Abstract
Purpose of review
An exciting advance in the field of neuroimaging is the acquisition and processing of very large data sets (so called ‘big data’), permitting large-scale inferences that foster a greater understanding of brain function in health and disease. Yet what we are clearly lacking are quantitative integrative tools to translate this understanding to the individual level to lay the basis for personalized medicine.
Recent findings
Here we address this challenge through a review on how the relatively new field of neuroinformatics modeling has the capacity to track brain network function at different levels of inquiry, from microscopic to macroscopic and from the localized to the distributed. In this context, we introduce a new and unique multiscale approach, The Virtual Brain (TVB), that effectively models individualized brain activity, linking large-scale (macroscopic) brain dynamics with biophysical parameters at the microscopic level. We also show how TVB modeling provides unique biological interpretable data in epilepsy and stroke.
Summary
These results establish the basis for a deliberate integration of computational biology and neuroscience into clinical approaches for elucidating cellular mechanisms of disease. In the future, this can provide the means to create a collection of disease-specific models that can be applied on the individual level to personalize therapeutic interventions.
Video abstract
Keywords: brain modeling, computational neuroscience, connectome, epilepsy, neural coupling, neuroinformatics, neurology, personalized medicine, stroke
INTRODUCTION
Clinical observations of patients with brain disease reveal a highly variable relationship between the nature of the lesion and the consequent functional deficit [1,2]. There currently exists no coherent framework for linking brain injury to functional deficit that sufficiently explains the observed variation or predicts recovery. We do know, however, that focal damage to the brain results in disruption of a distributed network of connections initiated in the damaged region(s), suggesting that some of the variations noted above may be understood as a network response [3–5].
Network analyses are thus useful in studying the widespread effect of brain injury. However, such methods have not yet achieved a personalized data-driven approach to treatment (i.e. precision medicine [6], wherein individual patient data, encompassing the gamut from genomics to demographics, are modeled and used for treatment decisions on a case-by-case basis). Because of this multidimensionality, using these data for personalized treatments will depend first on building computational models that integrate information from the clinic and from basic research to develop a mechanistic understanding of the disease process [6,7]. In other words, an essential characteristic of these models is that they need to encompass multiple scales of biological organization, from the molecular and cellular to the tissue, organ, and entire organism levels [7,8▪]. This type of multiscale approach can be especially challenging in clinical neurology as the variability among patients is high [9–11].
The current trajectory toward personalized medicine in neurology has been enabled by two components: the collection of ‘big data’ (data sets with large amounts of multimodal imaging data in a high number of patients [12]), and neurocomputational analyses able to process and interpret such large data sets. Although there has been a recent push to collect large data sets, as evidenced by such initiatives as the Human Connectome Project [13], CONNECT [14], Brain Connectome [15], the NIH Pediatric Database, and the Alzheimer’s Disease Neuroimaging Initiative, it is far less clear how to make useful diagnostic and therapeutic inferences from them.
NEUROCOMPUTATIONAL MODELING LINKS BRAIN NETWORK FUNCTION TO BEHAVIOR
Although big data provide the necessary empirical foundation to build a more complete picture of individual patients, interpreting such large volumes of data requires the use of extensive neuroinformatics processes involving connectionist methods linked to brain function and ultimately to behavior [16,17]. The current theoretical framework behind such processes is that a greater understanding of individual variability in brain disease can be achieved by integrating brain anatomy with function in a distributed network context [18,19]. This integration has proven difficult as it is not clear how static anatomical connections can generate fluctuating brain physiology and behavior [20]. That is, there is not a one-to-one relationship between structure and function. This discrepancy has been addressed with formal computational approaches applied at different scales of inquiry [21,22], from those with a focus on single neuron behavior (e.g. Hodgkin-and-Huxley-Model) to macroscopic models reflecting distributed networks [23▪▪]. Interestingly, many large-scale models include a large number of neurons (e.g. model of spiking activity of cortical columns; Human Brain Project [24]) rather than a large number of brain areas. However, in neurology and psychiatry, whole brain models suggest that some of the observed variability in clinical phenotype can be associated to network disruption [25,26▪▪]. Although highly informative, these modeling approaches have limited predictive value at the individual patient level as they lack a biological interpretation [27].
More recently, the development of multiscale brain network models is leading to a novel and increasingly popular approach [28–30,31▪] that produces biologically relevant interpretations. This new generation of brain models are high-dimensional systems that define the links between macroscale and microscale architectures in the brain as they include significant numbers of structural and biophysical parameters governing brain dynamics [32▪]. Crucially, these computational models operate via simulation of brain activity as a means to understand its properties [33]. In summary, the formal and mathematical link between different observational levels requires numerical simulations resulting from the integration of local and global dynamics [34].
