Abstract
The gradual accumulation of genetic mutations in human adult stem cells (ASCs) during life is associated with various age-related diseases, including cancer1,2. Extreme variation in cancer risk across tissues was recently proposed to depend on the lifetime number of ASC divisions, owing to unavoidable random mutations that arise during DNA replication1. However, the rates and patterns of mutations in normal ASCs remain unknown. Here we determine genome-wide mutation patterns in ASCs of the small intestine, colon and liver of human donors with ages ranging from 3 to 87 years by sequencing clonal organoid cultures derived from primary multipotent cells3–5. Our results show that mutations accumulate steadily over time in all of the assessed tissue types, at a rate of approximately 40 novel mutations per year, despite the large variation in cancer incidence among these tissues1. Liver ASCs, however, have different mutation spectra compared to those of the colon and small intestine. Mutational signature analysis reveals that this difference can be attributed to spontaneous deamination of methylated cytosine residues in the colon and small intestine, probably reflecting their high ASC division rate. In liver, a signature with an as-yet-unknown underlying mechanism is predominant. Mutation spectra of driver genes in cancer show high similarity to the tissue-specific ASC mutation spectra, suggesting that intrinsic mutational processes in ASCs can initiate tumorigenesis. Notably, the inter-individual variation in mutation rate and spectra are low, suggesting tissue-specific activity of common mutational processes throughout life.
It has not yet been possible to measure somatic mutation loads in ASCs from specific human tissues. However, such knowledge could be valuable in understanding tissue homeostasis and repair capacities as well as ASC vulnerabilities to extrinsic factors. The accumulation of mutations as life progresses is thought to underlie the genesis of age-related diseases such as cancer6 and organ failure2. Mutations acquired in the genomes of multipotent ASCs are believed to have the largest impact on the mutational load of tissues, owing both to their potential for self-renewal and capacity to propagate mutations to their daughter cells1,2. Consistently, cancer-initiating mutations in intestinal ASCs lead to tumour formation within weeks, whereas these mutations fail to drive intestinal adenomas when induced in differentiated cells7. Unavoidable random mutations that arise during DNA replication in normal ASCs have recently been proposed to impart a large influence on cancer risk1. Consequently, tissues with a high ASC turnover would show higher cancer incidence when compared to tissues with low ASC proliferation rates1,8. However, computational modelling has suggested that the variation in ASC proliferation rate alone cannot exclude extrinsic risk factors as important determinants of organ-specific cancer incidence9. Yet, the number of mutations that accumulate during the lifespan of normal human ASCs with different turnover rates has, to date, not been directly determined and compared. To understand tissue homeostasis and tissue-specific susceptibility to cancer and ageing-associated diseases it is important to assess mutation accumulation in ASCs of different tissues.
Here, we experimentally define ASCs as those cells that give rise to long-term organoid cultures and have the potential to differentiate into multiple tissue-specific cell types3–5. To catalogue the in vivo-acquired somatic mutations in individual normal human ASC genomes, we used an in vitro system to expand single ASCs into epithelial organoids, which reflect the genetic make-up of the original ASC (Extended Data Fig. 1a and Methods). This procedure allowed us to obtain sufficient DNA for accurate whole-genome sequencing (WGS) analysis, while circumventing the high noise levels associated with single-cell DNA amplification10. We assessed ASCs from the small intestine, colon and liver, tissues that differ greatly in proliferation rate and cancer risk1. Cancer incidence is much higher in the colon compared to the small intestine and liver1. We sequenced 45 independent clonal organoid cultures derived from 19 donors ranging in age from 3 to 87 years (Extended Data Table 1). In addition, we sequenced a blood or polyclonal biopsy sample of each donor to identify and exclude germline variants. Subclonal mutations, which must have been introduced in vitro after the single-cell step, were discarded based on their low variant-allele frequency (Extended Data Figs 1b–d, 2 and Methods). Overall, we identified 79,790 heterozygous clonal somatic point mutations and subsequent extensive validations showed an overall confirmation rate of approximately 91% (Extended Data Figs 1, 3).
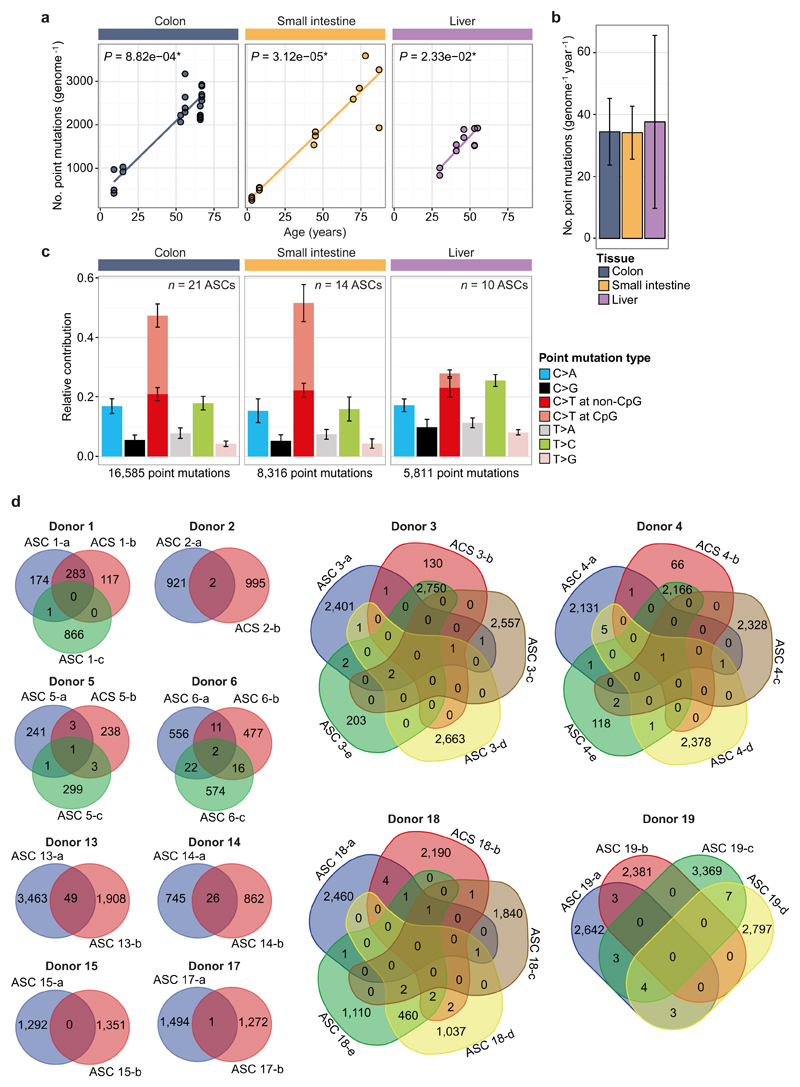
A positive correlation (t-test linear mixed model; P < 0.05) between the number of somatic point mutations and the age of the donor could be observed for all organs (Fig. 1a and Extended Data Fig. 4), indicating that ASCs gradually accumulate mutations with age, independent of tissue type. Notably, we found that the annual mutation rate in ASCs was in the same range for all assessed tissues, despite the dissimilar cancer incidence in these tissues; ASCs of the colon, small intestine and liver accumulate around 36 mutations per year (95% confidence intervals are 26.9–50.6, 25.8–43.6 and 11.9–60.1, respectively; Fig. 1b). The mutation spectra in small intestinal and colon ASCs were very similar, but differed markedly from liver (Fig. 1c). Notably, the mutation spectrum within tissues did not differ between young and elderly donors (Extended Data Fig. 5).
