Abstract
Gene coexpression networks are a useful tool for summarizing transcriptomic data and providing insight into patterns of gene regulation in a variety of species. Though there has been considerable interest in studying the evolution of network topology across species, less attention has been paid to the relationship between network position and patterns of molecular evolution. Here, we generated coexpression networks from publicly available expression data for seven flowering plant taxa (Arabidopsis thaliana, Glycine max, Oryza sativa, Populus spp., Solanum lycopersicum, Vitis spp., and Zea mays) to investigate the relationship between network position and rates of molecular evolution. We found a significant negative correlation between network connectivity and rates of molecular evolution, with more highly connected (i.e., “hub”) genes having significantly lower nonsynonymous substitution rates and dN/dS ratios compared to less highly connected (i.e., “peripheral”) genes across the taxa surveyed. These findings suggest that more centrally located hub genes are, on average, subject to higher levels of evolutionary constraint than are genes located on the periphery of gene coexpression networks. The consistency of this result across disparate taxa suggests that it holds for flowering plants in general, as opposed to being a species-specific phenomenon.
Introduction
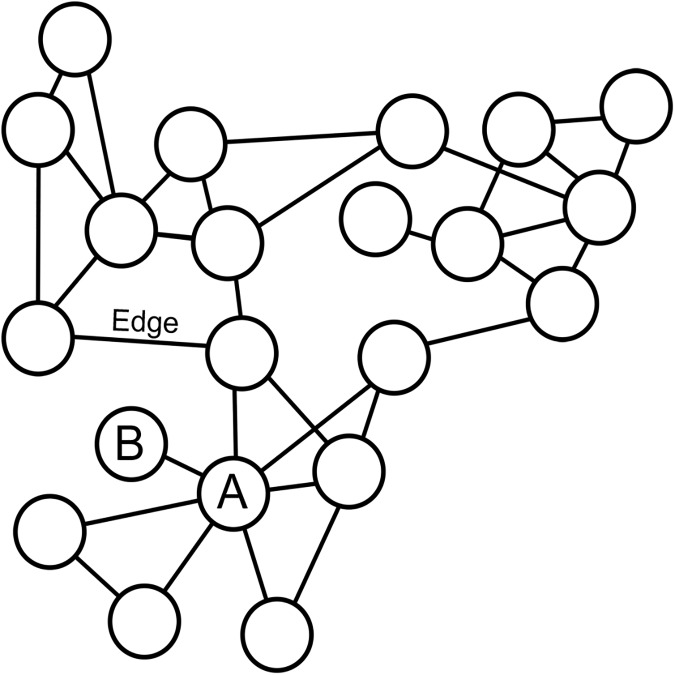
In recent years, transcriptomic analyses have become a standard tool in many laboratories (e.g., [1–3]). Driven by the availability of high-density arrays and the ongoing improvement of nucleotide sequencing platforms, massive amounts of transcriptomic data have thus been produced (e.g., [3,4]). While much attention has been paid to patterns of differential expression and instances of alternative splicing across experimental perturbations, many have sought to place transcriptomic data into a broader biological framework via the construction of gene coexpression networks (e.g., [5–11]). Coexpression networks are composed of a series of nodes (i.e., genes) and edges (i.e., connections) that reflect correlations in gene expression (Fig 1). These networks are often constructed from multiple tissue types, developmental stages, and/or experimental treatments, providing a holistic view of gene coexpression.
Fig 1. Simplified representation of a hypothetical coexpression network.
Node A represents a hub gene while node B represents a peripheral gene. Lines connecting nodes represent network edges, and reflect correlations in expression.