The Virtual Brain (TVB) falls into this category as a novel neuroinformatics platform that simulates individualized brain activity using empirical structural data [35,36]. Specifically, TVB creates virtual representations of an individual’s brain by generating person-specific functional data based on his/her empirically determined anatomical connectivity. The simulated functional data results from integrating global (between regions) and local (within regions) brain dynamics. The TVB platform offers a variety of local models containing biophysical parameters that produce different empirical brain states [37] and can be applied to different conditions. The biophysical parameters at the local level describe the properties and dynamics of small populations of neurons, and at the global level, they represent mechanisms governing dynamics between brain regions. The overall simulation integrates these two levels to express dynamics from the macroscopic to the microscopic levels. Thus, modeling in TVB involves a multiscale approach to brain dynamics allowing for the inference of internal states and processes.
THE VIRTUAL BRAIN CAPTURES BIOLOGICALLY REALISTIC LARGE-SCALE BRAIN DYNAMICS
Large-scale dynamic network models including TVB have three critical components: anatomical connectivity, neural mass models, and in brain disease, lesions. First, TVB uses T1w–MRI data to create a custom brain surface, and uses diffusion-weighted MRI data to infer the anatomical connections among brain areas. Current procedures to reconstruct connectivity from DTI data reliably extract structural network connectivity with high anatomical precision, and efforts are on the way to developing more advanced methods reproducible at the subject level [38]. Thus, there is hope that patient-specific connectomes may soon enter into routine clinical use. Second, neural mass models describing the network nodes (brain regions) are used to determine the dynamic nature of the interactions and it is in this dynamical sense that they are biologically realistic. Because TVB’s neuroinformatics architecture incorporates a library of models, which catalogues biophysical parameters not measurable by brain imaging devices, TVB behaves as a computational microscope. Finally, acquisition of functional data, whether from MRI (rsfMRI) or electroencephalography (EEG), permits testing the accuracy of the simulated signals generated by the model.
THE VIRTUAL BRAIN-DERIVED MODELS ARE SENSITIVE TO DISRUPTION OF BRAIN DYNAMICS ASSOCIATED WITH DISEASE
The software platform for TVB has already been established and applied to normative data [39] for learning and plasticity [40] and we have begun assessments in epilepsy and stroke as described later.
Epilepsy
Objective
Despite the heavy sequelae from medically refractory epilepsy, there is a potentially curative procedure — surgical resection of the epileptogenic zone — that depends on the accurate identification of epileptogenic zone and its separation from the propagation zone comprised by areas recruited during seizure evolution. A comprehensive presur-gical evaluation is necessary to pinpoint the epileptogenic zone along with the identification of the risks of postoperative neurologic morbidity. Based on structural anomalies as identified by MRI and the assessment of functional data such as EEG, estimation of TVB parameters (epileptogenicity, anomalies) are set in the network model to predict the propagation pattern of seizures in order to explore brain intervention strategies [41].
TVB modeling (Fig. 1)
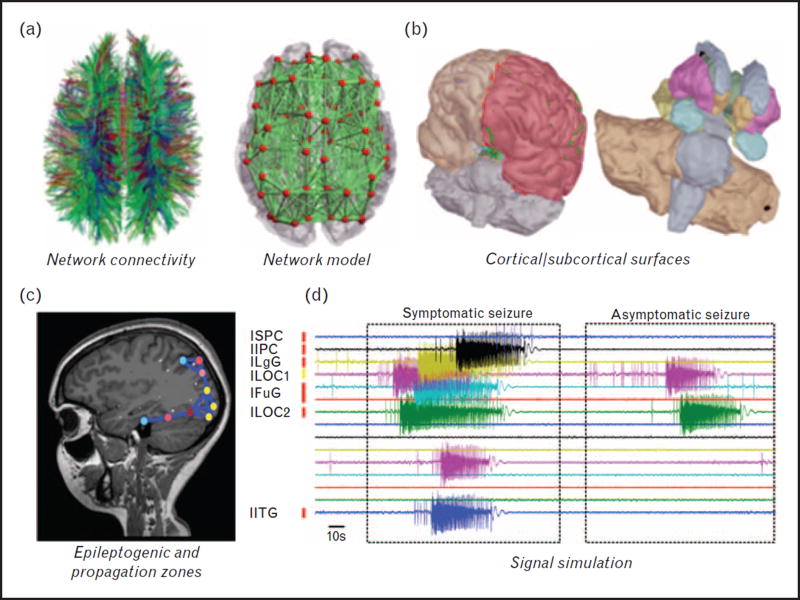
FIGURE 1.
