Abstract
Background and Purpose
von Willebrand Factor (vWF) plays an important role in thrombus formation during cerebrovascular damage. We sought to investigate the potential role of circulating vWF in recurrent cerebrovascular events and identify genetic contributors to variation in vWF level in an ischemic stroke population.
Methods
We analyzed the effect of circulating vWF on risk of recurrent stroke using survival models in the Vitamin Intervention for Stroke Prevention (VISP) trial as well as the utility of vWF in reclassification over traditional factors. We conducted a genome-wide association study (GWAS) with imputation, based upon 1000 Genomes Project data, for circulating vWF levels and then interrogated loci previously associated with vWF levels. We performed expression quantitative trait locus (eQTL) analysis for vWF across different tissues.
Results
Elevated vWF levels were associated with increased risk for recurrent stroke in VISP. Adding vWF to traditional clinical parameters also improved recurrent stroke risk prediction. We identified SNPs significantly associated with circulating vWF at the ABO locus (p < 5×10-8) and replicated findings from previous genetic associations of vWF levels in humans. eQTL analyses demonstrate that most associated ABO SNPs were also associated with vWF gene expression.
Conclusions
Elevated vWF levels are associated with recurrent stroke in VISP. In the VISP population, genetic determinants of vWF levels that impact vWF gene expression were identified. These data add to our knowledge of the pathophysiologic and genetic basis for recurrent stroke risk, and may have implications for clinical care decision-making.
Introduction
Many genetic and epidemiological studies have investigated factors affecting risk of first ischemic stroke. Fewer studies have focused on recurrent ischemic stroke as an outcome. After a first ischemic stroke, the risk for recurrence is increased 9-fold over the incident stroke risk for the general population.1 Recurrent strokes more frequently disable or kill individuals than first-ever strokes. Young and middle-aged individuals (<50 years) with recurrent stroke have an observed mortality approximately 14-fold higher than the expected mortality in age-matched non-stroke controls.2 Further, mortality resulting from stroke recurrence among older individuals (>65 years) is increased between 2- and 4-fold.1 Although established as a risk factor for first stroke3, the effect of elevated von Willebrand Factor (vWF) levels on recurrent stroke risk remains under-investigated. We hypothesize that expanding our knowledge of both vWF levels and genetic determinants of vWF levels influence recurrent stroke risk and could aid future clinical practice.
vWF is a multimeric glycoprotein expressed in endothelial cells and megakaryocytes.4,5 vWF has two major roles in hemostasis, though its role is thought to very broad6: First, it acts as a carrier of factor VIII (FVIII) in plasma and second, it aids in platelet function which includes adhesion to the subendothelium, platelet-to-platelet interactions, and platelet aggregation in vessels that have undergone elevated shear stress. Even though high plasma levels of vWF predict stroke risk7,8, the direct relationship between vWF and ischemic stroke is less well understood.3
Our research on vWF levels, associated genes, and recurrent stroke risk utilizes samples and data from the Vitamin Intervention for Stroke Prevention clinical trial (VISP). The VISP biomarker and genetic subsample9 consists of n = 1931 individuals over the age of 35 who had a non-disabling ischemic stroke and total homocysteine levels in the top quartile of the general population (Table 1).). For this subset, we merged data on vWF protein levels with genome-wide genotyping data (dbGaP Study Accession: phs000343.v2.p1). We aimed to explore a possible role for vWF in recurrent stroke risk, and investigate genomic loci that contribute to variation of vWF levels.
Table 1. Study Population Summary Statistics.
