Summary
Incidence and mortality for sex-unspecific cancers is higher among men and is largely unexplained1,2. Furthermore, age-related loss of chromosome Y (LOY) is frequent in normal haematopoietic cells3,4, but the phenotypic consequences of LOY have been elusive5–10. From analysis of 1153 elderly men, we report that LOY was associated with risks of all-cause mortality (HR=1.91, 95% CI=1.17-3.13, events=637) and non-haematological cancer mortality (HR=3.62, CI=1.56-8.41, events=132). LOY affected at least 8.2% of subjects in this cohort and median survival among men with LOY was 5.5 years shorter. Risk of all-cause mortality and LOY was validated in an independent cohort (HR=3.66), in which 20.5% of subjects displayed LOY. These results illustrate the impact of post-zygotic mosaicism on disease risk, could explain why males are more frequently affected by cancer and suggest that chromosome Y is important in processes beyond sex determination. LOY in blood could become a predictive biomarker of male carcinogenesis.
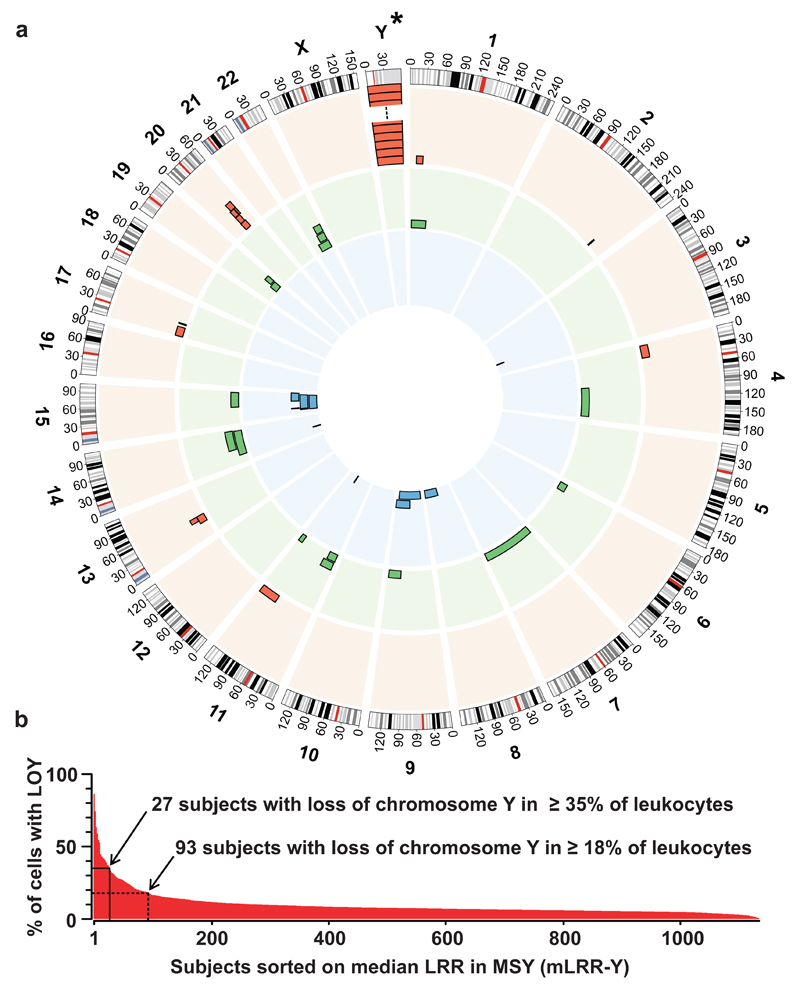
Peripheral blood DNA from 1153 participants of the Uppsala Longitudinal Study of Adult Men (ULSAM) was genotyped using high-resolution 2.5MHumanOmni SNP-beadchip from Illumina. The population-based ULSAM cohort has extensive phenotypic information of naturally aging men that were clinically followed for >40 years. We studied DNA sampled at an age window of 70.7-83.6 years. Scoring of structural genetic variants was focused on post-zygotic, acquired changes such as deletions, copy number neutral loss of heterozygozity (CNNLOH, also called acquired uniparental disomy, aUPD) and gains, as described previously11–13 with a minimum size of 2 Mb. Twelve subjects had a history of haematological malignancy before sampling and these were analyzed separately to avoid mixed analyses of normal blood and malignant clones (Supplementary Figs. 1 and 2). In the remaining 1141 participants, 40 autosomal somatic structural variants ≥2 Mb in size occurring in 37 subjects (3.2%) were uncovered, including 13 deletions, 16 CNNLOH and 11 gains (Fig. 1, Supplementary Table 1).
Figure 1.
Structural genetic variants found in phenotypically normal blood cells from 1141 elderly men with no prior record of haematological malignancy. Circular-plot in panel a shows position and frequency of 40 autosomal variants including 13 deletions (red outer circle), 16 CNNLOH regions (green middle circle) and 11 gains (blue inner circle). The asterisk (*) above chromosome Y in panel a indicates that the frequency of loss of chromosome Y (LOY) is not shown to scale with the autosomal variants in panel a. Panel b shows the frequency of LOY, with the percentage of cells affected in each participant, plotted on the y-axis after sorting subjects with descending mLRR-Y, i.e. the median Log R Ratio (LRR) for ~2560 SNP-probes in the male specific region of chromosome Y (MSY) (chrY:2694521-59034049, hg19/GRCh37). The percentage of cells affected in each participant was calculated as described in Supplementary Figure 3. Solid line in panel b indicates the threshold of LOY used in the survival analyses and the dotted line shows the threshold for estimation of the frequency of LOY in the studied cohort.