Components of the virtual epileptic patient brain. (a) Connectivity derived from DTI, large-scale network model. (b) Cortical and subcortical surfaces. (c). Brain areas in model with increased epileptogenicity. (d) Exemplary simulated times series of symptomatic/asymptomatic seizure (modified after Jirsa et al., 2016 [41]).
We used the Epileptor, which is a neural mass model derived from mathematical reasoning in which five state variables provide a complete taxonomy of epileptic seizures including onset, offset, and features of seizure evolution. The network model is composed of two neuronal populations, characterized by fast excitatory bursting neurons and regular spiking inhibitory neurons, embedded in a common extracellular environment represented by a slow variable. By systematically analyzing the parameter landscape via the simulation, it is possible to reproduce typical sequences of neural activity observed during status epilepticus [41,42].
Our results suggest a temporal shift of typical spike waves with fast discharges as synaptic strengths are varied. In particular, gap junction coupling plays the dominant role in synchronizing both neuronal types, whereas the slow variable aids in organizing the seizure progression. The model also confirms that large amplitude spike-wave components of interictal and ictal spikes arises from synchronized discharges of inhibitory interneurons (Fig. 2).
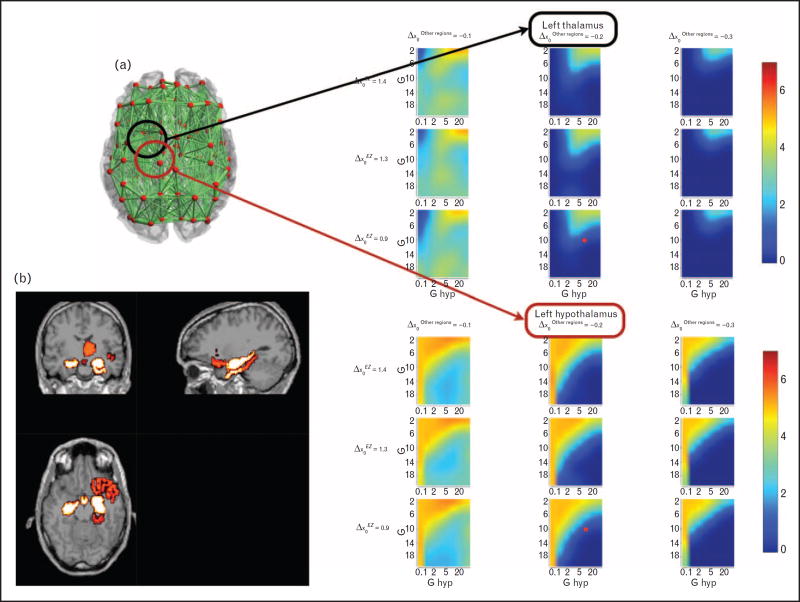
FIGURE 2.
The virtual epileptic patient model: the epileptogenic zone (EZ). (a) Parameter space exploration of a patient’s excitability. Excitability values x = x0C + Δx0 are expressed via their deviations from the critical value of seizure onset, x0C. Positive Δx0 indicates increased excitability. The color indicates the number of seizure-like events within a given region during simulation of fixed length in dependence of global coupling strength G (y-axis), local coupling strength for the hypothalamic connections Ghyp (x-axis), the excitability Δχ′ριγηιηιπποψαμπθσ of the right hippocampus (each row is a different value), and the excitability Δx0Otherregions of the regions not recruited in the propagation zone (each column is a different value). The red point indicates the working point in the parameter space chosen for the simulation. Parameter spaces are shown for left thalamus and left hypothalamus. (b) Clinical heat map of epileptogenicity. The distribution of excitability values is plotted over the anatomical MRI indicating the EZ (in yellow) and propagation zone (in red) (modified after Jirsa et al., 2016 [41]).
Our approach does not rule out other physiological organizations giving rise to the same dynamics. They nevertheless can give insight about experimental paradigms via simulations and analyses bridging the gap between neuronal spiking, network and abstract seizure evolution across large temporal scales.
Stroke
Objective
Although there are multiple therapeutic approaches to foster recovery after stroke [43], there is no cure. In our previous work using TVB modeling in stroke patients [44], we found consistent parameter changes associated with poor long-term motor recovery. These included local hyper-excitability, lower conduction velocities in cortico-cortical connectivity, and an imbalance between local and global dynamics. Here we hypothesize that the normalization of parameter values to those found in healthy controls will result in the reestablishment of healthier brain dynamics even in the presence of a damaged brain. That is, we are proposing a virtual therapy using TVB to determine the potential therapeutic value of restoring the affected biophysical parameters.