Figure 1. Age-associated accumulation of somatic point mutations in human ASCs.
a, Correlation of the number of somatic point mutations in each ASC type examined (extrapolated to the whole autosomal genome) with age of the donors per tissue. Each data point represents a single ASC. The P values of the age effects in the linear mixed model (two-tailed t-test) are indicated for each tissue. The sample sizes for colon, small intestine and liver ASCs are 6, 9 and, 5 donors, with, in total, 21, 14 and 10 ASCs, respectively. b, Somatic mutation accumulation rate per tissue as estimated by the linear mixed models in a. Error bars represent the 95% confidence intervals of the slope estimates. c, Relative contribution of the indicated mutation types to the point mutation spectrum for each tissue type. Data are represented as the mean relative contribution of each mutation type over all ASCs per tissue type (n = 21, 14 and 10 for colon, small intestine and liver, respectively) and error bars represent standard deviation. The total number of identified somatic point mutations per tissue is indicated.
Genome-wide mutation patterns in the ASCs provide insights into the mutational and DNA repair processes that are active in different organs11. Using non-negative matrix factorization12, we extracted three mutational process signatures (Fig. 2a and Methods). All of these signatures were previously described in a pan-cancer analysis11. Signature A (corresponding to signature 5 in ref. 11), characterized by T:A to C:G transitions, was the main contributor to the mutation spectrum observed in the liver and was also clearly present in the small intestine and colon (Fig. 2). Although the underlying mutational process remains unknown, the number of mutations attributed to this signature that accumulate with age resembles a linear trend in all tissues (Fig. 2b). This suggests that this signature represents a universal genomic ageing mechanism (that is, a chemical process acting on DNA molecules) independent of cellular function or proliferation rate.
Figure 2. Signatures of mutational processes in human ASCs and their tissue-specific contribution.
a, Contribution of context-dependent mutation types to the three mutational signatures that were identified by non-negative matrix factorization (NMF) analysis of the somatic mutation collection observed in the ASCs across all assessed tissues. The contribution of each trinucleotide (order is similar to that in ref. 11) to each signature is shown. For each signature, the presence of transcriptional-strand bias, DNA-replication-timing bias and/or association with the culture system is indicated. b, Absolute contribution of each mutational signature type (extrapolated to the whole autosomal genome) plotted against the age of the donors for each tissue. Each data point represents a single ASC. The P values of the age effects per tissue are shown (linear mixed model, two-tailed t-test). c, Signature-specific mutation rate per year per genome for each tissue as estimated by the linear mixed model in b. Error bars represent the 95% confidence intervals of the slope estimates.
The majority of the somatic mutations observed in small intestinal and colon ASCs could be attributed to signature B (corresponding to signature 1A in ref. 11), which is characteristic of spontaneous deamination of methylated cytosine residues into thymine at CpG sites (Fig. 2a). The resulting T:G mismatch can be effectively repaired, but the mutation is incorporated if DNA replication occurs before the repair is initiated13. In line with this, high rates of signature B mutations are observed in many cancer types of epithelial origin with high cell turnover13. This process showed a minimal contribution to the age-related mutational load in liver ASCs (Fig. 2c), which is likely to reflect the relatively low division rate of these cells during life. Finally, contribution of a third signature, signature C (corresponding to signature 18 in ref. 11), was minimal in all tissues and did not correlate with age (Fig. 2b). Sequential clonal ASC expansions in culture followed by WGS analysis showed that in vitro-induced mutations are predominantly characterized by this signature (Extended Data Fig. 6 and Methods).
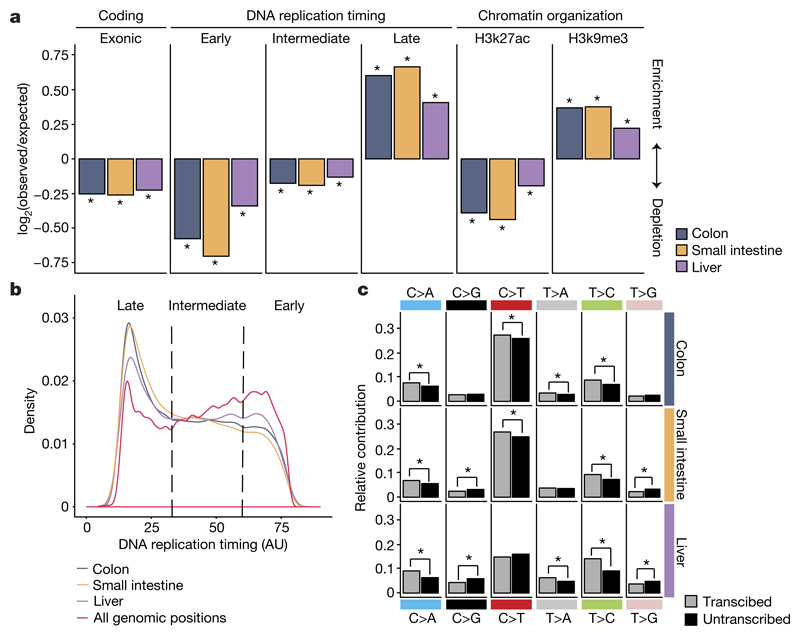
Signature B mutations were strongly associated with the timing of replication and predominantly present in late-replicating DNA (Extended Data Fig. 7) even though the majority of CpG dinucleotides are located in early-replicating DNA. This bias suggests that this mutagenic process is more active in late-replicating DNA or, alternatively, that replication-coupled repair shows reduced activity in late-replicating DNA14. Consequently, somatic mutations in small intestine and colon ASCs were strongly enriched in late-replicating DNA and depleted in early-replicating DNA (Fig. 3a, b). In addition, somatic point mutations in small intestine and colon ASCs were depleted in H3K27ac (histone H3 acetyl Lys27)-associated DNA and enriched in H3K9me3 (histone H3 trimethyl Lys9)-associated DNA (Fig. 3a), similar to patterns previously observed in cancer15. As genic regions are predominantly located in early-replicating DNA and open chromatin, we observed a depletion of mutations in exonic sequences (Fig. 3a). This demonstrates that genome-wide mutation rates and spectra cannot be reliably estimated using mutation discovery in reporter genes16, such as the T-lymphocyte HPRT cloning assay17, or by deep sequencing of genic regions18–21. To test whether the depletion of coding mutations was caused by selection against cells with damaging mutations, we calculated the ratio of non-synonymous to synonymous mutations (dN/dS) taking into account the mutation spectra and sequence composition (see Methods)18. We did not observe negative selection for non-synonymous mutations (Extended Data Fig. 7f), arguing against the negative selection of cells with damaging protein-coding mutations.