| N | |||
|---|---|---|---|
| Total Subjects Evaluated | 2100 | ||
| Male | 1335 (63.6%) | ||
| Female | 765 (36.4%) | ||
|
| |||
| Trait | N | Median | IQR1 |
|
| |||
| Age Male | 1335 | 68 | 16 |
| Age Female | 765 | 69 | 15 |
| Hcy (μmol/L) | 2029 | 12.2 | 4.6 |
| vWF (IU/L) | 2014 | 1074.1 | 729.5 |
| Prothrombin fragments F1+2 (nmol/L) | 1986 | 0.96 | 0.69 |
| Thrombin-antithrombin complex (μg/L) | 1484 | 3.7 | 3.2 |
| Thrombomodulin (ng/mL) | 2009 | 3.6 | 2.6 |
| Fibrinogen (mg/dL) | 2000 | 392 | 104 |
| Vitamin B6 (pm/mL) | 1793 | 33.4 | 37.3 |
| Vitamin B12 (pg/mL) | 1898 | 327 | 165 |
| Folate assay with L. casei (ng/mL) | 1897 | 22.7 | 15.6 |
| BMI | 2017 | 27.4 | 6.4 |
|
| |||
| Vascular Risk Factor | N | Events(%) | |
|
| |||
| Current Smoker | 2100 | 328(15.6) | |
| Prevalent Diabetes | 2100 | 570(27.1) | |
| Prevalent Hypertension (JNC-7 Stage 1 or greater) | 2096 | 1515(72.3) | |
IQR: interquartile range
Materials and Methods
Study population
The Vitamin Intervention for Stroke Prevention Trial (VISP) was a randomized, controlled clinical trial of B-vitamin supplementation to prevent recurrent stroke, myocardial infarction or death.9All participants enrolled in the VISP trial were diagnosed with a non-disabling ischemic stroke and had hyperhomocysteinemia. Subtype classification was not performed in the VISP trial. Detailed inclusion/exclusion criteria can be found in Toole et al. 2004.9 All participants in VISP provided informed consent for the proposed study, and the study was approved by site Institutional Review Boards.
von Willebrand Factor measurement
vWF activity was measured in citrated plasma using Enzyme-linked immunosorbent assay using standard manufacturer's protocols (ELISA) (IMUBIND vWF Activity, American Diagnostica Inc., Greenwich, CT).
Genetic analyses
A detailed description of genotyping methods can be found in previous reports.10,11 We conducted a genome-wide test of association between vWF level and single nucleotide polymorphism (SNPs) in 1931unrelated individuals included in the genetic subsample of VISP. Analyses were conducted with PLINK v1.0.7.12 using multivariable linear regression modeling. We adjusted for age, sex and population structure using the first 10 principal components derived from genotype data using KING software (Wei-Min Chen. KING: Kinship-based Inference for Gwas. http://people.virginia.edu/∼wc9c/KING/ [access date 09/01/16]).13
eQTL analysis
Expression quantitative trait loci (eQTL) analyses were performed using data from the Gene Tissue Expression Project (GTEx). eQTL analyses were performed by testing rs505922 against all mRNAs in all tissues (Tissue Expression Project (GTEx) http://gtexportal.org/home/snp/rs505922 [access date 01/15/17]). Next, we tested rs505922 specifically against vascular related tissues and ABO and vWF mRNA using the “Test your own” function found at Tissue Expression Project (GTEx) http://www.gtexportal.org/home/testyourown [access date 01/15/17].
Survival analyses of recurrent stroke and all-cause mortality
The associations between vWF levels and time to recurrent stroke/mortality during the trial, or SNPs and time to recurrent stroke/mortality during the trial, were performed using Cox proportional hazards models. There were 182 individuals with stroke recurrence during the study follow-up period and 108 individuals that died of all-causes (all-cause mortality) (total n = 2014). Time was measured from the time of randomization (high or low dose B-vitamins). Censoring occurred due to the loss of follow up, unexplained death, and/or early termination. SNPs were coded using additive genetic models. Clinical site was treated as a strata variable. We used two models: a basic model adjusting only for the intervention group (high or low dose B-vitamins), and a full model adjusting for the intervention group, age, race, sex, smoking, BMI, diabetes status, hypertension, and LDL.
Variance Analysis
Variance explained by SNP rs505922 was conducted using the statistical computing software R.14 The R code for this analysis can be found at https://osf.io/9y3k8/.
AUC, NRI, and IDI analyses
Area under the receiver operating characteristic curve (AUC) was used to assess the ability of demographic variables and the vWF level to predict for recurrent stroke. This value was estimated using the entire cohort as well as 100 bootstrapped samples to reduce the possibility of overfitting. Reclassification of recurrent stroke risk between the two nested models (demographic variables with or without vWF level) was calculated based on predicted values from regression models. Net reclassification improvement (NRI) was used to measure the degree to which recurrent stroke risk was appropriately reclassified.15 Each risk result was classified into low (<0.06), intermediate (0.06-<0.09), or high (≥0.09) risk categories. Since the cut of points may affect the result for the categorical NRI. The continuous NRI was also calculated.16 The integrated discrimination improvement (IDI) calculated the difference in discrimination slopes between the two nested models.17 The SAS macro can be found here: Nancy Cook's Risk Prediction Modeling. Division of Preventative Medicine. http://ncook.bwh.harvard.edu/sas-macros.html [access date 02/03/2017].