TVB modeling (Fig. 3)
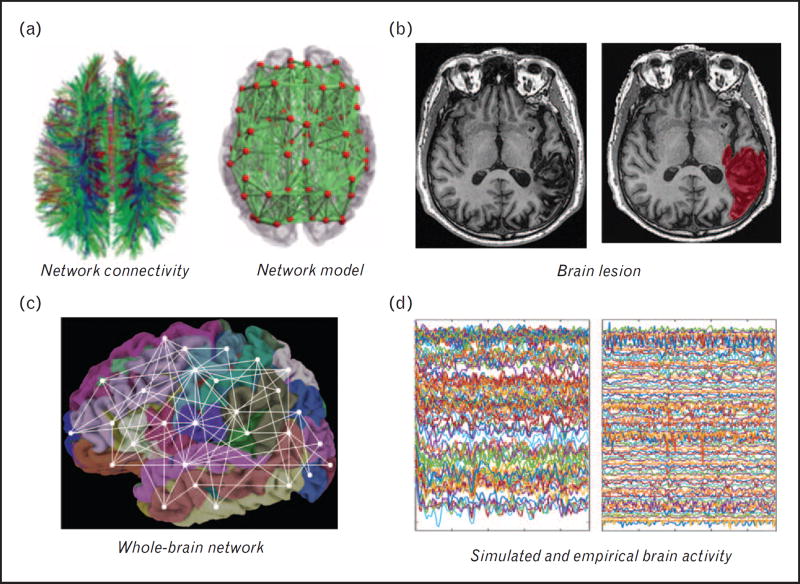
FIGURE 3.
Components of the virtual stroke patient brain. Connectivity derived from (a) DTI (input to the modeling system), large-scale network model (multiscale including global and local parameters) and (b) lesion maps. The modeling done included all brain regions (c, whole brain modeling) that generated the simulated rsfMRI signals (d, left). These are then compared to the empirical rsfMRI signals (d, right) fitting parameters until both are signals are similar.
In stroke, local dynamics are modeled via the Stefanescu Jirsa 3D model, a neural mass model based on the Hindmarsh—Rose model [45], developed at the single neuronal level but expanded to populations of neurons with different membrane excitability [46]. Hence, the model’s parameters determine interactions between neuronal groups including coupling between excitatory and inhibitory populations. This model was chosen because it enables the characterization of the association between neural mass responses and rsfMRI [47,48], and because it does not rely heavily on synaptic delays, making it particularly suitable for simulating fMRI signals, which have notoriously poor temporal resolution [49].
In contrast to epilepsy, a focal lesion in stroke is easily identified and can be considered in the modeling process through its direct physical effect on the anatomical connectome. Because of this dependency, our modeling approach started by determining the impact of the damage on the structural connectivity in regions affected by the stroke. We performed a regression analysis, with percentage of damage in brain regions as the independent variable and the number of tracts associated with each of them as the dependent variable. This latter variable is referred to as ‘degree centrality’ in graph theoretic analysis [18]. Here we provide two examples from damage to the precentral gyrus and putamen, common sites of injury in our patients with upper limb impairments. In both regions there was a linear relationship between percentage of damage and degree centrality, where more damage results in fewer detectable tracts (precentral gyrus: t = —4.35; P = 0.01; putamen: t = —6.6; P = 0.003). This linear relationship had more variability in the precentral gyrus where half of the cases had a disproportionate loss of tracts in the presence of limited damage to the region. Because these patients had large cortical strokes (i.e. heavily affecting adjacent regions), this apparent discrepancy may reflect transneuronal degeneration [50,51].
The Virtual Therapy (TVT)
To test the effects of restored parameter values on brain dynamics, the following steps were performed:
Baseline (Cbl) identification: Cbl was defined as the Pearson’s correlation between the average empirical functional connectivity matrix from controls and the individual simulated functional matrices of each patient.
Virtual therapy: Parameters that changed after stroke [44] were adjusted for each patient to match the average control value (global coupling, conduction velocity, coupling between inhibitory and excitatory populations).
Resimulation: Brain signals (rsfMRI) were resimulated with the modified parameter values and the consequent functional connectivity matrices were generated.
Correlation post-virtual therapy (Cvt): Cvt was defined as the Pearson’s correlation between the average empirical functional connectome from controls with the individual functional connectomes derived post-virtual therapy.
- Therapeutic effect (TE) of TVT: TE was calculated by subtracting the baseline correlation from that obtained after TVT (TE = Cvt − Cbl). We then calculated the percentage final gain (G) from baseline using the following algorithm:
After normalizing the parameters to control levels (Fig. 4), 17 of 20 patients showed an increased gain as dynamics got closer to those in healthy controls. The percentage gain ranged from 5 to 85.7% and the parameters that produced the largest improvement in brain dynamics included global coupling and local excitatory on inhibitory coupling. In other words, the change in these parameters produced normalization of the balance between global and local dynamics and excessive excitation. Notably, these two parameters were those that correlated with long-term motor recovery in our previous work [44].