Figure 3. Non-random genomic distribution of somatic point mutations in ASCs.
a, Enrichment and depletion of somatic point mutations in the indicated genomic regions for each tissue. The log2 ratio of the number of observed and expected point mutations indicates the effect size of the enrichment or depletion in each region. *P < 0.05, one-sided binomial test. b, Distribution of DNA replication timing for all genomic positions and the somatic point mutations detected in human ASCs per tissue. c, Relative contribution of each point-mutation type on the transcribed and untranscribed strand for each tissue. *P < 0.05, two-sided Poisson test.
In liver ASCs, somatic mutations are more randomly distributed throughout the genome and are less associated with replication timing or chromatin status (Fig. 3a). Nevertheless, a comparable depletion of exonic mutations was observed in all tissues (Fig. 3a), suggesting that liver ASCs use different mechanisms to maintain genetic integrity in functionally relevant regions. Signature A, the most predominant in liver ASCs, shows little bias towards DNA-replication-timing dynamics, but a pronounced transcriptional-strand bias11 (Extended Data Fig. 7), consistent with activity of transcription-coupled repair22. In line with this, point mutations in the genic regions of the assessed ASCs showed a significant transcriptional strand bias, exemplified by the more frequent occurrence of T:A to C:G transitions on the transcribed strand compared to the untranscribed strand (Fig. 3c).
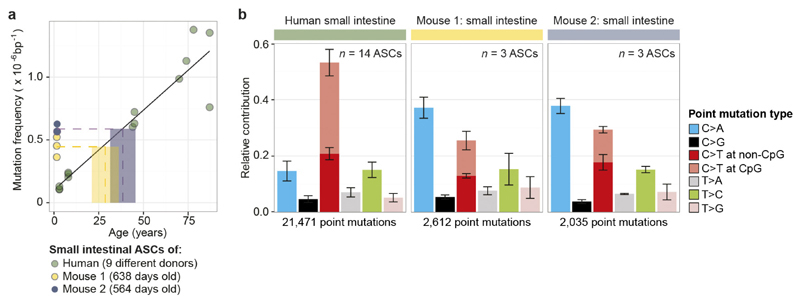
Our results indicate that a stable balance between the degree of DNA damage and the subsequent repair is maintained throughout life in various ASC types, since mutations accumulate steadily and display a constant mutation spectrum. Earlier work in mice using mutation-discovery in a LacZ reporter gene, showed major age-related changes in mutation spectra in different tissues23. The difference between these observations could be explained by the comprehensive genome-wide analysis applied here to ASCs, whereas reporter assays assess specific genes predominantly in differentiated cells. Although variation in tissue-specific mutation spectra in mice has been reported previously23–25, we observed a difference in both mutation rate and spectrum in human cells (Extended Data Fig. 8). This indicates that mutation data derived from mice are not necessarily suitable for interpreting mutational processes and their consequences in humans.
Although we analysed cells from many different donors without controlling for lifestyle differences or gender, the point-mutation rate and spectrum were highly similar between individuals within organs. This suggests that incidental exposure to environmental mutagenic factors has minimal effect on the point-mutation landscapes in normal ASCs of the organs we assessed. Cell-intrinsic mutational processes, such as deamination-induced mutagenesis in rapidly cycling ASCs, seem to be more important determinants of point-mutation load. Indeed, many colorectal cancer mutations in the driver genes APC, TP53, SMAD4 and CTNNB1 are C:G to T:A transitions at CpG dinucleotides, whereas liver cancer driver mutations in the same genes have a completely different spectrum (Fig. 4a). However, ASCs of the colon and small intestine show very similar age-related mutation characteristics, although cancer incidence is extremely low in the human small intestine1,9. In addition to somatic point mutations, we evaluated the presence of somatic structural variants (Fig. 4b–e and Extended Data Table 2). We detected small deletions (91–443 kb) in 3 out of 14 small intestinal ASCs and a larger deletion (2 Mb) in one ASC. Notably, colon ASCs showed complex and larger chromosomal instability in 4 out of 15 colon ASCs, including a complex translocation (Fig. 4d) and a trisomy (Fig. 4e). These events are characteristic of segregation errors that can occur during cell division, and are a hallmark of many colorectal cancers26. In addition, other factors, such as tissue clonality or external agents may also contribute to the difference in cancer incidence between colon and small intestine.
Figure 4. Cancer-associated mutation spectra in driver genes and structural variation in normal ASCs.
a, Spectrum of point mutations in cancer driver genes APC, TP53, SMAD4 and CTNNB1 identified in colorectal and liver cancer. The total number of somatic point mutations per gene per cancer type is indicated. b, Read-depth analysis indicating a relatively small deletion (~90 kb) located within a common fragile site (FRA2I) in intestinal ASC 11-a. Each point represents the log2 value of the GC-corrected read-depth ratio per 5-kb window. Dashed lines indicate breakpoint regions; a schematic representation of the identified structural variant with associated genomic and breakpoint features is depicted below. c, Two large (>1 Mb) tandem duplications identified in liver ASC 14-a with microhomology at the breakpoints; duplications are indicated in the schematic representation of the identified structural variants below the graph. d, A complex structural variation (an unbalanced translocation involving 3 chromosomes) identified in colon ASCs 4-b and 4-e. Coloured lines in the schematic below show the predicted derivative chromosomes. e, Read-depth analysis indicating a trisomy of chromosome 13 in colon ASC 3-c. Each data point represents the median chromosome copy number per 500-kb bin plotted over the genome, with alternating colours for each successive chromosome.
Here we have shown that ASCs of organs with different cancer incidences gradually accumulate mutations at similar rates, but that the mutation profiles are tissue-specific. In the ASCs of the tissues assessed here, mutation accumulation is primarily driven by a combination of proliferation-dependent mutation incorporation following spontaneous deamination of methylated cytosine residues and another process with a currently unknown underlying molecular mechanism. Notably, the former intrinsic, unavoidable mutational process can cause the same types of mutation as those observed in cancer driver genes. We have shown that, at least in colon ASCs, this class of mutations could have a role in driving tumorigenesis.
Methods
No sample-size estimate was calculated before the study was executed. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Human tissue material
Endoscopic, colorectal and duodenal biopsy samples were obtained from individuals of different ages that had been admitted for suspected inflammation. One individual (donor 1) showed no inflammation during colonoscopy, but was later diagnosed with microscopic colitis. The other individuals were found to be healthy based on standard histological examination. Endoscopic biopsies were performed at the University Medical Center Utrecht and the Wilhelmina Children’s Hospital. The patients’ informed consent was obtained and this study was approved by the ethical committee of University Medical Center Utrecht. Additionally, normal tissue was isolated from resected colon segments at >5 cm distance from a tumour in three colorectal cancer patients (donors 3, 4 and 19). The colonic tissues were obtained at The Diakonessen Hospital Utrecht with informed consent and the study was approved by the ethical committee. Liver biopsies (0.5–1 cm3) were obtained from donor livers during transplantations performed at the Erasmus Medical Center, Rotterdam. Both liver and colon biopsies were obtained from donor 18. The Medical Ethical Council of the Erasmus MC approved the use of this material for research purposes, and informed consent was provided by all donors and/or relatives.
