would like to thank Ana María Rodríguez, Ph.D. and Kimberly Macellaro, Ph.D., members of the Baylor College of Medicine Michael E. DeBakey Department of Surgery Research Core Team, for their editorial assistance during the preparation of this manuscript.
Footnotes
Accession codes
Data have been deposited into NCBI Gene Expression Omnibus (GEO): sncRNA expression microarrays (GSE55857) and mRNA gene expression microarrays (GSE55856).
Author Contributions
H.J.J., H.S.L., Y.S.L., B.M.B. and J.S.L. conceptualized and planned the study.
H.J.J., H.S.L., M.S.K., J.M.L., J.S.Y., K.J.N., Y.L.C., J.I.Z., and Y.M.S. contributed to collection of surgical samples and associated clinical information.
G.K.L. conducted the pathology assessment.
H.J.J., H.S.L, Y.S.L., and J.S.L. coordinated the data generation and led to the data analysis.
D.M. and J.K.B. generated the mass cytometry data.
L.H. and H.L. supported to generate sncRNA data from TCGA database.
H.J.J., H.S.L., Y.Y.P., B.H.S., and S.B.K. generated the gene expression data.
H.J.J., H.S.L., K.Y.Y., S.B.K., J.J, and S.C.K processed, analyzed and participated in discussions related to the genomics data.
H.J.J., H.S.L., and J.J. conducted the statistical analysis of the clinical data.
H.J.J., H.S.L, B.M.B., J.S.L., J.L.P., S.Y.K., Y.S.L., and D.J.S. participated in discussions, provided critical scientific input, analysis suggestions and logistical support toward the project.
H.J.J., H.S.L., B.M.B., Y.S.L., and J.S.L. wrote the manuscript.
References
- 1.Ingelfinger JR, Rustgi AK, El-Serag HB. Esophageal Carcinoma. N Engl J Med. 2014;371:2499–509. doi: 10.1056/NEJMra1314530. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Arnold M, Soerjomataram I, Ferlay J, et al. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381–7. doi: 10.1136/gutjnl-2014-308124. [DOI] [PubMed] [Google Scholar]
- 4.Mathé EA, Nguyen GH, Bowman ED, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192–200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo Y, Chen Z, Zhang L, et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 6.Kano M, Seki N, Kikkawa N, et al. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127:2804–14. doi: 10.1002/ijc.25284. [DOI] [PubMed] [Google Scholar]
- 7.Hong L, Han Y, Zhang H, et al. The prognostic and chemotherapeutic value of miR-296 in esophageal squamous cell carcinoma. Ann Surg. 2010;251:1056–63. doi: 10.1097/SLA.0b013e3181dd4ea9. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Chen Z, Tian L, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63:1700–10. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Bio. 2002;3:research0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–52. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mestdagh P, Van Vlierberghe P, De Weer A, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bargaje R, Hariharan M, Scaria V, et al. Consensus miRNA expression profiles derived from interplatform normalization of microarray data. RNA. 2010;16:16–25. doi: 10.1261/rna.1688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang KH, Mestdagh P, Vandesompele J, et al. MicroRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer. BMC Cancer. 2010;10:173. doi: 10.1186/1471-2407-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nat. 2010;464:1071–76. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K, Kunkeaw N, Jeon SH, et al. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA. 2011;17:1076–89. doi: 10.1261/rna.2701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee HS, Lee K, Jang HJ, et al. Epigenetic silencing of the non-coding RNA nc886 provokes oncogenes during human esophageal tumorigenesis. Oncotarget. 2014;5:3472–81. doi: 10.18632/oncotarget.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budczies J, Klauschen F, Sinn BV, et al. Cutfoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS ONE. 2012;7:e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineagesurvival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–42. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui Y, Morgenstern H, Greenland S, et al. Polymorphism of Xeroderma Pigmentosum group G and the risk of lung cancer and squamous cell carcinomas of the oropharynx, larynx and esophagus. Int J Cancer. 2006;118:714–20. doi: 10.1002/ijc.21413. [DOI] [PubMed] [Google Scholar]