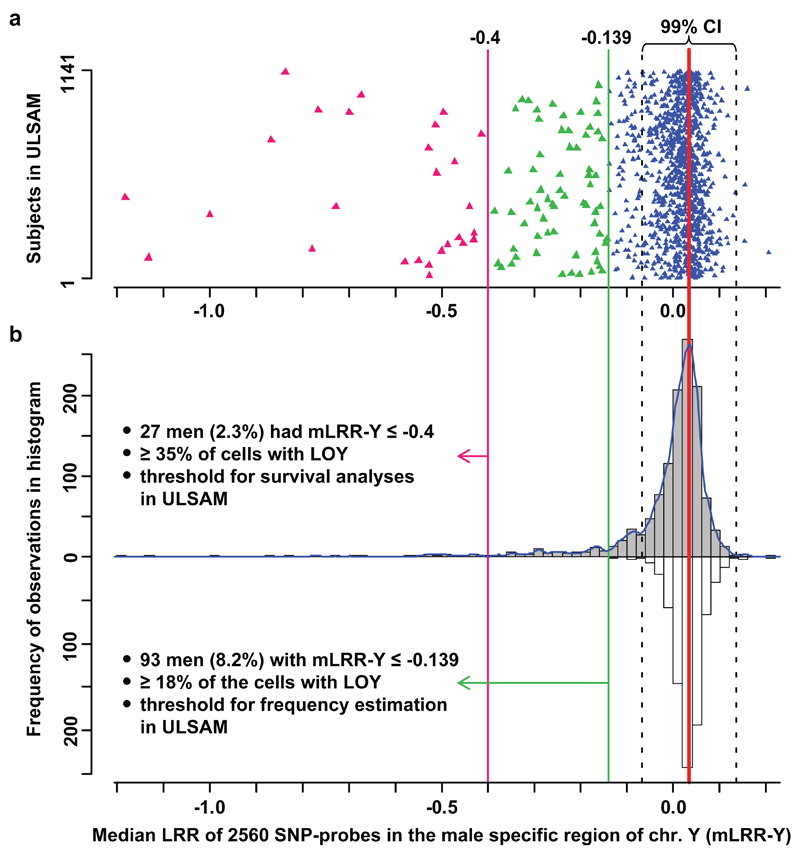
Strikingly, the most frequent somatic variant was loss of chromosome Y (LOY) (Figs. 1 and 2). The degree of LOY was calculated for each subject from the median Log R Ratio (measure of copy number) for approx. 2560 probes in the male specific region of chromosome Y (mLRR-Y) and suggested considerable inter-individual differences regarding the proportion of cells with nullisomy Y. A conservative estimate of the frequency of LOY in the ULSAM cohort at 8.2% (93/1141) was based on the lowest value (-0.139) in a simulated distribution of experimental variation of mLRR-Y (Fig. 2). At this threshold, ≥18% of cells in affected participants would be expected to have nullisomy Y. For calculating the fraction of cells affected with nullisomy Y we implemented a novel approach, using B-allele frequency (BAF)-values in the pseudo-autosomal region 1 (PAR1) on chromosomes X/Y from SNP-array data, which is explained in Supplementary Figure 3.
Figure 2.
LOY frequency estimation after accounting for experimental variation. Panel a show the median Log R Ratio (LRR) in the male specific part of chromosome Y (mLRR-Y) observed in 1141 men with no history of haematological malignancies prior to blood sampling. Each triangle represents one participant. Panel b show the distribution of the mLRR-Y (grey bars) and the experimental noise (white bars) that were used to find the threshold for estimation of LOY frequency. The latter distribution was generated as described in methods. The dotted black lines represent the 99% confidence intervals (CI) of the distribution of expected experimental background noise (white bars). Among the 1141 men we found that 168 subjects (14.7%) had a lower median LRR than the lower 99% CI representing LOY in ~13.1% of cells. For the frequency of LOY reported here, we used the lowest value in the noise-distribution as threshold (green line at -0.139).
Aberrations detected with 2.5M-arrays were validated using low coverage (~5x) whole genome next generation sequencing (NGS), performed in 100 random participants. Among 93 subjects with LOY in ≥18% of cells, sequencing was performed in six and all these cases of LOY were validated (Supplementary Figs. 4 - 7). Among 37 subjects with autosomal events ≥2 Mb, whole genome and/or exome NGS were performed in four subjects with deletions and all were validated (Supplementary Fig. 8 - 10). Moreover, the SNP-array data suggested that some participants could have a gain of chromosome Y (GOY) (Fig. 2). However, NGS failed to confirm these observations in the three suspected cases of GOY that were sequenced (Supplementary Fig. 11). In summary for autosomes and sex chromosomes among 1141 subjects, we scored 133 somatic structural variants in 128 men (11.2%). Furthermore, participants with LOY did not show a difference in frequencies of autosomal structural variants when compared with the rest of the cohort (details not shown) and we did not observe evidence for region-specific deletions on chromosome Y (Supplementary Figs. 5 - 7).
The effects from the above described structural variants on all-cause mortality, cancer mortality and non-cancer-related mortality were examined by Cox proportional hazards regression using the R package survival14. In survival analyses, 982 participants free from cancer diagnosis prior to sampling were studied (Supplementary Fig. 1) and survival was adjusted for nine potential confounders: age, hypertension, exercise, smoking, diabetes, BMI, LDL and HDL and education (Table 1, Supplementary Table 2). The study entry was the date of DNA sampling, age was used as timeline and the median follow-up time was 8.7 years (range 0-20.2 years). The structural genetic variants were analyzed in three categories. LOY was analyzed separately as a continuous variable (i.e. mLRR-Y). Other deletions and CNNLOH were grouped as autosomal loss of heterozygozity variants (LOH), reflecting their similar biological effects15. The third category was the autosomal gains. The primary survival analysis using the continuous estimator of LOY (mLRR-Y) showed that men with a higher degree of LOY had an increased risk of all-cause mortality (HR=2.13, p=0.029, Table 1). Further tests with the continuous estimator showed that LOY was an important risk factor for cancer mortality in the ULSAM cohort (HR=3.76, p=0.022, Table 1). However, LOY was not significantly associated with non-cancer-related mortality (p=0.245) (Supplementary Fig. 1).