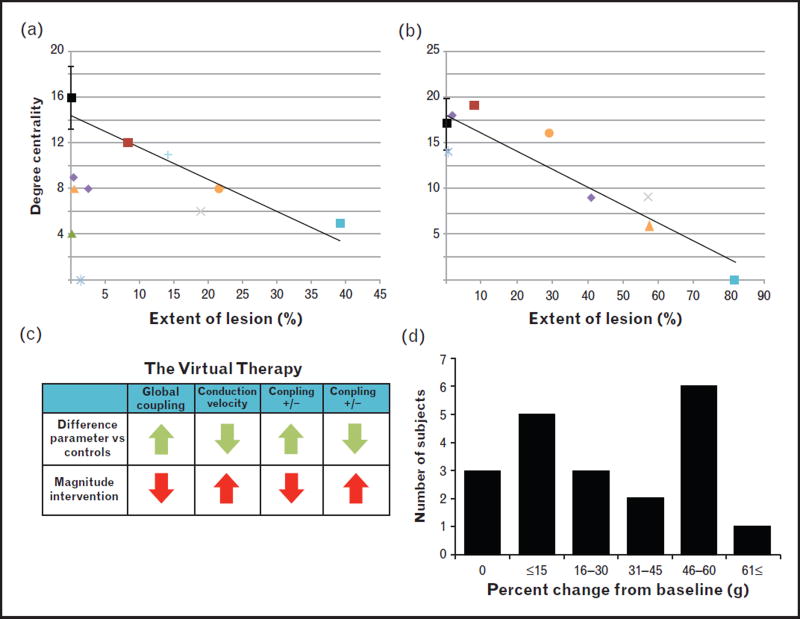
FIGURE 4.
Brain connectivity and virtual therapy on the virtual stoke patient brain. The upper row shows the effect of damage and the corresponding anatomical tracts. These scatter plots show a linear relationship between the percentage of damage in individual regions and the number of tracts associated with them (degree centrality). Two examples are shown. (a, precentral gyrus) The linear trend is clear for most cases. Some of them, however, showed a disproportionate reduction in the number of tracts despite the small percentage of lesion suggesting transneuronal degeneration originated in the large cortical strokes in these cases. (b, putamen) Note the clear linear trend seen in subcortical stroke. The lower row shows the process and effect of parameter normalization on brain dynamics. (c) Example of The Virtual Therapy implementation for one stroke case. Parameters were normalized from the baseline value to the healthy control average. (d) Frequency histogram showing the number of stroke cases stratified by percentage of change in the correlation of functional connectomes from baseline after The Virtual Therapy in relation to healthy controls. In all but three cases, the functional connectivity matrices after the therapy were closer to those in healthy controls. Changing the parameter values in the stroke patients to mimic those in controls resulted in healthier brain dynamics even when the damage did not change.
From the three patients that showed no change in correlation with healthy controls post-virtual therapy, two had baseline global coupling values equal to the control averages, thus it was not manipulated for TVT. The third individual had a large, widespread lesion including both cortical and subcortical regions.
Although the global coupling parameter has a sizable effect on brain dynamics after TVT, its biological significance warrants exploration. Our previous work using graph analysis [52] has shown that it is negatively correlated with global efficiency, or a network’s capacity for communication [18]. Because this still does not provide a clear biological interpretation, our current efforts are focusing on possible biophysical factors to provide better grounding to this parameter. Current proposals are that it is associated with an overall change in connection strengths [53], and/or with the regulation of local feedback inhibition [54].
GENERAL SUMMARY
TVB provides a number of unique features for contributing to the development of precision neurology, as illustrated here. First, using TVB, complex brain dynamics are generated by a data-constrained mechanistic model utilizing physiological parameters (multiscale integration). Specific model parameters may then be identified to specific disease states (e.g. epilepsy, stroke), with the TVB model thus acting as a compact generator of dynamics-based biomarkers. In epilepsy, the structural network alteration is not obvious (location of the epileptogenic zone), but the functional network dynamics are accessible; in stroke, the structural network alteration is evident, but its functional consequences are not. For both cases, TVB offers a good framework due to the network context where both diseases may be framed. The modeling of network dynamics may provide novel biomarkers that otherwise would not be accessible.
Second, because these models are individualized, TVB can be used to test the validity and utility of candidate biomarkers by modeling brain dynamics in response to interventions that target specific parameters correlated with disease. Therefore, an important clinical application of TVB is that the model of an individual patient’s brain can act as an indicator of pathogenic processes and responses to therapeutic intervention [55]. The integration of this modeling to new technological advances in biomedical imaging can yield unprecedented information on brain structure and function, providing a promising application of data that are currently severely under-utilized for clinical decision support.