- 21.Tan W, Chen GF, Xing DY, et al. Frequency of CYP2A6 gene deletion and its relation to risk of lung and esophageal cancer in the Chinese population. Int J Cancer. 2001;95:96–101. doi: 10.1002/1097-0215(20010320)95:2<96::aid-ijc1017>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 22.Yang A, Kaghad M, Wang Y, et al. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–16. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 23.Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–61. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nat. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 26.Blankenstein T, Coulie PG, Gilboa E, et al. The determinants of tumour immunogenicity. Nat Rev Cancer. 2012;12:307–13. doi: 10.1038/nrc3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amir E-aD, Davis KL, Tadmor MD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–52. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu P. Inferring phenotypic properties from single-cell characteristics. PLoS One. 2012;7:e37038. doi: 10.1371/journal.pone.0037038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu P, Simonds EF, Bendall SC, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–91. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamanishi J, Mandai M, Iwasaki M, et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc Natl Acad Sci U S A. 2007;104:3360–5. doi: 10.1073/pnas.0611533104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueda T, Volinia S, Okumura H, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–46. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu N, Chen N-Y, Cui R-X, et al. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol. 2012;13:633–41. doi: 10.1016/S1470-2045(12)70102-X. [DOI] [PubMed] [Google Scholar]
- 33.Yu S-L, Chen H-Y, Chang G-C, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Streppel MM, Pai S, Campbell NR, et al. MicroRNA 223 is upregulated in the multistep progression of Barrett’s esophagus and modulates sensitivity to chemotherapy by targeting PARP1. Clin Cancer Res. 2013;19:4067–78. doi: 10.1158/1078-0432.CCR-13-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bu P, Wang L, Chen K-Y, et al. miR-1269 promotes metastasis and forms a positive feedback loop with TGF-β. Nat Commun. 2015;6:6879. doi: 10.1038/ncomms7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee K-S, Park J-L, Lee K, et al. nc886, a non-coding RNA of anti-proliferative role, is suppressed by CpG DNA methylation in human gastric cancer. Oncotarget. 2014;5:3944–55. doi: 10.18632/oncotarget.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Candi E, Amelio I, Agostini M, et al. MicroRNAs and p63 in epithelial stemness. Cell Death Differ. 2015;22:12–21. doi: 10.1038/cdd.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su X, Chakravarti D, Cho MS, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nat. 2010;467:986–90. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non–small-cell lung cancer. N Engl J Med. 2015;373:123–35. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao Y-B, Chen Z-L, Li J-G, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46:1097–102. doi: 10.1038/ng.3076. [DOI] [PubMed] [Google Scholar]
- 41.Kim Y, Hammerman PS, Kim J, et al. Integrative and comparative genomic analysis of lung squamous cell carcinomas in East Asian patients. J Clin Oncol. 2014;32:121–8. doi: 10.1200/JCO.2013.50.8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin D-C, Hao J-J, Nagata Y, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet. 2014;46:467–73. doi: 10.1038/ng.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Network, C.G.A.R. Comprehensive genomic characterization of squamous cell lung cancers. Nat. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Network, C.G.A.R. Comprehensive molecular characterization of urothelial bladder carcinoma. Nat. 2014;507:315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song Y, Li L, Ou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nat. 2014;509:91–5. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- 46.Drilon A, Rekhtman N, Ladanyi M, et al. Squamous-cell carcinomas of the lung: emerging biology, controversies, and the promise of targeted therapy. Lancet Oncol. 2012;13:e418–26. doi: 10.1016/S1470-2045(12)70291-7. [DOI] [PubMed] [Google Scholar]
- 47.Wang Z, Zang C, Cui K, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–31. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woods DM, Sodré AL, Sarnaik AA, et al. HDAC Inhibition Upregulates PD-1 Ligands in Melanoma and Augments Immunotherapy with PD-1 Blockade. Cancer Immunol Res. 2015:1–11. doi: 10.1158/2326-6066.CIR-15-0077-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.