Results
We investigated the influence of vWF levels on recurrent stroke and all-cause mortality. Elevated vWF increases (per standard deviation) risk for stroke recurrence in both our unadjusted (p=0.0007, HR = 1.26) and fully adjusted (p=0.018, HR=1.19) models. Elevated vWF was also found to be associated with all-cause mortality in the unadjusted (p=0.0122, HR=1.21) but not the adjusted model (p=0.263, HR=1.10) in the VISP stroke population (Table 1). The positive association between vWF and recurrent stroke prompted us to investigate the genetic determinants of vWF variation in VISP.
Our genome wide association analysis (GWAS) identified 146 SNPs within or near the ABO locus that reached our predefined significance cutoff of p=5×10-8 (Figure 1; Supplemental Table I). Our most strongly associated SNP, rs505922 (p=2.32×10-30, variance explained = 6%), was previously associated with vWF levels in the general population (Table 2; Supplemental Table I).18
Figure 1. Global and regional genome-wide association study results.
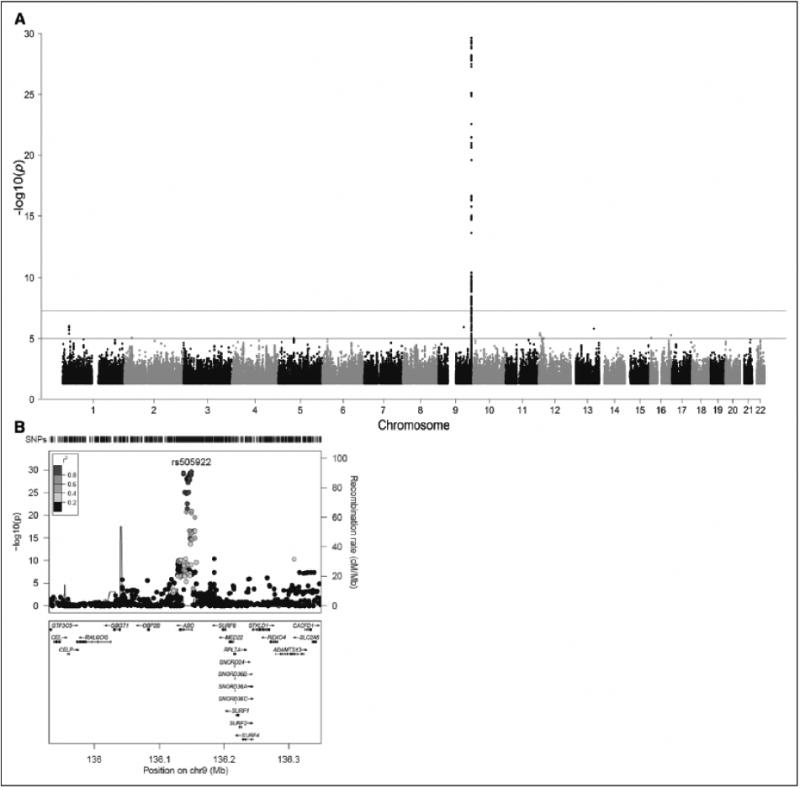
A, Manhattan plot of von Willibrand Factor GWAS results. Topline represents genome wide significance cutoff (p = 5x10-8). Bottomline represents genome wide suggestive line (p = 1x10-5) B, Regional association plot of chromosome 9 vWF associated locus surrounding ABO.
Table 2. Replicated SNPs across other GWAS studies.