Establishment of clonal ASC cultures
Dissociated colon and small intestinal crypts were isolated from the biopsies and cultured for 1 - 2 weeks under conditions that are optimal for stem-cell proliferation, as previously described5. Liver cells were isolated from human liver biopsies and cultured as previously described3. From these cultures, single cells were sorted by flow cytometry and clonally expanded (Extended Data Fig. 1a). Clonal ASC cultures were subsequently established by manual picking of individual organoids derived from single cells and in vitro expansion for a period of ~6 weeks.
Whole-genome sequencing and read alignment
DNA libraries for Illumina sequencing were generated using standard protocols (Illumina) from 200 ng - 1 μg of genomic DNA isolated from the clonally expanded ASC cultures with genomic tips (Qiagen). The libraries were sequenced with paired-end (2 × 100 bp) runs using Illumina HiSeq 2500 sequencers to a minimal depth of 30× base coverage. Samples of donors 1, 2, 3, 4, 10, 12, 13, 15, 16, 18 and 19 were sequenced using Illumina HiSeq X Ten sequencers to equal depth. The reference samples, blood or biopsy, were sequenced similarly. Sequence reads were mapped against human reference genome GRCh37 using Burrows–Wheeler Aligner v0.5.9 mapping tool27 with settings ‘bwa mem -c 100 –M’. Sequence reads were marked for duplicates using Sambamba v0.4.7 (ref. 28) and realigned per donor using Genome Analysis Toolkit (GATK) IndelRealigner v2.7.2 and sequence read-quality scores were recalibrated with GATK BaseRecalibrator v2.7.2. Alignments from different libraries of the same ASC culture were combined into a single BAM file.
Point mutation calling
Raw variants were multi-sample (per donor) called using the GATK UnifiedGenotyper v2.7.2 (ref. 29) and GATK-Queue v2.7.2 with default settings and additional option ‘EMIT_ALL_CONFIDENT_SITES’. The quality of variant and reference positions was evaluated using GATK VariantFiltration v2.7.2 with options ‘–filterExpression “MQ0 ≥4 && ((MQ0 / (1.0 * DP)) > 0.1)”–filterName “HARD_TO_VALIDATE”–filterExpression “QUAL < 30.0 “–filterName “VeryLowQual”–filterExpression “QUAL > 30.0 && QUAL < 50.0 “–filterName “LowQual”–filterExpression “QD < 1.5 “–filterName “LowQD”’.
Point mutation filtering
To obtain high-quality catalogues of somatic point mutations, we applied a comprehensive filtering procedure (Extended Data Fig. 1b). We considered variants that were passed by VariantFiltration and had a GATK phred-scaled quality score ≥100. Subsequently, for each ASC culture, we considered the positions with a base coverage of at least 20× in both the culture and the reference sample (blood or biopsy). Furthermore, we only regarded variants at autosomal chromosomes. We excluded variant positions that overlapped with single-nucleotide polymorphisms (SNPs) in the SNP database (dbSNP) v137.b37 (ref. 30). Furthermore, we excluded all positions that were found to be variable in at least two of three unrelated individuals (that is, donor 5, 6 and X (not in study)) to exclude recurrent sequencing artefacts. To obtain somatic point mutations, we filtered out all variants with any evidence of the alternative allele in the reference sample. We validated the clonal origin of the sequenced ASC cultures by analysing the variant allele frequencies (VAFs) of the somatic mutations. Two cultures (donor 14, cell b and donor 17, cell c) showed a shift in the peak of the somatic heterozygous mutations to the left, indicating that they did not arise from a single stem cell, and were therefore excluded from the analysis (Extended Data Fig. 2). Finally, for all cultures we excluded point mutations with a VAF < 0.3 to exclude mutations that were potentially induced in vitro after the (first) clonal step (Extended Data Fig. 1b–d). The number of mutations that passed each filtering step for the samples of donor 5 and 6 is depicted in Extended Data Fig. 1c. The overlap of the point mutations between ASCs of the same donor is depicted in Extended Data Fig. 4d.
Validations of point mutations
We evaluated our mutation filtering procedure by independent validations of 374 pre-selected positions that were either discarded or passed during filtering using amplicon-based next-generation sequencing. To this end, primers were designed ~250 nucleotides 5′ and 3′ from the candidate point mutations to obtain amplicons of ~500 bp (primer sequences available upon request). These regions were PCR-amplified for both the organoid cultures and reference samples of donor 5 and 6, using 5 ng genomic DNA, 1× PCR Gold Buffer (Life Technologies), 1.5 mM MgCl2, 0.2 mM of each dNTP and 1 unit of AmpliTaq Gold (Life Technologies) in a final volume of 10 μl. This which was held at 94 °C for 60 s followed by 15 cycles at 92 °C for 30 s, 65 °C for 30 s (with a decrement of 0.2 °C per cycle) and 72 °C for 60 s; followed by 30 cycles of 92 °C for 30 s, 58 °C for 30 s and 72 °C for 60 s; with a final extension at 72 °C for 180 s. The PCR products were pooled and barcoded per culture. Illumina sequence libraries were generated according to the manufacturer’s protocol. Subsequently, the libraries were pooled and sequenced using the MiSeq platform (2 × 250 bp) to an average depth of ~100×. Alignment and variant-calling was performed as described above. For each ASC we evaluated those positions with at least 20× coverage for both culture and reference sample, and defined positive positions as those with a call in culture, with a VAF ≥ 0.3 and no call in the reference sample. Subsequently, we determined the number of confirmed negatives of the positions that were filtered out for each filter step (Extended Data Fig. 1d). Moreover, we determined the number of confirmed positive of the positions that passed all filters (Extended Data Fig. 1e, f).
Assessment of effects of in vitro culturing on ASC mutation load
We expanded 10 initial clonal organoid cultures from small intestine and liver for a further 3–5 months (equivalent to ~20 weekly passages), upon which we isolated single cells and subjected them to clonal expansion to obtain sufficient DNA for WGS (Extended Data Fig. 6a). This approach allowed us to catalogue the mutations that accumulated in single ASCs during the culturing period between the two clonal steps. To this end, we selected the somatic point mutations that were unique to the sub-clonal cultures and not present in the corresponding original clonal cultures and therefore acquired during the in vitro expansion. We evaluated the specificity of our mutation-discovery procedure by determining the confirmation rate of the mutations identified in the original clone in the corresponding subclone. Only positions that had a coverage of ≥20× in both the original clonal and corresponding subclonal culture as well as in the reference sample were evaluated. On average, 91.1% ± 4.87 (mean ± s.d). of these point mutations were confirmed in the subclonal cultures (Extended Data Fig. 3).