Table 1. Cox proportional hazards regression evaluating effects from LOY on mortality.
| All-cause mortality (no of events=637) |
Cancer mortality (no of events=172) |
|||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Genotyping age | 1.09 | 1.06-1.13 | <0.0001 * | 1.06 | 1.00-1.12 | 0.039 * |
| Hypertension | 1.68 | 1.35-2.08 | <0.0001 * | 1.30 | 0.88-1.93 | 0.191 |
| Exercise habits | 0.49 | 0.32-0.74 | <0.0001 * | 1.12 | 0.35-3.53 | 0.852 |
| Smoking | 1.38 | 1.14-1.68 | <0.0001 * | 1.37 | 0.94-2.00 | 0.105 |
| Diabetes | 1.43 | 1.09-1.86 | 0.010 * | 0.98 | 0.55-1.76 | 0.949 |
| Body mass index (BMI) | 0.99 | 0.96-1.02 | 0.425 | 1.01 | 0.96-1.07 | 0.674 |
| LDL-cholesterol | 0.97 | 0.88-1.07 | 0.501 | 0.95 | 0.79-1.16 | 0.625 |
| HDL-cholesterol | 0.89 | 0.69-1.15 | 0.359 | 0.64 | 0.38-1.09 | 0.099 |
| Education level | 0.95 | 0.88-1.02 | 0.151 | 0.98 | 0.85-1.13 | 0.784 |
| Autosomal Gain (>2 Mb) | 0.94 | 0.42-2.12 | 0.880 | 1.41 | 0.34-5.82 | 0.635 |
| Autosomal LOH (>2 Mb) | 1.41 | 0.86-2.32 | 0.172 | 1.19 | 0.43-3.26 | 0.742 |
| LOY | 2.13 | 1.08-4.19 | 0.029 * | 3.76 | 1.21-11.67 | 0.022 * |
Notes: HR – hazard ratio. 95% CI – 95% confidence interval. Autosomal LOH (>2 Mb) – autosomal loss of heterozygozity; this category was composed of deletions and CNNLOH events larger than 2 million bp. The median follow-up time was 8.7 years (range 0-20.2 years). A continuous explanatory variable was used as a proxy for loss of chromosome Y (mLRR-Y, see text and Fig. 2). None of the participants included in analyses (n=982) had any history of cancer before sampling.
indicates statistically significant effects with 0.05 alpha value.
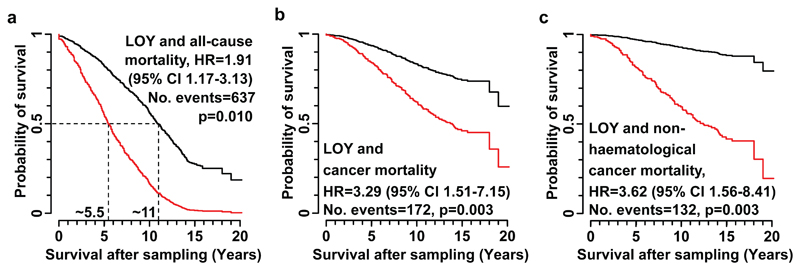
To plot the above results and to perform further exploratory tests, participants were scored based on a defined threshold of mLRR-Y. Specifically, individuals with mLRR-Y ≤ -0.4 (Fig. 2) were scored as 1 and other subjects as 0, as explained in Supplementary Figure 12. These tests confirmed the effect of LOY on risk of all-cause mortality (HR=1.91, p=0.010, Fig. 3a) and showed that median survival (50% probability of survival) in the group of men with LOY was 5.5 years shorter compared to controls, which is half the survival time. The effect from LOY on risk of cancer mortality was also confirmed (HR=3.29, p=0.003, Fig. 3b). We further found that risk for non-haematological cancer mortality was clearly associated with LOY (HR=3.62, p=0.003, Fig. 3c). Moreover, the risk of any cancer diagnosis as well as risk of non-haematological cancer diagnosis, was higher in participants with mLRR-Y ≤ -0.4 (HR=2.47, p=0.014; HR=2.68, p=0.008, respectively) (Supplementary Fig. 1). A valid test of the risk of mortality in haematological malignancies as an effect of LOY could not be performed since only one participant scored with LOY died from such cancer.
Figure 3.
LOY and its effect on mortality. Panels a, b and c show impact of LOY on all-cause mortality, cancer mortality and mortality from non-haematological cancers, respectively, in 982 men with no history of cancer prior to sampling. Hazard ratios (HR), 95% confidence intervals (CI), number of events and p-values are shown for each model. Results are derived from Cox proportional hazards regression models, with subjects classified into groups 1 and 0 based on their level of LOY. Individuals in the affected group (red curves) had LOY in ≥35% of nucleated blood cells (Fig. 2).