| Ch. | SNP | BP1 | P-value VISP | Gene | Replication2 | Found in Vascular Trait2,3 | Found in Other Trait(s)2,4 |
|---|---|---|---|---|---|---|---|
|
| |||||||
| vWF | |||||||
| 9 | rs2519093 | 136142120 | 1.53E-21 | ABO | - | + | 1 |
| 9 | rs495828 | 136155117 | 2.88E-17 | ABO | - | + | 1 |
| 9 | rs4962153 | 136324004 | 5.54E-08 | ABO | - | - | 1 |
| 9 | rs657152 | 136139515 | 7.22E-26 | ABO | - | 1 | |
| 9 | rs505922 | 136149479 | 2.32E-30 | ABO | + | + | 4 |
| 9 | rs507666 | 136149649 | 2.36E-21 | ABO | - | - | 1 |
| 9 | rs579459 | 136154418 | 2.46E-17 | ABO | - | + | 7 |
| 9 | rs612169 | 136143692 | 7.92E-29 | ABO | - | - | 1 |
| 9 | rs644234 | 136142467 | 1.35E-25 | ABO | - | - | 1 |
| 9 | rs649129 | 136154554 | 2.76E-17 | ABO | - | - | 1 |
| 9 | rs651007 | 136154125 | 2.60E-17 | ABO | - | - | 3 |
| 9 | rs657152 | 136139515 | 7.22E-26 | ABO | - | + | 5 |
| 9 | rs687289 | 136137356 | 3.93E-30 | ABO | + | - | - |
| 9 | rs687621 | 136137315 | 6.83E-30 | ABO | - | + | 1 |
| 9 | rs8176743 | 136131665 | 1.84E-10 | ABO | + | - | - |
| 9 | rs8176749 | 136131438 | 9.24E-11 | ABO | + | - | 2 |
| 9 | rs8176746 | 136131322 | 4.00E-8 | ABO | + | - | 1 |
Human genome Feb. 2009 (GRCh37/hg19) Assembly.
NHGRI catalog of published genome-wide association studies(http://www.genome.gov/gwastudies/ [access date 09/01/16) replication of vWF levels.
Significant in GWAS of vascular trait (i.e. cardiovascular disease, venous thrombosis).
Significant in GWAS of other clinical phenotypes. Expanded in Supplemental Table III.
Further, we assessed the utility of vWF levels and ABO SNP rs505922 in recurrent stroke prediction over conventional risk factors using those taken from the Essen risk score using NRI, IDI, ROC curves. The results of these analyses can be found in Supplemental table II and Supplemental figure I. We confirmed that in all conditions (conventional risk factor (CF) vs CF +vWF, CF vs CF+ rs505922, and CF vs CF+vWF+ rs505922) vWF remains significantly associated with increased risk for recurrent stroke over traditional risk measures. The traditional model adjusted for treatment group, diabetes status, hypertension, smoking, age, and prior history of stroke. When vWF was added to the risk model, it improved discrimination; the C statistic increased from 0.635 to 0.648 based on logistic regression models. A significantly higher percentage of participants were reclassified appropriately than inappropriately, with the net reclassification benefit measured by net reclassification improvement (NRI) of 10.6% (p=0.0028). The integrated discrimination improvement (IDI) was 0.004 (p=0.0388). With 100 bootstrap samples, the category-free NRI was 19.7% (p=0.0113) indicating that there was a 19.7% net improvement in the predictions of recurrent stroke. The IDI was 0.004 (95% CI 0.0004-0.0087 from bootstrap samples; p=0.1014 assumes normality). We further investigated the prediction ability using a survival model. With the 100 bootstrap samples, the average difference of the two C statistics was 0.006 (p<0.0001). Overall, this shows the addition of vWF does improve the ability to predict recurrent stroke risk. The results of using the whole sample and bootstrap samples were consistent.
To limit confounding factors in these analyses, we also performed a comprehensive interaction analysis and shown that there is no significant interaction between tHCy or treatment arm in 7 different survival models which include both genetic and non-genetic covariates (all p >0.05). These include model parameters: Original model + thcy + thcy*vWF, Original model + treatment arm*vWF, Original model + thcy + treatment arm*vWF, Original model + thcy + rs505922 + treatment arm*vWF, Original model + thcy + rs505922 + thcy*vWF, Original model + thcy + rs505922 + treatment arm* rs505922, and Original model + thcy + rs505922 + thcy* rs505922. Data not shown.
We sought to determine whether our remaining most significantly associated SNPs had also been reported in previous genetic studies of vWF levels and other vascular traits as defined by the NHGRI-EBI GWAS catalogue (http://www.ebi.ac.uk/gwas/ [access date 09/01/16]). Five of our top SNPs replicated previous findings in genetic studies of vWF levels and six were found to have been significantly associated with vascular traits (Table 2). Ten of our significantly associated SNPs had been previously reported as associated with one or more traits (Supplemental Table III).