Correlation between ASC somatic point mutation accumulation and age
The surveyed area per ASC was calculated as the number of positions coverage ≥20× in both culture and the reference sample. The percentage of the whole non-N autosomal genome (GCRh37: 2,682,655,440 bp) that is surveyed in each ACS is depicted in Extended Data Table 1. For each ASC the total number of identified somatic point mutations was extrapolated to the whole non-N autosomal genome using its surveyed area. Subsequently, a linear mixed-effects regression model was fitted to estimate the effect of age on the number of somatic point mutations for each tissue using the nlme R package31,32, in which ‘donor’ is modelled as a random effect to resolve the non-independence that results from having multiple measurements per donor. A two-tailed t-test was performed to test whether the slope is significantly different from zero (that is to say, whether the fixed age effect in the linear mixed model is statistically significant). The intercept of the regression lines with the y axis represents the somatic mutations present at birth (that have accumulated in the tissue lineage during prenatal development) plus the noise levels in the data and the mutations that have accumulated during the first week(s) of culturing proceeding the clonal step (see above). Since all cells were assessed in a similar manner, noise levels will be comparable and therefore will not bias the mutation rate (slope) estimates. The slope of the regression line was used to estimate the fixed age effect on somatic point mutation rate per tissue.
To exclude the possibility that differences in surveyed areas between ASCs bias our results, we performed the age correlation and spectrum analyses on a subset of mutations that are located in genomic regions that are surveyed (≥20×) in all samples in this study. This consensus surveyed area comprises 38.2% of the autosomal non-N genome and both the mutation rate and spectra were highly similar to those in Fig. 1c (Extended Data Fig. 4a–c), indicating that the differences in surveyed areas between the clones do not bias our conclusions.
Definition of genomic regions
To generate a conserved DNA replication timing profile for the human genome, we downloaded 16 Repli-seq data sets from the ENCODE project33 at the University of California, SantaCruz (UCSC) genome browser34 (GRCh37/hg19). The data consisted of Wavelet-smoothed values per 1-kb bin throughout the genome for 15 different cell lines (BJ, BG02ES, GM06990, GM12801, GM12812, GM12813, GM12878, HeLa-S3, HepG2, HUVEC, IMR90, K562, MCF-7, NHEK and SK-N-SH). We considered the median values of all cell lines per bin, thereby excluding cell-specific values. We arbitrarily divided the genome into early- (≥60), intermediate- (>33 & <60) and late- (≤33) replicating bins (Fig. 3b). To generate a conserved chromatin-association profile for the human genome, we downloaded data containing the H3K9me3 signal per 25-nucleotide bin throughout the genome for 22 different cell lines (A549, AG04450, DND41, GM12878, H1-hESC, HeLa-S3, HepG2, HMEC, HSMM, HSMMt, HUVEC, K562, monocytes-CD14+_RO1746, NH-A, NHDF-Ad, NHEK, NHLF, osteoblasts, MCF-7, NT2-D1, PBMC and U2OS) and the H3K27ac signal for 9 different cell lines (CD20+_RO01794, DND41, H1-hESC, HeLa-S3, HSMM, monocytes-CD14+_RO1746, NH-A, NHDF and osteoblasts). Data were downloaded from the ENCODE project33 at the UCSC browser34 (GRCh37/hg19) and the median values of all cell lines per bin were calculated. Next, we determined the distribution of the fractions of all bins (genome-wide). According to the shape of the resulting graph, we considered bins with an H3K9me3 value ≥4, or an H3K27ac value ≥2, as associated with that chromatin mark. Finally, exonic sequences were defined as all exonic regions reported in Ensembl v75 (GCRh37)35.
Enrichment or depletion of point mutations in genomic regions
We determined whether somatic point mutations were enriched or depleted in the genomic regions described above. To this end, we determined how many point mutations were observed in each genomic region for each donor. Next, we calculated the number of bases that were surveyed in each genomic region and calculated the expected number of point mutations by multiplying this surveyed length with the genome-wide point-mutation frequency. The log2(observed/expected) of the mutations in the genomic regions was used as a measure of the effect size of the depletion or enrichment. One-tailed binomial tests were performed to calculate the statistical significance of deviations from the expected number of mutations in the genomic regions using pbinom31; P < 0.05 was considered significant.
Mutational signatures
The occurrences of all 96-trinucleotide changes were counted for each ASC and averaged per donor. Three mutational signatures were extracted using NMF36. To determine the replication bias of signatures, we determined whether the point mutations were located in an intermediate, early or late replicating region (as defined above) using GenomicRanges37 and repeated the NMF on a 288 count matrix (96 trinucleotides × 3 replication timing regions). Similarly, we looked at transcriptional strand bias by performing NMF on a 192 count matrix (96 trinucleotides × 2 strands). To this end, we selected all point mutations that fall within gene bodies and checked whether the mutated C or T was located on the transcribed or non-transcribed strand. We defined the transcribed units of all protein coding genes based on Ensembl v75 (GCRh37)35 and included introns and untranslated regions.
Selection analysis (dN/dS)
The dN/dS ratio was determined as described previously18. In brief, we used 192 rates, one for each of the possible trinucleotide changes in both strands. For each substitution type, we counted the number of potential synonymous and non-synonymous mutations in the protein-coding sequences of the human genome, using the longest DNA coding sequence as the reference sequence for each gene. Poisson regression was used to obtain maximum-likelihood estimates and confidence intervals of the normalized ratio of non-synonymous versus synonymous mutations (dN/dS ratio). The dN/dS ratio was tested against neutrality (dN/dS = 1) using a likelihood-ratio test.
Comparison of mouse and human intestinal ASCs mutation loads
Intestinal ASCs were isolated from the proximal part of the small intestine of randomly chosen ~2-year-old mice (one male and one female) carrying the Lgr5-EGFP-Ires-CreERT2 allele (mice were C57BL/6 background) by sorting for GFPhigh cells. Subsequently, three Lgr5-positive cells per animal were clonally expanded as described4. All experiments were approved by the Animal Care Committee of the Royal Dutch Academy of Sciences according to the Dutch legal ethical guidelines. DNA isolated from the intestinal ASC cultures isolated from mouse 1 were sequenced with paired-end (75 and 35 bp) runs using SOLiD 5500 sequencers (Life Technologies) to an average depth of ~18× base coverage. Intestinal ASC cultures of mouse 2 were sequenced using Illumina HiSeq 2500 sequencers as described above. Sequence reads were aligned using Burrows–Wheeler Aligner to the mouse reference genome (NCBIM37) and point mutations were called using the GATK UnifiedGenotyper v2.7.2 as described above. Post-processing filters for the intestinal ASCs of mouse 1 (analysed by SOLiD sequencing) were as follows: a minimum depth of 10×, variant uniquely called in one intestinal stem cell without more than one alternative allele found at the same position in the other ASCs of the same mouse, a GATK a phred-scaled quality score ≥100, variant absent in mouse 2, variant position absent in the dbSNP (build 128) and a VAF ≥ 0.25. Post-processing filters for the intestinal ASCs of mouse 2 (analysed by Illumina sequencing) were as described above for the human mutation data.