We replicated the result that LOY was associated with increased risk of all-cause mortality in an independent cohort of slightly younger males (age range 69.8-70.7 years) from the PIVUS (Prospective Investigation of the Vasculature in Uppsala Seniors) study. The longest and the median follow-up time was >10 and 7.0 years, respectively. Blood DNA from 488 men was genotyped using Illumina Omni-Express chip and aberrations on autosomes and sex chromosomes were scored as described above for ULSAM. The experimental noise of SNP genotyping was considerably lower in PIVUS cohort and scoring of LOY revealed that 100 males (20.5%) had LOY in ≥13% of nucleated blood cells (Supplementary Fig. 13). Furthermore, 12 autosomal somatic aberrations ≥2 Mb were scored in PIVUS including six gains, three deletions and three CNNLOH events (Supplementary Table 3). Cox proportional hazards regression with the same nine confounders were performed in PIVUS as described above for ULSAM (Supplementary Table 4). The analysis using the continuous estimator of LOY (mLRR-Y) confirmed that men with a higher degree of LOY have an increased risk of all-cause mortality (HR=5.24, p=0.022,). Furthermore, after scoring of subjects as 1 or 0 depending on LOY-status (Supplementary Fig. 14), the survival analysis showed that the men with LOY had a higher risk for all-cause mortality (HR=3.66, p=0.016, 95% CI=1.27-10.54, number of events=59). In summary, the results from ULSAM of LOY increasing the risk for all-cause mortality were validated in the independent PIVUS cohort. It has recently been reported that somatic structural variants of autosomes in blood cells are connected with risks of haematological cancers, ranging from 5.5 to 35.4-fold12,13,16. Thus, the question of whether LOY is also connected with increased risk of haematological malignancies needs to be further investigated. Furthermore, Jacobs et al. 2012 found that autosomal somatic aberrations were more frequent in subjects with solid tumors (odds ratio=1.25)12, which is in agreement with results from chromosome Y reported here.
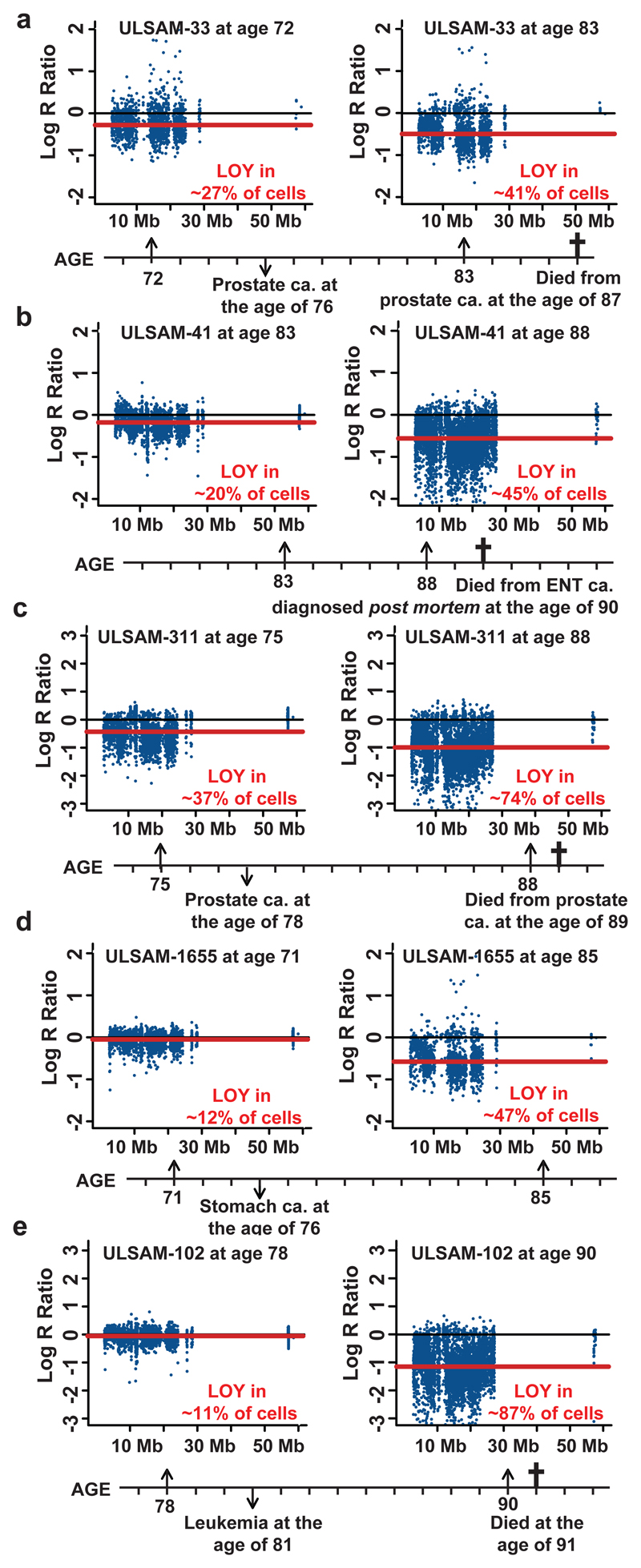
Chromosome Y is recognized for its role in sex determination and normal sperm production, but it has long been considered as a genetic wasteland and, for several reasons, its characterization has lagged behind the rest of the genome. However, recent studies in humans and in a few other primates have shown that human chromosome Y contains a large number of genes17,18, indicating that it is not a gene desert. Furthermore, variation on chromosome Y has been associated with expression of hundreds of autosomal and X-linked genes in Drosophila19,20. Our results support the concept that human chromosome Y is important for biological processes beyond sex determination and reproduction. It has been known for more than half a century that elderly males frequently lose chromosome Y in normal haematopoietic cells3,4. The clinical consequences of this aneuploidy have been unclear and the prevailing consensus suggests that this mutation should be considered phenotypically neutral and related to normal aging5–10. Furthermore, nullisomy Y has also been shown to occur in human cancer cells and the literature provides a list of up to 20 different malignancies with LOY in combination with numerous other structural aberrations. Various methods were employed in these studies showing that LOY is often occurring in high frequencies of tumors21–24. Recent comprehensive analysis shows that chromosome Y is one of the most commonly deleted chromosomes in human cancer genomes25. Moreover, azoospermia is associated with higher cancer risk26 suggesting a connection between chromosome Y and cancer. Also, reintroduction of chromosome Y into prostate cancer cell line suppresses its tumorigenicity in nude mice27. In summary, the above indicate that chromosome Y has a role in tumor suppression and our results regarding expanding cell clones with LOY are in agreement with this hypothesis. Previously published data has shown that clonal expansions are common among elderly, as a result of autosomal structural aberrations11,12,15 ,16. We addressed the question of dynamics of changes in the proportion of cells with LOY by longitudinal analyses of five ULSAM subjects collected at two times, 6-14 years apart (Fig. 4). Longitudinal LOY analyses in these subjects showed a progressive accumulation of cells containing LOY with time. All five subjects had cancer diagnoses and a high percentage of cells with LOY, ranging 41-87%. This illustrates a need for further longitudinal analyses of larger number of subjects, aiming at better characterization of the critical level of LOY mosaicism in blood that is connected with cancer risk.