Next, using a candidate gene approach, we identified regions that were previously associated with vWF levels, or vWF with venous thrombosis, and interrogated the current analysis for significance. We found positive associations with rs732505,19 identified in a combined analysis of two GWAS reports of vWF (Table 3). Furthermore, we replicated vWF associations in ADAMTS13, (rs4962153, p = 5.54×10-8), OBP2B (rs11244035, p = 2.33 × 10-6), C9orf96 (rs28602591, p = 1.52 × 10-4), and vWF (rs1063856, p =5.26×10-6)20 (Table 3; a priori Bonferroni corrected significance threshold p = 6.25 × 10-3).
Table 3. Significant loci from previous studies looked up in current study data.
| vWF | CHR | SNP | BP | Gene | P VISP | Publication | Publication P | Trait |
|---|---|---|---|---|---|---|---|---|
| 19 | rs732505 | 5582535 | SAFB2 | 0.05441 | Antoni G19 | 9.38×10-6 | vWF | |
| 9 | rs4962153 | 136323754 | ADAMTS13 | 5.54×10-8 | Desch KC20 | 1.20E-24 | vWF | |
| 9 | rs11244035 | 136081319 | OBP2B | 2.33×10-6 | Desch KC20 | 3.40E-17 | vWF | |
| 9 | rs28602591 | 136246666 | C9orf96 | 0.0001517 | Desch KC20 | 4.80E-16 | vWF | |
| 9 | rs554710 | 136181848 | SURF6 | 0.00222 | Desch KC20 | 1.80E-07 | vWF | |
| 12 | rs1063856 | 6153534 | VWF | 5.26×10-6 | Desch KC20 | 8.50E-14 | vWF | |
|
| ||||||||
| vWF and Venous Thrombosis | CHR | SNP | BP | Gene | P VISP | Publication | Publication P | Trait |
|
| ||||||||
| 6 | rs1039084 | 147635413 | STXBP5 | 0.03577 | Smith NL21 | 5.20E-03 | vWF and Venous Thrombosis | |
| 9 | rs687621 | 136137065 | ABO | 6.83E-30 | Smith NL21 | 2.40E-06 | vWF and Venous Thrombosis | |
| 12 | rs1063856 | 6153534 | VWF | 5.26E-06 | Smith NL21 | 3.60E-02 | vWF and Venous Thrombosis | |
To determine whether our significantly associated SNPs influence tissue-specific mRNA expression of ABO and vWF we performed expression quantitative trait loci (eQTL) analyses. Using the Gene Tissue Expression Project (GTEx V6) dataset, we found that rs505922 was significantly associated with ABO expression levels in 15 tissues, with the highest effect size in skeletal muscle (p=2.7×10-32, effect size = 0.80) when tested against all mRNA transcripts (Table 4). Interestingly, rs505922 was also associated with vWF mRNA levels in lung (p = 0.04, effect size = 0.12). Given the source of vWF measures in VISP (whole blood), we tested rs505922 specifically for an effect on ABO expression in only whole blood and found the association remained, although with an effect size in the opposite direction of skeletal muscle (p = 7×10-4, effect size = -0.27).
Table 4. eQTL results between rs505922 and ABO. GTEx (V6p).
| Gencode Id | Gene Symbol | SNP Id | P-Value1 | Effect Size | Tissue |
|---|---|---|---|---|---|
| ENSG00000271875.1 | ABO | rs505922 | 2.70E-32 | 0.8 | Muscle - Skeletal |
| ENSG00000271875.1 | ABO | rs505922 | 1.30E-28 | 0.67 | Esophagus - Mucosa |
| ENSG00000175164.9 | ABO | rs505922 | 6.20E-28 | 0.73 | Esophagus - Mucosa |
| ENSG00000175164.9 | ABO | rs505922 | 1.80E-20 | 0.67 | Muscle - Skeletal |