Cancer-associated mutation spectra analysis in driver genes
Mutations identified in the indicated genes in colorectal or liver cancers were downloaded from cBioPortal (http://www.cbioportal.org/). Only point mutations that resulted in a missense, nonsense or splice-site mutation were considered.
CNV detection
To detect copy-number variations (CNVs), BAM files were analysed for read-depth variations by CNVnator v0.2.7 (ref. 38) with a bin size of 1 kb and Control-FREEC v6.739 with a bin size of 5 kb. Highly variable regions, defined as harbouring germline CNVs in at least three control samples, were excluded from the analysis. To obtain somatic CNVs, we excluded CNVs for which there was evidence in the reference sample (blood/biopsy) of the same individual. Resulting candidate CNV regions were assessed for additional structural variants on the paired-end and split-read level through DELLY v0.3.3 (ref. 40). Based on these results, we excluded five candidate CNV regions as mapping artefacts on the read-depth level and acquired base-pair accuracy of the involved breakpoints for the other events. This also revealed the tandem orientation of the duplication events and the complex structural variation in the colon sample.
Reported gene definitions (Extended Data Table 2) are based on Ensembl v75 (GCRh37)35. Common fragile sites overlapping the events were detected using existing definitions41. LINE/SINE elements within 100 bp of the breakpoints were determined with the repeat element annotation42 from the UCSC genome browser34 GCRh37 (retrieved 26 October 2015).
Code availability
All code and filtered vcf files are freely available under a MIT License at https://wgs11.op.umcutrecht.nl/mutational_patterns_ASCs/ and https://github.com/CuppenResearch/MutationalPatterns/.
Extended Data
Extended Data Figure 1. Cataloguing somatic mutation loads in human ASCs.
a, Schematic overview of the experimental setup to determine somatic mutations in individual human ASCs. Colon, small intestine and liver biopsies were cultured in bulk for 1-2 week(s) before single cells were sorted and clonally expanded until enough DNA could be isolated for WGS analysis. WGS of the clonal organoid culture allows for cataloguing of somatic variants in the original ASCs that gave rise to the clonal cultures that were acquired during life and the first 7–14 days of culturing. Biopsy or blood was sequenced as a reference sample. b, Filter steps to obtain somatic mutations in ASCs. c, Number of point mutations that pass each corresponding filter step in a for each ASC culture of donors 5 and 6. d, Independent validations of mutations that were filtered out by amplicon-based resequencing. The asterisk indicates a position that is not located in the surveyed areas of the assessed ASCs in the original experiment, which is corrected for in all analyses. e, Independent validations of mutations that passed all filters by amplicon-based re-sequencing. Confirmed positions are defined as those with a call in the indicated ASC with a VAF ≥ 0.3 and without a call in the corresponding reference sample. f, Qualification of unconfirmed positions based on manual inspection. True-positive positions are positions that were correctly called, but for which the VAF threshold was not met in the validation experiment. False-positive positions are positions without evidence in the validation experiment or are noisy. ‘Missed in germline’ are positions that were called in the reference sample in the validation experiment.
Extended Data Figure 2. Variant allele frequency distribution plot for each assessed ASC.
A distribution plot of the VAFs of all somatic mutations that remain before filtering for the VAF in filter step 6 (Extended Data Fig. 1b). Clonal heterozygous somatic mutations form a peak around VAF = 0.5. A threshold of VAF ≥ 0.3 was used to obtain somatic mutations that were clonal in the organoid cultures and therefore present in the original cloned ASCs (see Methods). Mutations acquired after the single ASC expansion step are subclonal (that is, not present in all cells of the clonal culture) and have lower VAFs. Two samples (donor 14, ASC 14-b and donor 17, ASC 17-c) showed a shift in the main VAF peak to the left, indicating that these cultures did not arise from a single ASC and were therefore excluded from the study.
Extended Data Figure 3. Confirmation rate of somatic point mutations.
a, Overlap of somatic point mutations between the clonal organoid cultures and corresponding subcloned cultures depicted in Extended Data Fig. 6. b, Confirmation rate of point mutations, which were observed in the original cloned culture, in the corresponding subcloned culture. Data are represented as the mean percentage of confirmed point mutations over all clone–subclone pairs indicated in a (n = 10) and error bars represent s.d.
Extended Data Figure 4. Somatic mutation loads in consensus-surveyed area and overlap of point mutations between ASCs from the same donor.
a, Correlation of the number of somatic point mutations per ASC, which were observed in the genomic regions that were surveyed (for example, a base coverage of at least 20× in both the clonal culture and the reference sample; Methods) in all the ASCs, with the age of the donors per tissue indicated. This consensus-surveyed area comprises 38.2% of the non-N autosomal genome. Each data point represents a single ASC. Indicated are the P values of the age effects in the linear mixed model (two-tailed t-test) for each tissue. The sample sizes for colon, small intestine and liver are 6, 9 and 5 donors and 21, 14 and 10 ASCs, respectively. b, Somatic mutation accumulation rate per tissue as estimated by the linear mixed models in a. Error bars represent the 95% confidence intervals of the slope estimates. c, Relative contribution of the indicated mutation types to the point mutation spectra in the consensus-surveyed area per tissue type. Data are represented as the mean relative contribution of each mutation type over all ASCs per tissue type (n = 21, 14 and 10 for colon, small intestine and liver, respectively); error bars represent s.d. The total number of identified somatic point mutations per tissue is shown. d, Overlap of the somatic point mutations between ASCs of the same donor. The number of point mutations, observed in the total surveyed area per ASC, that are shared between the assessed ASCs of the same donor is indicated.
Extended Data Figure 5. Point-mutation spectrum per donor.
Relative contribution of the different types of point mutation to the spectrum of each donor. Data are represented as the mean relative contribution of each mutation type when multiple ASCs were measured per donor (the number n of ASC per donor is depicted for each donor) and error bars represent standard deviation. Indicated are the age of the donors, the total number of point mutations used to determine each spectrum and the tissue type.
Extended Data Figure 6. Mutation patterns associated with long-term in vitro expansion of ASCs.
a, Schematic overview of the experimental setup to catalogue mutations associated with the organoid culture system. Clonal small intestinal and liver organoid cultures (Extended Data Fig. 1a) were cultured for 3–5 months. A second clonal expansion step was subsequently performed, followed by WGS analysis, to catalogue all the mutations that were present in the subcloned ASCs. To obtain mutations that were specifically acquired during culturing, mutations in the original clonal cultures were subtracted from those observed in the corresponding second subcloned cultures. b, Relative contribution of the indicated mutation types to the point mutation spectra specifically observed in vitro per tissue type. Data are represented as the mean relative contribution of each mutation type over all subcloned ASCs per tissue type (n = 6 and 4 for small intestine and liver, respectively) and error bars represent s.d. Indicated are the total number of identified somatic point mutations, which were specifically acquired between the two clonal expansion steps indicated in a, per tissue. c, Relative contribution of the mutational signatures depicted in Fig. 2a, which explain the mutation spectra depicted in b.