Figure 4.
Longitudinal LOY analyses in five elderly men showing progressive accumulation of cells containing LOY with increasing age. Panels a-e show ULSAM subjects 33, 41, 311, 1655 and 102, respectively. Each subject was analyzed at two different ages and the lower part of each panel shows the time axis with genotyping ages, time point of cancer diagnosis and cancer type as well as age and cause of death, when applicable. The red line in each panel shows the median Log R Ratio of the male specific region of chromosome Y (mLRR-Y) as estimated from SNP-array experiments performed on blood collected at different ages in each subject. The text in red color indicates the estimated percentage of nucleated blood cells affected with LOY. This number was calculated using MAD-software from analysis of SNP-array data for the pseudoautosomal region 1 (PAR1) of chromosomes X/Y (see Supplementary Fig. 3 and Online Methods).
One of our most important findings is that men with LOY in peripheral blood are at risk for cancer outside the haematopoietic system (Fig. 3). These results could be explained by two non-mutually exclusive hypotheses. First, a normal feature of the immune system includes immunosurveillance, suppressing tumor development in other tissues28. In participants with a high degree of leukocytes with LOY, this normal function could be disturbed, leading to higher cancer risk. This explanation is supported by a higher frequency of cancer in patients with history of treatment with immunosuppressive drugs29,30. Another hypothesis is that LOY also occurs in additional tissues, in parallel with the Y-aneuploidy in blood, stimulating neoplastic proliferation of cancer progenitors outside the haematopoietic system. This hypothesis is in agreement with the above briefly described frequent LOY in a wide range of cancers. We investigated the spectrum of cancer diagnoses in men scored with LOY, in comparison to the rest of the cohort, and there were no apparent differences (Supplementary Fig. 15). Regardless of the underlying mechanism(s) for the increased cancer risk among men with LOY, our results demonstrate the importance of post-zygotic, acquired during lifetime variation in normal cells, for the risk of cancer development. As mentioned above, men show a higher incidence and higher mortality for most sex-unspecific cancers, which has been largely unexplained by known risk factors1,2. Thus, our findings could help to explain why males are more frequently affected by cancer. We further anticipate that extension of our study regarding post-zygotic LOY in blood cells, and possibly also in other tissues, could in the future become a useful predictive biomarker of male carcinogenesis.
Online Methods
Studied cohorts
The ULSAM study (Uppsala Longitudinal Study of Adult Men) was initiated in 197031, where 2322 men, born in Uppsala in 1920-1924, participated at the age of 50 (http://www2.pubcare.uu.se/ULSAM/invest/indexinv.htm). The study is investigating a wide range of phenotypes, including cancer history from the National Cancer Registry and the Swedish Civil Registry. Major re-examinations have been made at ages 60, 70, 77, 82 and 88 years. Here, we have used DNA sampled at the age window of 70.7-83.6 years and the longest follow-up time was >20 years. The study was approved by the research ethics committee. The DNA was controlled for quality as described11.
The PIVUS study (Prospective Investigation of the Vasculature in Uppsala Seniors) (http://www.medsci.uu.se/pivus) started in 2001 with the primary aim to investigate the predictive power of various measurements of endothelial function and arterial compliance. Eligible were all subjects aged 70 living in the community of Uppsala, Sweden. The subjects were randomly chosen from the community register and 1016 men and women participated. Two reinvestigations of the cohort were undertaken, starting in the spring 2006 and in the spring of 2011 at the age of 75 and 80 years, respectively32. Here, we have used 488 male DNA samples collected at the age of 70 years which were successfully genotyped. The longest and the median follow-up time was >10 and 7.0 years, respectively. The survival analysis included the same nine confounders, as described for ULSAM cohort (age at sampling, hypertension, exercise habits, smoking, diabetes, BMI, LDL, HDL and education level). All above factors, except smoking, were analyzed as for the ULSAM cohort. In PIVUS two classes of smokers were defined, current smokers and former smokers/non-smokers. The ULSAM and PIVUS studies were approved by the local research ethics committees and participants have given their informed consent.