| ENSG00000175164.9 | ABO | rs505922 | 6.40E-18 | 0.86 | Adrenal Gland |
| ENSG00000271875.1 | ABO | rs505922 | 8.20E-18 | 0.62 | Adipose - Visceral (Omentum) |
| ENSG00000271875.1 | ABO | rs505922 | 1.40E-17 | 0.48 | Thyroid |
| ENSG00000271875.1 | ABO | rs505922 | 2.80E-17 | 0.39 | Lung |
| ENSG00000271875.1 | ABO | rs505922 | 4.20E-16 | 0.62 | Nerve - Tibial |
| ENSG00000271875.1 | ABO | rs505922 | 2.20E-15 | 0.83 | Adrenal Gland |
| ENSG00000271875.1 | ABO | rs505922 | 9.90E-15 | 0.62 | Esophagus - Muscularis |
| ENSG00000175164.9 | ABO | rs505922 | 1.10E-14 | 0.58 | Adipose - Visceral (Omentum) |
| ENSG00000271875.1 | ABO | rs505922 | 1.80E-14 | 0.72 | Pituitary |
| ENSG00000271875.1 | ABO | rs505922 | 3.40E-14 | 0.51 | Heart - Left Ventricle |
| ENSG00000271875.1 | ABO | rs505922 | 3.60E-14 | 0.6 | Skin - Sun Exposed (Lower leg) |
| ENSG00000271875.1 | ABO | rs505922 | 1.10E-13 | 0.46 | Testis |
| ENSG00000271875.1 | ABO | rs505922 | 1.50E-13 | 0.36 | Artery - Tibial |
| ENSG00000175164.9 | ABO | rs505922 | 2.60E-12 | 0.38 | Lung |
| ENSG00000175164.9 | ABO | rs505922 | 2.60E-12 | 0.48 | Thyroid |
| ENSG00000271875.1 | ABO | rs505922 | 1.40E-11 | 0.45 | Adipose - Subcutaneous |
| ENSG00000175164.9 | ABO | rs505922 | 6.60E-11 | 0.82 | Pancreas |
| ENSG00000175164.9 | ABO | rs505922 | 1.80E-10 | 0.69 | Heart - Atrial Appendage |
| ENSG00000271875.1 | ABO | rs505922 | 1.80E-10 | 0.78 | Vagina |
| ENSG00000271875.1 | ABO | rs505922 | 1.90E-10 | 0.28 | Colon - Transverse |
| ENSG00000271875.1 | ABO | rs505922 | 2.20E-10 | 0.62 | Skin - Not Sun Exposed (Suprapubic) |
| ENSG00000271875.1 | ABO | rs505922 | 2.30E-10 | 0.8 | Pancreas |
| ENSG00000175164.9 | ABO | rs505922 | 5.90E-10 | 0.75 | Vagina |
| ENSG00000175164.9 | ABO | rs505922 | 8.10E-10 | 0.51 | Nerve - Tibial |
| ENSG00000175164.9 | ABO | rs505922 | 1.20E-09 | 0.42 | Adipose - Subcutaneous |
| ENSG00000271875.1 | ABO | rs505922 | 2.10E-09 | 0.61 | Heart - Atrial Appendage |
| ENSG00000271875.1 | ABO | rs505922 | 2.10E-09 | 0.41 | Artery - Coronary |
| ENSG00000175164.9 | ABO | rs505922 | 3.10E-09 | 0.44 | Testis |
| ENSG00000175164.9 | ABO | rs505922 | 3.80E-09 | 0.45 | Skin - Sun Exposed (Lower leg) |
| ENSG00000175164.9 | ABO | rs505922 | 5.00E-08 | 0.27 | Colon - Transverse |
| ENSG00000175164.9 | ABO | rs505922 | 8.10E-08 | 0.42 | Heart - Left Ventricle |
| ENSG00000175164.9 | ABO | rs505922 | 9.50E-08 | 0.58 | Pituitary |
| ENSG00000175164.9 | ABO | rs505922 | 1.10E-07 | 0.47 | Esophagus - Muscularis |
| ENSG00000175164.9 | ABO | rs505922 | 1.80E-07 | 0.54 | Skin - Not Sun Exposed (Suprapubic) |
| ENSG00000175164.9 | ABO | rs505922 | 2.00E-07 | 0.45 | Artery - Coronary |
| ENSG00000271875.1 | ABO | rs505922 | 4.50E-07 | 0.71 | Liver |
| ENSG00000175164.9 | ABO | rs505922 | 8.60E-07 | 0.7 | Ovary |
| ENSG00000271875.1 | ABO | rs505922 | 1.4E-06 | 0.43 | Colon - Sigmoid |
| ENSG00000271875.1 | ABO | rs505922 | 0.000002 | 0.45 | Breast - Mammary Tissue |
| ENSG00000175164.9 | ABO | rs505922 | 4.8E-06 | 0.43 | Breast - Mammary Tissue |
Corrected p-value.