CONCLUSION
We therefore believe the results presented here hold promise for providing the basis for a more deliberate integration of computational neuroscience into clinical approaches for diagnosis and treatment of brain disorders. The potential for secondary prevention is obvious, where computational models based on a patient’s own data can help constrain diagnoses and the choices of individualized therapeutic interventions. There is, however, an even more exciting potential application in primary, and perhaps tertiary prevention, whereby the combination of informatics approaches on big data and computational models of early detection can help an individual monitor his or her own brain health and introduce effective mitigation strategies in the case of elevated risk of disease.
KEY POINTS.
The Virtual Brain (TVB) is a novel multiscale neuroinformatics modeling platform linking brain dynamics to biophysical parameters.
TVB modeling is individualized as it is based on individualized anatomical connectomes.
The virtual epileptic patient model can assist in surgical planning for intractable epilepsy.
The virtual stroke patient model can be used to test individualized therapies.
TVB modeling can be applied to any disease state, providing avenues for individualized interventions of various neurologic diseases.
Acknowledgments
The authors thank Dr A.R. McIntosh, Dr P. Ritter, and Dr S.L. Small for helpful discussions.
Financial support and sponsorship
This work was supported by the James McDonnell Foundation (NRG Group), the National Institutes of Health (NIH RO1-NS-54942), and the European Union Seventh Framework Programme [FP7-ICT BrainScales and Human Brain Project (grant no. 60402)].
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
- 1.Fridriksson J, Holland AL, Coull BM, et al. Aphasia severity: association with cerebral perfusion and diffusion. Aphasiology. 2002;16:859–871. doi: 10.1080/02687030244000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Page SJ, Gauthier LV, White S. Size doesn’t matter: cortical stroke lesion volume is not associated with upper extremity motor impairment and function in mild, chronic hemiparesis. Arch Phys Med Rehabil. 2013;94:817–821. doi: 10.1016/j.apmr.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breakspear M, Jirsa VK. Neuronal dynamics and brain connectivity. In: Jirsa VK, McIntosh AR, editors. Handbook of brain connectivity. Berlin Heidelberg: Springer; 2007. pp. 3–64. [Google Scholar]
- 4.Cabral J, Kringelbach ML, Deco G. Exploring the network dynamics underlying brain activity during rest. Prog Neurobiol. 2014;114:102–131. doi: 10.1016/j.pneurobio.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Grefkes C, Fink GR. Connectivity-based approaches in stroke and recovery of function. Lancet Neurol. 2014;13:206–216. doi: 10.1016/S1474-4422(13)70264-3. [DOI] [PubMed] [Google Scholar]
- 6.Duffy DJ. Problems, challenges and promises: perspectives on precision medicine. Brief Bioinform. 2015:bbv060. doi: 10.1093/bib/bbv060. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 7.Wolkenhauer O, Auffray C, Brass O, et al. Enabling multiscale modeling in systems medicine. Genome Med. 2014;6:21. doi: 10.1186/gm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8▪.Petersen SE, Sporns O. Brain networks and cognitive architectures. Neuron. 2015;88:207–219. doi: 10.1016/j.neuron.2015.09.027. This relevant review written by two pioneers in brain network analyses discusses the relationship between a network approach and cognition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews PM, Edison P, Geraghty OC, Johnson MR. The emerging agenda of stratified medicine in neurology. Nat Rev Neurol. 2014;10:15–26. doi: 10.1038/nrneurol.2013.245. [DOI] [PubMed] [Google Scholar]
- 10.Jung K-H, Lee K-H. Molecular imaging in the era of personalized medicine. J Pathol Transl Med. 2015;49:5–12. doi: 10.4132/jptm.2014.10.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamburg MA, Collins FS. The path to personalized medicine. N Engl J Med. 2010;363:301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 12.Poldrack RA, Gorgolewski KJ. Making big data open: Data sharing in neuroimaging. Nat Neurosci. 2014;17:1510–1517. doi: 10.1038/nn.3818. [DOI] [PubMed] [Google Scholar]
- 13.Sotiropoulos SN, Jbabdi S, Xu J, et al. Advances in diffusion MRI acquisition and processing in the Human Connectome Project. Neuroimage. 2013;80:125–143. doi: 10.1016/j.neuroimage.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Assaf Y, Alexander DC, Jones DK, et al. The CONNECT project: combining macro- and micro-structure. Neuroimage. 2013;80:273–282. doi: 10.1016/j.neuroimage.2013.05.055. [DOI] [PubMed] [Google Scholar]