Scoring of structural aberrations
The SNP genotyping in ULSAM and PIVUS cohorts were performed using 2.5MHumanOmni-and HumanOmniExpress-beadchips, respectively, according to the recommendations of the manufacturer. Illumina genotyping results passed strict quality control, as described11. Output files were analyzed using Nexus-Copy-Number-6.1 (BioDiscovery, CA, USA), which applies the “Rank Segmentation” algorithm, based on the Circular Binary Segmentation33, as described11. The applied version, “SNPRank Segmentation,” an extended algorithm in which Log R Ratio (LRR) as well as B allele frequency (BAF) are included in the segmentation process, generated both copy-number and allelic-event calls. The size cut-off for scoring of autosomal aberrations was set to ≥2 Mb to minimize the false classification of constitutional structural variants as somatic events12. All CNV calls made by the software were manually inspected by two investigators, prior to entering the data into a MySQL database. Circos-plots were used for visualizations (Fig. 1 and Supplementary Fig. 2)34.
Estimations of LOY frequency in the cohorts and the fraction of cells with nullisomy-Y
In ULSAM cohort, the LOY was scored from the median Log R Ratio of approx. 2560 SNP-probes (2.5M-chip) in the male-specific region of chromosome Y (mLRR-Y) in the 56 Mb region between the pseudoautosomal regions 1 and 2 (PAR1 and PAR2) on chromosome Y (chrY:2694521-59034049, hg19/GRCh37)17,18. In PIVUS cohort, an analogous scoring was done using the 1690 SNP-probes in the region on the HumanOmniExpress-chip. The total variation of mLRR-Y in the cohorts (Fig. 2 and Supplementary Fig. 13) consists of a signal from LOY and a signal from experimental variation. In order to estimate the LOY frequencies, the contribution from experimental factors was estimated. We assumed that the experimental variation in mLRR-Y was distributed in a non-skewed fashion and that the variation of the positive tail of the mLRR-Y distribution was all experimentally induced (grey bars in Fig. 2 and Supplementary Fig. 13). The latter assumption was reasonable since validation experiments failed to confirm all cases of suspected cases of gain of Y (Supplementary Fig. 11). The experimental noise (white bars in Fig. 2 and Supplementary Fig. 13) was generated by imposing the observed variation in the positive tail into a reflected negative tail in two steps. First, the peak (thick red line in Fig. 2 and Supplementary Fig. 13) in the histogram of mLRR-Y (grey bars in Fig. 2 and Supplementary Fig. 13) was determined using a kernel density estimation method (kernel medial), using the function density in R. Specifically, the bandwidth “SJ” was used and the bin with the highest value (i.e. distribution peak) in the smoothed distribution was selected as local median35. Next, the observations in the positive tail were mirrored over the kernel-median to create the negative tail. Finally, for the most conservative estimate of frequency of LOY, the lowest value in the simulated noise-distribution was used as threshold (mLRR-Y of -0.139 in ULSAM and -0.154 in PIVUS).
The percentage of cells containing a structural variant was calculated using MAD-software36. MAD is included in the R-package R-GADA37 and is used in SNP data analysis to identify regions containing deletions, gains and CNNLOH. R-GADA is detecting allelic imbalances caused by genetic abnormalities using a CBS-based BAF-segmentation algorithm38 producing calls from the BAF typical for heterozygous probes (0.5). The extent of the deviation (Bdev) is a measure of the percentage of mosaic cells, calculated using the published formula39.
For calculating the fraction of nullisomy-Y cells, we implemented a novel approach, using BAF from the pseudo-autosomal region 1 (PAR1) on chromosomes X/Y, which is explained in Supplementary Figure 3. We took advantage of LRR- and BAF-values derived from pseudoautosomal region 1 (PAR1) of chromosomes Y and X. This was possible because chromosome X was never involved in any region-specific aberrations among 1153 participants (Fig. 1). It was not feasible to use MAD-algorithm for calculating the percentage of cells in the PIVUS as was done in the ULSAM because of a few SNP-probes in the PAR1-region on the platform used (HumanOmniExpress). This platform has 33 SNP-probes in the PAR1 in comparison to the 1111 SNP-probes in the same region on the 2.5MHumanOmni. However, after adjusting for the difference in median LRR between the two cohorts the percentages of cells at the thresholds in the PIVUS cohort (i.e. ≥36% at -0.5 and >13% at -0.154) could be estimated, as it was performed in the ULSAM cohort (Supplementary Fig. 3).
Validations using next-generation sequencing
Sequencing libraries were prepared according to the TruSeq DNA v2 sample preparation EUC #15026489 revA using reagents from the TruSeq DNA v2 sample prep kit set A and set B (Illumina). A 14 pM solution of 5-6 DNA libraries pooled in equimolar amounts was subjected to cluster generation on the cBot instrument (Illumina Inc.) using the TruSeq PE cluster kit v3. Paired-end sequencing was performed for 100 cycles using a HiSeq2000 instrument (Illumina Inc.) using TruSeq SBS chemistry v3, according to the manufacturer’s protocols. Base calling was done on the instrument by RTA 1.13.48 and the resulting bcl files were filtered, demultiplexed, allowing for one mismatch base, and converted to fastq format with tools provided by CASAVA-1.8 (Illumina Inc.). Copy number profiles were computed from NGS-data using the Control-FREEC software40. The sequence read depth was used to calculate an equivalent to the Log R Ratio using a sliding window approach. We applied a fixed window size of 5 kb and the other settings were default. The NGS dataset was composed of 100 whole genomes with 5x average coverage and additional exomes, with an average coverage of 17x.