Finally, we investigated whether there was shared genetic variation at the ABO locus that not only contributed to vWF levels but also recurrent stroke. Even though elevated vWF protein levels increased the risk for stroke recurrence, and ABO SNPs influence vWF mRNA levels, there was no independent relationship between significant ABO GWAS SNPs and recurrent stroke prediction.
Discussion
In the current study, we investigated the relationship between vWF and recurrent stroke risk in the VISP population. Individuals with high vWF levels in VISP have an increased risk for stroke recurrence using both basic and more comprehensively adjusted survival analyses. vWF may play a role in all-cause mortality but when assessed in the fully adjusted model these findings become unclear. These results suggest that vWF levels may represent a novel and independent risk factor for recurrent ischemic stroke, even after accounting for known risk factors such as smoking, BMI, and hypertension. This raises the question as to whether measuring vWF following an acute ischemic stroke might provide meaningful risk stratification of stroke recurrence in the clinical setting.
Using a GWA approach for recurrent ischemic stroke, we identified a previously known locus for vWF at the ABO locus, including rs505922 which has been previously identified as associated with ischemic stroke, venous thromboembolism, and factor VIII levels (Table 3, Supplemental Table I).19–21 The EuroCLOT study identified this same SNP as associated with vWF, stroke, large vessel, and cardioembolic stroke.18 Additional ABO SNPs have been associated with vascular outcomes including venous thromboembolism, ischemic stroke, and coronary heart disease (Table 3) (Supplemental Table III) indicating a major role for this gene in the pathophysiology of vascular disease.
Based on a comprehensive literature search, we investigated loci other than ABO that influence vWF levels. Although these loci did not reach genome-wide significance in our initial analyses, using a post hoc candidate SNP lookup approach, we detected associations with reported loci in the VISP population. We not only replicated previous finding of an association of vWF levels with the VWF gene but also confirmed association between vWF levels and ADAMTS13, OBP2B, C9orf96, and SURF6 (Table 3), suggesting that these loci may influence vWF levels even in the setting of post-acute ischemic stroke even though our analysis might simply have been underpowered to identify these associations in the primary investigation. However, we must also investigate the possibility that these genes simply are not associated with vWF in the VISP study participants for the tissue investigated.
Although this study has identified a potentially useful biomarker in stratification of patients with risk for stroke recurrence, it is not without limitations. For example, our study includes a low proportion of individuals of non-European descent, potentially limiting generalizability. Stroke subtype information is not available for VISP; however, given that atrial fibrillation was an exclusion criterion, we speculate that the majority of qualifying strokes in VISP were caused by small-vessel disease, intracranial atherosclerosis, or cryptogenic mechanisms.11 The exclusion of most extracranial large artery and cardioembolic strokes from VISP contrasts with the EuroCLOT study which found an association between an ABO SNP and these stroke mechanisms.18 Also, while the GTEx project is the largest resource of its kind, numbers of individuals per tissue are limited and further analyses should and will be performed. Finally, there may truly be no association with VISP participants and the loci that failed to replicate which provides important information to the community given the homogenous nature of this population.
We show a novel association between vWF levels and recurrent stroke risk, and confirmed reported genetic associations with vWF in an ischemic stroke population. Finally, we found that several of the associated variants in the ABO gene appear to be tissue-specific drivers of gene expression levels, furnishing a plausible mechanism of association. In summary, we believe these data warrant further investigation to determine whether vWF and its determinants might help in stratifying risk of stroke recurrence in ischemic stroke patients. We strongly believe this warrants further investigation and could influence clinical care if validated.
Supplementary Material
Acknowledgments
The authors would like to thank all individuals who volunteered, participated, and continue to participate in the VISP studies.