Extended Data Figure 7. Non-random distribution of mutational signatures throughout the genome.
a, Context- and replication-timing-dependent mutation spectrum of the three mutational signatures depicted in Fig. 2a. Indicated is the contribution of each trinucleotide to the signatures (order is similar as in ref. 11), subdivided into the fraction of the trinucleotide-change present in early, intermediate or late replicating genomic regions. b, log2 ratio of the observed and expected number of mutations per indicated base substitution (summed over all trinucleotides) in early-, intermediate- and late-replicating genomic regions for each of the signatures depicted in a. log2 ratio indicates the effect size of the bias and asterisks indicate significant DNA-replication-timing bias (P < 0.05, binomial test). c, log2 ratio of the total number of observed and expected mutations in early-, intermediate- and late-replicating genomic regions for each signature depicted in a. log2 ratio indicates the effect-size of the bias and asterisks indicate significant DNA replication timing bias (P < 0.05, binomial test). d, Context- and transcriptional-strand-dependent mutation spectrum of the three mutational signatures depicted in Fig. 2a. Indicated is the contribution of each trinucleotide to the signatures (order is similar to that in ref. 11), subdivided into the fraction of the trinucleotide-change present on the transcribed and untranscribed strand. e, log2 ratio of the number of mutations on the transcribed and untranscribed strand per indicated base substitution for each signature depicted in d. log2 ratio indicates the effect size of the bias and asterisks indicate significant transcriptional strand bias (P < 0.05, binomial test). f, The dN/dS ratio for all protein-coding somatic point mutations observed in all ASCs per tissue type. Error bars indicate 95% confidence intervals (likelihood ratio test).
Extended Data Figure 8. Comparison of mutation loads between intestinal ASCs derived from human and mouse.
a, Mutation frequency in mouse intestinal ASCs is compared to the linear fit, describing the relationship between the mutation frequency in human intestinal ASCs and age of the donor. Indicated by the dotted lines are the mean mutation frequencies over all ASCs per mouse (n = 3) and the corresponding age of human linear fit. b, Relative contribution of the indicated mutation types to the point mutation spectra for all assessed human intestinal ASCs and for each mouse. Data are represented as the mean relative contribution of each mutation type over all the ASCs per indicated category (n = 14, 3 and 3 for human, mouse 1 and mouse 2, respectively), error bars indicate s.d.
Extended Data Table 1. Overview of somatic point mutations detected in ASCs.
| ASC | Donor | Age (years) | Gender | Tissue | Surveyed genome (%)* | No. point mutations† |
|---|---|---|---|---|---|---|
| 1-a | 1 | 9 | Female | Colon | 93.8 | 458 |
| 1-b | 1 | 9 | Female | Colon | 91.2 | 400 |
| 1-c | 1 | 9 | Female | Colon | 97.6 | 867 |
| 2-a | 2 | 15 | Male | Colon | 96.8 | 923 |
| 2-b | 2 | 15 | Male | Colon | 96.8 | 997 |
| 18-a | 18 | 53 | Male | Colon | 98.5 | 2,468 |
| 18-b | 18 | 53 | Male | Colon | 98.1 | 2,201 |
| 19-a | 19 | 56 | Male | Colon | 97.7 | 2,655 |
| 19-b | 19 | 56 | Male | Colon | 97.1 | 2,384 |
| 19-c | 19 | 56 | Male | Colon | 98.2 | 3,383 |
| 19-d | 19 | 56 | Male | Colon | 97.8 | 2,811 |
| 4-a | 4 | 66 | Female | Colon | 90.7 | 2,140 |
| 4-b | 4 | 66 | Female | Colon | 95.3 | 2,234 |
| 4-c | 4 | 66 | Female | Colon | 95.7 | 2,332 |
| 4-d | 4 | 66 | Female | Colon | 93.9 | 2,386 |
| 4-e | 4 | 66 | Female | Colon | 96.1 | 2,289 |
| 3-a | 3 | 67 | Female | Colon | 91.8 | 2,409 |
| 3-b | 3 | 67 | Female | Colon | 91.8 | 2,882 |
| 3-c | 3 | 67 | Female | Colon | 91.9 | 2,561 |
| 3-d | 3 | 67 | Female | Colon | 92.0 | 2,667 |
| 3-e | 3 | 67 | Female | Colon | 92.0 | 2,957 |
| 5-a | 5 | 3 | Female | Small intestine | 89.0 | 246 |
| 5-b | 5 | 3 | Female | Small intestine | 85.5 | 245 |
| 5-c | 5 | 3 | Female | Small intestine | 88.5 | 304 |
| 6-a | 6 | 8 | Female | Small intestine | 97.1 | 591 |
| 6-b | 6 | 8 | Female | Small intestine | 93.5 | 506 |
| 6-c | 6 | 8 | Female | Small intestine | 96.2 | 614 |
| 7-a | 7 | 44 | Male | Small intestine | 91.1 | 1,461 |
| 8-a | 8 | 45 | Male | Small intestine | 95.5 | 1,838 |
| 9-a | 9 | 45 | Male | Small intestine | 93.9 | 1,571 |
| 10-a | 10 | 70 | Female | Small intestine | 94.8 | 2,497 |
| 11-a | 11 | 74 | Male | Small intestine | 87.3 | 2,613 |
| 12-a | 12 | 78 | Female | Small intestine | 95.6 | 3,516 |
| 13-a | 13 | 87 | Male | Small intestine | 97.7 | 3,512 |
| 13-b | 13 | 87 | Male | Small intestine | 97.0 | 1,957 |
| 14-a | 14 | 30 | Male | Liver | 81.3 | 771 |
| 14-b | 14 | 30 | Male | Liver | 85.2 | 888 |
| 15-a | 15 | 41 | Female | Liver | 93.5 | 1,292 |
| 15-b | 15 | 41 | Female | Liver | 95.1 | 1,351 |
| 17-a | 17 | 46 | Female | Liver | 79.4 | 1,495 |
| 17-b | 17 | 46 | Female | Liver | 73.7 | 1,273 |
| 18-c | 18 | 53 | Male | Liver | 97.9 | 1,845 |
| 18-d | 18 | 53 | Male | Liver | 98.5 | 1,504 |
| 18-e | 18 | 53 | Male | Liver | 98.2 | 1,577 |
| 16-a | 16 | 55 | Male | Liver | 97.5 | 1,919 |
Percentage of the non-N autosomal genome with ≥20× coverage in both ASC culture and reference sample.
Number of somatic point mutations detected within surveyed genome.