Survival and statistical analysis in ULSAM cohort
Cox proportional hazards regression models were performed in R using the package survival14. The effects on survival from three types of structural genetic variants were tested, adjusting also for nine confounders (Supplementary Table 2). LOY was the first category. In the primary survival analysis a continuous explanatory variable was used as a proxy for LOY using the median Log R Ratio (mLRR-Y). To plot the results and perform further exploratory tests, subjects were scored based on a defined threshold of mLRR-Y. Specifically, individuals with mLRR-Y ≤ -0.4 (Fig. 2) were scored as 1 and other subjects as 0, as explained in Supplementary Figure 12. The second type of structural variants investigated in the survival analyses contained the autosomal deletions and CNNLOH, grouped together as autosomal loss of heterozygozity mutations (LOH). The third category was the autosomal gains. LOHs and gains were analyzed as binary variables. The study entry was the date of DNA sampling, age was used as timeline and the median follow-up time was 8.7 years, range 0-20.2 years. All participants with cancer history (n=171) prior to sampling were excluded from survival analyses as well as samples not passing quality control. The remaining 982 subjects were included in analyses using various endpoints (Supplementary Fig. 1). We used the statistical software R (versions 2.15.0-3.0.1)41 for data mining and statistical analyses. The above described survival and statistical analyses in the ULSAM cohort were performed in an analogous way in the independent PIVUS cohort (Supplementary Figures 13 and 14, Supplementary Tables 3 and 4).
Supplementary Material
Acknowledgements
We thank G. Arnqvist and K. Lindblad-Toh for advice and F. Steele for review of the manuscript. This study was sponsored by the Olle Enqvist Byggmästare Foundation to L.A.F., by the Swedish Cancer Society, the Swedish Research Council, the Swedish Heart-Lung Foundation and Sci-Life-Lab-Uppsala to J.P.D. Genotyping and next-generation sequencing was performed by the SNP&SEQ Technology Platform in Uppsala, Sweden, and supported by Wellcome Trust Grants WT098017, WT064890, WT090532, Uppsala University, Uppsala University Hospital, the Swedish Research Council and the Swedish Heart-Lung Foundation. The SNP&SEQ Technology Platform is part of Science for Life Laboratory at Uppsala University and supported as a national infrastructure by the Swedish Research Council. C.M.L. is a Wellcome Trust Research Career Development Fellow (086596/Z/08/Z). A.P.M is a Wellcome Trust Senior Research Fellow (grant numbers WT098017, WT090532, and WT064890).
Footnotes
URLs:
ULSAM (Uppsala Longitudinal Study of Adult Men), http://www2.pubcare.uu.se/ULSAM/invest/indexinv.htm
PIVUS (Prospective Investigation of the Vasculature in Uppsala Seniors) (http://www.medsci.uu.se/pivus)
Accession codes. dbVar accession ID is nstd92.
Author Contributions
L.A.F and J.P.D conceived the study; L.A.F, C.R., E.T.J., E.I., L.L. and J.P.D. were involved in study design; D.A., L.La., V.G., C.M.L., A.P.M., E.I., and L.Li. provided materials, genotyping data and epidemiologic data; H.D., S.P., G.P., A.Z. and J.Sc. performed wet-lab analyses; C.R., L.A.F., N.M., J.Sa. and T.D.d.S., and J.Sc. implemented bioinformatic analyses; L.A.F., performed statistical analyses; L.A.F., C.R., E.I. and L.Li. were involved in survival analyses; L.A.F., N.C.P.C. and J.P.D. analyzed data; L.A.F. and J.P.D. coordinated the work and wrote the manuscript; All authors discussed the results and commented on the manuscript.
Competing Financial Interests
L.A.F and J.P.D has filed for a patent protecting the commercial applications of LOY for the assessment of cancer risk.
References
- 1.Cook MB, McGlynn KA, Devesa SS, Freedman ND, Anderson WF. Sex disparities in cancer mortality and survival. Cancer epidemiology, biomarkers & prevention. 2011;20:1629–37. doi: 10.1158/1055-9965.EPI-11-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edgren G, Liang L, Adami HO, Chang ET. Enigmatic sex disparities in cancer incidence. European journal of epidemiology. 2012;27:187–96. doi: 10.1007/s10654-011-9647-5. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs PA, Brunton M, Court Brown WM, Doll R, Goldstein H. Change of human chromosome count distribution with age: evidence for a sex differences. Nature. 1963;197:1080–1. doi: 10.1038/1971080a0. [DOI] [PubMed] [Google Scholar]
- 4.Pierre RV, Hoagland HC. Age-associated aneuploidy: loss of Y chromosome from human bone marrow cells with aging. Cancer. 1972;30:889–94. doi: 10.1002/1097-0142(197210)30:4<889::aid-cncr2820300405>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 5.Nowinski GP, et al. The frequency of aneuploidy in cultured lymphocytes is correlated with age and gender but not with reproductive history. Am J Hum Genet. 1990;46:1101–11. [PMC free article] [PubMed] [Google Scholar]
- 6.Loss of the Y chromosome from normal and neoplastic bone marrows. United Kingdom Cancer Cytogenetics Group (UKCCG) Genes, chromosomes cancer. 1992;5:83–8. doi: 10.1002/gcc.2870050112. [DOI] [PubMed] [Google Scholar]
- 7.Wiktor A, et al. Clinical significance of Y chromosome loss in hematologic disease. Genes, chromosomes & cancer. 2000;27:11–6. [PubMed] [Google Scholar]
- 8.Wong AK, et al. Loss of the Y chromosome: an age-related or clonal phenomenon in acute myelogenous leukemia/myelodysplastic syndrome? Archives of pathology & laboratory medicine. 2008;132:1329–32. doi: 10.5858/2008-132-1329-LOTYCA. [DOI] [PubMed] [Google Scholar]