Sources of Funding: VISP was supported by the National Human Genome Research institute and the Genomics and Randomized Trials Network (U01HG00516-03, http://www.genome.gov/27541119). G.D. and J.R.3 were supported by NIH R25 HL088724. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dennis MS, Burn JPS, Sanderrock DM, Bamford JM, Wade DT, Warlow CP. Long-term survival after first-ever stroke: the Oxfordshire Community Stroke Project. Stroke. 1993;24:796–800. doi: 10.1161/01.str.24.6.796. [DOI] [PubMed] [Google Scholar]
- 2.Aarnio K, Haapaniemi E, Melkas S, Kaste M, Tatlisumak T, Putaala J. Long-term mortality after first-ever and recurrent stroke in young adults. Stroke. 2014;45:2670–2676. doi: 10.1161/STROKEAHA.114.005648. [DOI] [PubMed] [Google Scholar]
- 3.De Meyer SF, Stoll G, Wagner DD, Kleinschnitz C. Von Willebrand factor: An emerging target in stroke therapy. Stroke. 2012;43:599–606. doi: 10.1161/STROKEAHA.111.628867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Meyer SF, De Maeyer B, Deckmyn H, Vanhoorelbeke K. Von Willebrand factor: drug and drug target. Cardiovasc Hematol Disord Drug Targets. 2009;9:9–20. doi: 10.2174/187152909787581327. [DOI] [PubMed] [Google Scholar]
- 5.Sadler JE. Biochemistry and genetics of von Willebrand factor. Annu Rev Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 6.Peyvandi F, Garagiola I, Baronciani L. Role of von Willebrand factor in the haemostasis. Blood Transfus. 2011;9(2):s3–8. doi: 10.2450/2011.002S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flossmann E, Schulz UGR, Rothwell PM. Systematic review of methods and results of studies of the genetic epidemiology of ischemic stroke. Stroke. 2004;35:212–227. doi: 10.1161/01.STR.0000107187.84390.AA. [DOI] [PubMed] [Google Scholar]
- 8.Roldán V, Marín F, García-Herola A, Lip GYH. Correlation of plasma von Willebrand factor levels, an index of endothelial damage/dysfunction, with two point-based stroke risk stratification scores in a trial fibrillation. Thromb Res. 2005;116:321–325. doi: 10.1016/j.thromres.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Toole JF, Malinow MR, Chambless LE, Spence JD, Pettigrew LC, Howard VJ, et al. Lowering Homocysteine in Patients With Ischemic Stroke to Prevent Recurrent Stroke, Myocardial Infaction, and Death. JAMA. 2004;291:565–575. doi: 10.1001/jama.291.5.565. [DOI] [PubMed] [Google Scholar]
- 10.Williams SR, Yang Q2, Chen F3, Liu X4, Keene KL5, Jacques P6, et al. Genome-Wide Meta-Analysis of Homocysteine and Methionine Metabolism Identifies Five One Carbon Metabolism Loci and a Novel Association of ALDH1L1 with Ischemic Stroke. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams SR, Hsu FC, Keene KL, Chen WM, Nelson S, Southerland AM, et al. Shared genetic susceptibility of vascular-related biomarkers with ischemic and recurrent stroke. Neurology. 2016;86:351–359. doi: 10.1212/WNL.0000000000002319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Core Team. R: A Language and Environment for Statistical Computing. [Accessed 09/01/16];The R Project for Statistical Computing. https://www.r-project.org/
- 15.Pencina MJ, D'Agostino RB, D'Agostino RB, Vasan RS. Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 16.Pencina MJ, Steyerberg EW, DA RB., Sr Net reclassification index at event rate: properties and relationships. Stat Med. 2016:1097–0258. doi: 10.1002/sim.7041. [DOI] [PubMed] [Google Scholar]
- 17.Pencina MJ, Fine JP, D'Agostino RB. Discrimination slope and integrated discrimination improvement - properties, relationships and impact of calibration. Stat Med. 2016:1097–0258. doi: 10.1002/sim.7139. [DOI] [PubMed] [Google Scholar]
- 18.Williams FMK, Carter AM, Hysi PG, Surdulescu G, Hodgkiss D, Soranzo N, et al. Ischemic stroke is associated with the ABO locus: the Euro CLOT study. Ann Neurol. 2013;73:16–31. doi: 10.1002/ana.23838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antoni G, Oudot-Mellakh T, Dimitromanolakis A, Germain M, Cohen W, Wells P, et al. Combined analysis of three genome-wide association studies on vWF and FVIII plasma levels. BMC Medical Genetics. 2011;12:102. doi: 10.1186/1471-2350-12-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desch KC, Ozel AB, Siemieniak D, Kalish Y, Shavit JA, Thornburg CD, et al. Linkage analysis identifies a locus for plasma von Willebrand factor undetected by genome-wide association. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:588–593. doi: 10.1073/pnas.1219885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith NL, Rice KM, Bovill EG, Cushman M, Bis CJ, McKnight B, et al. Genetic variation associated with plasma von Willebrand factor levels and the risk of incident venous thrombosis. Blood. 2011;117:6007–6011. doi: 10.1182/blood-2010-10-315473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