- 15.Jiang T. Brainnetome: a newome to understand the brain and its disorders. Neuroimage. 2013;80:263–272. doi: 10.1016/j.neuroimage.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 16.McIntosh AR. Towards a network theory of cognition. Neural Netw. 2000;13(8–9):861–870. doi: 10.1016/s0893-6080(00)00059-9. [DOI] [PubMed] [Google Scholar]
- 17.Hannawi Y, Smirnakis SM. Emerging subspecialties: neuroinformatics. Neurology. 2013;80:e166–e168. doi: 10.1212/WNL.0b013e31828c2f2e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullmore E, Sporns O. Complex brain networks: Graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 19.Hartman D, Hlinka J, Palus M, et al. The role of nonlinearity in computing graph-theoretical properties of resting-state functional magnetic resonance imaging brain networks. Chaos. 2011;21:013119. doi: 10.1063/1.3553181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park HJ, Friston K. Structural and functional brain networks: from connections to cognition. Science. 2013;342:1238411. doi: 10.1126/science.1238411. [DOI] [PubMed] [Google Scholar]
- 21.Sporns O. Contributions and challenges for network models in cognitive neuroscience. Nat Neurosci. 2014;17:652–660. doi: 10.1038/nn.3690. [DOI] [PubMed] [Google Scholar]
- 22.Deco G, Tononi G, Boly M, Kringelbach ML. Rethinking segregation and integration: Contributions of whole-brain modelling. Nat Rev Neurosci. 2015;16:430–439. doi: 10.1038/nrn3963. [DOI] [PubMed] [Google Scholar]
- 23▪▪.Yuste R. From the neuron doctrine to neural networks. Nat Rev Neurosci. 2015;16:487–497. doi: 10.1038/nrn3962. This outstanding review describes the fundamentals of multiscale studies. This is an essential read for those interested in understanding the links between the different levels of inquiry (from the microscopic to the macroscopic) in the context of brain networks. [DOI] [PubMed] [Google Scholar]
- 24.Reimann MW, Anastassiou CA, Perin R, et al. A biophysically detailed model of neocortical local field potentials predicts the critical role of active membrane currents. Neuron. 2013;79:375–390. doi: 10.1016/j.neuron.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffa A, Baumann PS, Thiran J-P, Hagmann P. Structural connectomics in brain diseases. Neuroimage. 2013;80:515–526. doi: 10.1016/j.neuroimage.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 26▪▪.Fornito A, Zalesky A, Breakspear M. The connectomics of brain disorders. Nat Rev Neurosci. 2015;16:159–172. doi: 10.1038/nrn3901. This excellent review establishes the relationship between brain connectivity and brain disorders. Importantly, it provides a global view of the advantages and limitations of the now popular approach of connectomics. [DOI] [PubMed] [Google Scholar]
- 27.Smith SM, Vidaurre D, Beckmann CF, et al. Functional connectomics from resting-state fMRI. Trends Cogn Sci. 2013;17:666–682. doi: 10.1016/j.tics.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grillner S. Megascience efforts and the brain. Neuron. 2014;82:1209–1211. doi: 10.1016/j.neuron.2014.05.045. [DOI] [PubMed] [Google Scholar]
- 29.Sompolinsky H. Computational neuroscience: beyond the local circuit. Curr Opin Neurobiol. 2014;25:13–18. doi: 10.1016/j.conb.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Eliasmith C, Trujillo O. The use and abuse of large-scale brain models. Curr Opin Neurobiol. 2014;25:1–6. doi: 10.1016/j.conb.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 31▪.Stephan KE, Iglesias S, Heinzle J, Diaconescu AO. Translational perspectives for computational neuroimaging. Neuron. 2015;87:716–732. doi: 10.1016/j.neuron.2015.07.008. This excellent review establishes the relationship between medical imaging and generative computational modeling applied to psychiatry. Specifically, it describes different modeling approaches and their limitations. [DOI] [PubMed] [Google Scholar]
- 32▪.Sanz-Leon P, Knock SA, Spiegler A, Jirsa VK. Mathematical framework for large-scale brain network modeling in The Virtual Brain. Neuroimage. 2015;111:385–430. doi: 10.1016/j.neuroimage.2015.01.002. This article details the neuroinformatics architecture of modeling in TVB. Hence, it is a useful reference for those interested in understanding the mathematical bases of TVB and its flexibility and applicability to different conditions. [DOI] [PubMed] [Google Scholar]
- 33.Gerstner W, Sprekeler H, Deco G. Theory and simulation in neuroscience. Science. 2012;338:60–65. doi: 10.1126/science.1227356. [DOI] [PubMed] [Google Scholar]