Extended Data Table 2. Identified somatic structural variations in ASCs.
| Copy Number Variants | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Tissue | Chr | Start | Stop | Size | Type | No. genes | Fragile site | Microhomology | Genes at breakpoint | LINE/SINE |
| ASC 14-a | Liver | 3 | 94,491,729 | 95,651,811 | 1,160,082 | gain | 5 | - | 5 bp | -|- | L1MC1|- |
| ASC 14-a | Liver | 3 | 111,726,406 | 113,471,637 | 1,745,231 | gain | 46 | - | 2 bp | TAGLN3|ATP6V1A | L1MC1|L1M5 |
| ASC 16-a | Liver | 9 | 50,763,759 | 141,213,431 | 90,449,672 | gain | 1,472 | - | NA | NA | NA |
| ASC 18-e | Liver | 7 | 132,751,706 | 133,009,202 | 257,496 | gain | 2 | - | 0 bp | CHCHD3|EXOC4 | MIR|L1PA6 |
| ASC 18-d | Liver | 5 | 59,125,105 | 59,718,364 | 593,259 | loss | 1 | - | 0 bp | PDE4D|PDE4D | -|L1PA6 |
| ASC 8-a | Small intestine | 5 | 3,815,936 | 3,908,819 | 92,883 | loss | 0 | - | 2 bp | -|- | -|- |
| ASC 11-a | Small intestine | 2 | 205,420,067 | 205,511,877 | 91,810 | loss | 1 | FRA2I | 1 bp | PARD3B|PARD3B | AluSx|L1ME3B |
| ASC 13-a | Small intestine | 11 | 63,974,352 | 66,222,668 | 2,248,316 | loss | 163 | - | 3 bp | FERMT3|- | -|L1M4b |
| ASC 13-b | Small intestine | 1 | 5,878,566 | 6,321,750 | 443,184 | loss | 13 | FRA1A | 1 bp | -|- | THE1B|- |
| ASC 1-c | Colon | 3 | 60,700,662 | 61,199,328 | 498,666 | loss | 4 | FRA3B | 1 bp | FHIT|FHIT | L1PA3|L1PA3 |
| ASC 3-c | Colon | 13 | 0 | 115,169,878 | 115,169,878 | gain | 1,217 | - | NA | NA | NA |
| ASC 4-b&e | Colon | 14 | 102,805,595 | 104,172,376 | 1,366,781 | loss | 57 | - | NA | NA | NA |
| ASC 4-b&e | Colon | 17 | 2,429,169 | 2,572,747 | 143,578 | loss | 5 | - | CTTG ins | -|PAFAH1B1 | AluJo|AluSq |
| ASC 4-b&e | Colon | 17 | 2,634,433 | 2,927,007 | 292,574 | loss | 4 | - | NA | NA | NA |
| Unbalanced Translocations | |||||||||||
| Sample | Tissue | Chr (1) | Position (1) | Chr (2) | Position (2) | Type | No. genes | Fragile site | Microhomology | Genes at breakpoint | LINE/SINE |
| ASC 4-b&e | Colon | 14 | 102,805,595 | 17 | 2,634,145 | translocation | NA | - | 4 bp | ZNF839|- | -|- |
| ASC 4-b&e | Colon | 14 | 104,172,376 | 18 | 18,518,987 | translocation | NA | - | 0 bp | XRCC3|- | -|ALR Alpha |
| ASC 4-b&e | Colon | 17 | 2,927,007 | 18 | 18,518,987 | translocation | NA | - | 0 bp | RAF1GAP2|- | L1PA8|ALR Alpha |
No. genes, number of genes overlapping the event; fragile site, common fragile sites overlapping the event; microhomology, number of bases of microhomology observed at breakpoints; genes at breakpoint, gene bodies affected by the breakpoint; LINE/SINE elements, observed elements within 100 bp of the breakpoint.
Acknowledgements
The authors would like to thank the gastroenterologists of the UMCU/Wilhelmina Children’s Hospital and Diakonessen Hospital for obtaining human duodenal and colon biopsies and R. Eijkemans for his advice on the statistical analyses. This study was financially supported by a Zenith grant of the Netherlands Genomics Initiative (935.12.003) to E.C., the NWO Zwaartekracht program Cancer Genomics.nl and funding of Worldwide Cancer Research (WCR no. 16-0193) to R.B. We declare no competing financial interests.
Footnotes
Author Contributions C.L.W., S.M. and E.E.S.N. obtained duodenal biopsies. N.S., M.M., E.E.S.N., M.M.A.V. and J.J. obtained colon biopsies. M.M.A.V., L.J.W.L., J.J. and J.N.M.I. obtained human liver biopsies. M.J., V.S., N.S., M.H., E.K., C.L.W., T.S., G.S. and R.B. performed ASC culturing. M.W. performed cell sorting. S.R., M.R.S., E.C. and R.B. performed sequencing. F.B., J.L., S.B., P.P., I.J.N., I.M. and R.B. performed bioinformatic analyses. F.B, R.G.V., H.C., E.C. and R.B. were involved in the conceptual design of the study. F.B., H.C., E.C. and R.B. wrote the manuscript.
Author Information The human sequencing data have been deposited at the European Genome-phenome Archive (http://www.ebi.ac.uk/ega/) under accession numbers EGAS00001001682 and EGAS00001000881. The mouse sequencing data have been deposited at the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) under accession number ERP005717. Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of the paper.
The authors declare no competing financial interests.
Reviewer Information Nature thanks G. Pfeifer, L. Vermeulen, J. Vijg and the other anonymous reviewer(s) for their contribution to the peer review of this work.
References
- 1.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossi DJ, Jamieson CHM, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 3.Huch M, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 5.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 6.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 8.Milholland B, Auton A, Suh Y, Vijg J. Age-related somatic mutations in the cancer genome. Oncotarget. 2015;6:24627–24635. doi: 10.18632/oncotarget.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu S, Powers S, Zhu W, Hannun YA. Substantial contribution of extrinsic risk factors to cancer development. Nature. 2016;529:43–47. doi: 10.1038/nature16166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 11.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Reports. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexandrov LB, et al. Clock-like mutational processes in human somatic cells. Nat Genet. 2015;47:1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Supek F, Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015;521:81–84. doi: 10.1038/nature14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuster-Böckler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 16.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finette BA, et al. Determination of HPRT mutant frequencies in T-lymphocytes from a healthy pediatric population: statistical comparison between newborn, children and adult mutant frequencies, cloning efficiency and age. Mutat Res. 1994;308:223–231. doi: 10.1016/0027-5107(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 18.Martincorena I, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie M, et al. Age-related cancer mutations associated with clonal hematopoietic expansion. Nat Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dollé MET, Snyder WK, Dunson DB, Vijg J. Mutational fingerprints of aging. Nucleic Acids Res. 2002;30:545–549. doi: 10.1093/nar/30.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dollé ME, Snyder WK, Gossen JA, Lohman PH, Vijg J. Distinct spectra of somatic mutations accumulated with age in mouse heart and small intestine. Proc Natl Acad Sci USA. 2000;97:8403–8408. doi: 10.1073/pnas.97.15.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behjati S, et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014;513:422–425. doi: 10.1038/nature13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R Core Team. R: A language and environment for statistical computing. 2015 http://www.r-project.org/
- 32.Pinheiro J, et al. nlme: Linear and Nonlinear Mixed Effects Models. 2016 https://cran.r-project.org/web/packages/nlme/nlme.pdf.
- 33.ENCODE Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2013;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenbloom KR, et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cunningham F, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence M, et al. Software for computing and annotating genomic ranges. PLOS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boeva V, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Tallec B, et al. Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Reports. 2013;4:420–428. doi: 10.1016/j.celrep.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]