- 9.Wiktor AE, Van Dyke DL, Hodnefield JM, Eckel-Passow J, Hanson CA. The significance of isolated Y chromosome loss in bone marrow metaphase cells from males over age 50 years. Leukemia research. 2011;35:1297–300. doi: 10.1016/j.leukres.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs PA, et al. Male breast cancer, age and sex chromosome aneuploidy. Br J Cancer. 2013;108:959–63. doi: 10.1038/bjc.2012.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forsberg L, et al. Age-related somatic structural changes in the nuclear genome of human blood cells. Am J Hum Genet. 2012;90:217–28. doi: 10.1016/j.ajhg.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs KB, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651–8. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schick UM, et al. Confirmation of the reported association of clonal chromosomal mosaicism with an increased risk of incident hematologic cancer. PLoS ONE. 2013;8:22. doi: 10.1371/journal.pone.0059823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Therneau T. A Package for Survival Analysis in S. R package version 2.36-12. 2012 ( http://cran.r-project.org/web/packages/survival/index.html)
- 15.Forsberg LA, Absher D, Dumanski JP. Non-heritable genetics of human disease: spotlight on post-zygotic genetic variation acquired during lifetime. Journal of medical genetics. 2013;50:1–10. doi: 10.1136/jmedgenet-2012-101322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laurie CC, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44:642–50. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Carvalho CM, Santos FR. Human Y-chromosome variation and male dysfunction. J Mol Genet Med. 2005;1:63–75. doi: 10.4172/1747-0862.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes JF, Rozen S. Genomics and genetics of human and primate y chromosomes. Annu Rev Genomics Hum Genet. 2012;13:83–108. doi: 10.1146/annurev-genom-090711-163855. [DOI] [PubMed] [Google Scholar]
- 19.Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319:91–3. doi: 10.1126/science.1148861. [DOI] [PubMed] [Google Scholar]
- 20.Lemos B, Branco AT, Hartl DL. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci U S A. 2010;107:15826–31. doi: 10.1073/pnas.1010383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vijayakumar S, et al. Detection of recurrent copy number loss at Yp11.2 involving TSPY gene cluster in prostate cancer using array-based comparative genomic hybridization. Cancer Res. 2006;66:4055–64. doi: 10.1158/0008-5472.CAN-05-3822. [DOI] [PubMed] [Google Scholar]
- 22.Zhang LJ, Shin ES, Yu ZX, Li SB. Molecular genetic evidence of Y chromosome loss in male patients with hematological disorders. Chin Med J (Engl) 2007;120:2002–5. [PubMed] [Google Scholar]
- 23.Bianchi NO. Y chromosome structural and functional changes in human malignant diseases. Mutation research. 2009;682:21–7. doi: 10.1016/j.mrrev.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Veiga LCS, Bergamo NA, Reis PP, Kowalski LP, Rogatto SR. Loss of Y-chromosome does not correlate with age at onset of head and neck carcinoma: a case-control study. Brazilian Journal of Medical and Biological Research. 2012;45:172–178. doi: 10.1590/S0100-879X2012007500004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duijf PH, Schultz N, Benezra R. Cancer cells preferentially lose small chromosomes. Int J Cancer. 2013;132:2316–26. doi: 10.1002/ijc.27924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisenberg ML, Betts P, Herder D, Lamb DJ, Lipshultz LI. Increased risk of cancer among azoospermic men. Fertil Steril. 2013 doi: 10.1016/j.fertnstert.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vijayakumar S, et al. The human Y chromosome suppresses the tumorigenicity of PC-3, a human prostate cancer cell line, in athymic nude mice. Genes, chromosomes & cancer. 2005;44:365–72. doi: 10.1002/gcc.20250. [DOI] [PubMed] [Google Scholar]
- 28.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–8. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 29.Vial T, Descotes J. Immunosuppressive drugs and cancer. Toxicology. 2003;185:229–40. doi: 10.1016/s0300-483x(02)00612-1. [DOI] [PubMed] [Google Scholar]
- 30.Vajdic CM, van Leeuwen MT. Cancer incidence and risk factors after solid organ transplantation. Int J Cancer. 2009;125:1747–54. doi: 10.1002/ijc.24439. [DOI] [PubMed] [Google Scholar]
- 31.Hedstrand H. A study of middle-aged men with particular reference to risk factors for cardiovascular disease. Ups J Med Sci Suppl. 1975;19:1–61. [PubMed] [Google Scholar]
- 32.Lind L, Fors N, Hall J, Marttala K, Stenborg A. A comparison of three different methods to evaluate endothelium-dependent vasodilation in the elderly: the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study. Arterioscler Thromb Vasc Biol. 2005;25:2368–75. doi: 10.1161/01.ATV.0000184769.22061.da. [DOI] [PubMed] [Google Scholar]
- 33.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 34.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheather S, Jones M. A Reliable Data-Based Bandwidth Selection Method for Kernel Density Estimation. Journal of the Royal Statistical Society. 1991;53:683–690. [Google Scholar]
- 36.Gonzalez JR, et al. A fast and accurate method to detect allelic genomic imbalances underlying mosaic rearrangements using SNP array data. BMC bioinformatics. 2011;12:166. doi: 10.1186/1471-2105-12-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pique-Regi R, Caceres A, Gonzalez JR. R-Gada: a fast and flexible pipeline for copy number analysis in association studies. BMC Bioinformatics. 2010;11:380. doi: 10.1186/1471-2105-11-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pique-Regi R, et al. Sparse representation and Bayesian detection of genome copy number alterations from microarray data. Bioinformatics. 2008;24:309–18. doi: 10.1093/bioinformatics/btm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez-Santiago B, et al. Mosaic uniparental disomies and aneuploidies as large structural variants of the human genome. American journal of human genetics. 2010;87:129–38. doi: 10.1016/j.ajhg.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boeva V, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–5. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R_Development_Core_Team. R: A language and environment for statistical computing. Vienna, Austria: 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.