- 34.Deco G, Jirsa VK, Robinson PA, et al. The dynamic brain: From spiking neurons to neural masses and cortical fields. PLoS Comput Biol. 2008;4:e1000092. doi: 10.1371/journal.pcbi.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanz Leon P, Knock SA, Woodman MM, et al. The Virtual Brain: a simulator of primate brain network dynamics. Front Neuroinform. 2013;7:10. doi: 10.3389/fninf.2013.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritter P, Schirner M, McIntosh AR, Jirsa VK. The virtual brain integrates computational modeling and multimodal neuroimaging. Brain Connect. 2013;3:121–145. doi: 10.1089/brain.2012.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritter P, Schirner M, McIntosh AR, Jirsa V. The virtual brain integrates computational modelling and multimodal neuroimaging. Brain Connect. 2013;49:1–65. doi: 10.1089/brain.2012.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Besson P, Lopes R, Leclerc X, et al. Intra-subject reliability of the high-resolution whole-brain structural connectome. Neuroimage. 2014;102(Pt 2):283–293. doi: 10.1016/j.neuroimage.2014.07.064. [DOI] [PubMed] [Google Scholar]
- 39.Deco G, Senden M, Jirsa V. How anatomy shapes dynamics: a semi-analytical study of the brain at rest by a simple spin model. Front Comput Neurosci. 2012;6:68. doi: 10.3389/fncom.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy D, Sigala R, Breakspear M, et al. Using the virtual brain to reveal the role of oscillations and plasticity in shaping brain’s dynamical landscape. Brain Connect. 2014;4:791–811. doi: 10.1089/brain.2014.0252. [DOI] [PubMed] [Google Scholar]
- 41.Jirsa VK, Proix T, Perdikis D, et al. The virtual epileptic patient: individualized whole-brain models of epilepsy spread. Neuroimage. 2016 doi: 10.1016/j.neuroimage.2016.04.049. (in press) [DOI] [PubMed] [Google Scholar]
- 42.Jirsa VK, Stacey WC, Quilichini PP, et al. On the nature of seizure dynamics. Brain. 2014;137(Pt 8):2210–2230. doi: 10.1093/brain/awu133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobkin BH, Dorsch A. New evidence for therapies in stroke rehabilitation. Curr Atheroscler Rep. 2013;15:331. doi: 10.1007/s11883-013-0331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Falcon MI, Riley JD, Jirsa VK, et al. Functional mechanisms of recovery after chronic stroke: modeling with The Virtual Brain. eNeuro. 2016 doi: 10.1523/ENEURO.0158-15.2016. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hindmarsh JL, Rose RM. A model of neuronal bursting using three coupled first order differential equations. Proc R Soc B Biol Sci. 1984;221:87–102. doi: 10.1098/rspb.1984.0024. [DOI] [PubMed] [Google Scholar]
- 46.Stefanescu RA, Jirsa VK. A low dimensional description of globally coupled heterogeneous neural networks of excitatory and inhibitory neurons. PLoS Comput Biol. 2008;4:e1000219. doi: 10.1371/journal.pcbi.1000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magri C, Schridde U, Murayama Y, et al. The amplitude and timing of the BOLD signal reflects the relationship between local field potential power at different frequencies. J Neurosci. 2012;32:1395–1407. doi: 10.1523/JNEUROSCI.3985-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan WJ, Thompson GJ, Magnuson ME, et al. Infraslow LFP correlates to resting-state fMRI BOLD signals. Neuroimage. 2013;74:288–297. doi: 10.1016/j.neuroimage.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim S, Richter W, Ug˘urbil K. Limitations of temporal resolution in functional MRI. Magn Reson. 1997;37:631–636. doi: 10.1002/mrm.1910370427. [DOI] [PubMed] [Google Scholar]
- 50.Park H-YL, Park YG, Cho A-H, Park CK. Transneuronal retrograde degeneration of the retinal ganglion cells in patients with cerebral infarction. Ophthalmology. 2013;120:1292–1299. doi: 10.1016/j.ophtha.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 51.Lai C, Zhou H-C, Ma X-H, Zhang H-X. Quantitative evaluation of the axonal degeneration of central motor neurons in chronic cerebral stroke with diffusion tensor imaging. Acta Radiol. 2014;55:114–120. doi: 10.1177/0284185113492456. [DOI] [PubMed] [Google Scholar]
- 52.Falcon MI, Riley JD, Jirsa V, et al. The Virtual Brain: modeling biological correlates of recovery after chronic stroke. Front Neurol. 2015;6:228. doi: 10.3389/fneur.2015.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang GJ, Murray JD, Repovs G, et al. Altered global brain signal in schizophrenia. Proc Natl Acad Sci U S A. 2014;111:7438–7443. doi: 10.1073/pnas.1405289111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deco G, Ponce-Alvarez A, Hagmann P, et al. How local excitation-inhibition ratio impacts the whole brain dynamics. J Neurosci. 2014;34:7886–7898. doi: 10.1523/JNEUROSCI.5068-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Group BDW. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]