Once a coexpression network has been generated, it can be subdivided into ‘modules,’ which are suites of highly interconnected, or coexpressed genes. Within modules, new candidate genes for a trait of interest can be identified based on associations with genes previously linked to these traits, an approach known as ‘guilt-by-association’ [12–14]. Such analyses have thus emerged as a common approach for functional prediction (e.g., [15–18]), and have been performed in a wide variety of species (e.g., Arabidopsis thaliana: [14,19–23]; Homo sapiens: [24]; Mus musculus: [25]; Oryza sativa: [9,26,27]; and Zea mays: [27]). There has also been substantial interest in the evolution of network topologies across taxonomic groups and over time (e.g., [28–32]). Less attention has, however, been paid to the influence of network topology on the evolution of genes contained within such networks.
Interestingly, analyses of protein-protein interaction networks have revealed that the deletion of more centrally located genes is often lethal and that connectivity in such networks is negatively correlated with rates of protein evolution (e.g., [33–40]). These findings suggest that more centrally located proteins, which have more direct molecular interactions (i.e., connections), are more likely to be essential and/or subject to stronger purifying selection than those located on the periphery [6,41]. This pattern may reflect the involvement of such genes in more biological processes, thereby placing them under greater pleiotropic constraint, though the biological significance of this correlation has been debated [42]. Similarly, the position of a gene within a biochemical pathway has been found to influence rates of molecular evolution, with genes found earlier in pathways exhibiting evidence of greater functional constraint than their downstream counterparts (e.g., [43–48]. Once again, this pattern may be due to differences in pleiotropic constraint, with mutations in genes acting earlier in a pathway having more potential downstream consequences than those in genes later in the pathway. It has been suggested that a similar relationship should also hold in coexpression networks [6], with more centrally located genes experiencing greater selective constraint. Two recent studies in plants [49,50] have supported this prediction, though the extent to which this pattern holds across plant species in general remains an open question.
Here, we investigate the relationship between the position of a gene in a coexpression network and its rate of molecular evolution in seven disparate angiosperms. We hypothesize that more centrally located (i.e., hub) genes (e.g., gene A in Fig 1) will, on average, experience greater levels of functional constraint due to their potential involvement in a larger number of biological processes as compared to genes located on the periphery. As such, these genes should exhibit lower nonsynonymous substitution rates as compared to more peripheral genes (e.g., gene B in Fig 1). To test this hypothesis, we leveraged existing datasets to analyze genome-wide patterns of coexpression in Arabidopsis thaliana, Glycine max, Oryza sativa, Populus spp., Solanum lycopersicum, Vitis spp., and Zea mays and used the resulting networks to examine patterns of molecular evolution as a function of connectivity.
Materials and methods
Raw microarray datasets were downloaded from the Gene Expression Omnibus (GEO), ArrayExpress, and/or The Arabidopsis Information Resource (TAIR, for A. thaliana only) corresponding to 2501, 1163, 1572, 1020, 385, 517, and 627 experiments for A. thaliana, G. max, O. sativa, Populus spp., S. lycopersicum, Vitis spp., and Z. mays, respectively. The poplar and grape arrays were utilized at the genus level, and represent 13 and 4 species, respectively. As is common with coexpression networks, these arrays sample a large breadth of experimental data spanning multiple tissues types, developmental stages, and stress treatments (both abiotic and biotic). A complete list of selected experiments and arrays, including their GEO or ArrayExpress IDs along with a brief description, is provided in S1 Appendix. Expression intensities were extracted from all selected microarray experiments and normalized for each taxon using the Robust Multichip Average (RMA) method implemented through the Bioconductor package Affy in R [51]. After data normalization, unique probe-to-gene matches were identified, where the gene corresponds to the GenBank or Phytozome unique identifier provided in the array. If multiple array probes matched a given gene, one of these probes was selected at random to eliminate duplicate data. Genes with variable expression were identified by assessing the distribution of the coefficient of variation (CV) of expression, and only those genes with a CV found within the upper 95% confidence interval of CV were retained. A complete list of retained genes and probes can be found in S2 Appendix.
To determine gene connectivity, the R package Weighted Gene Co-expression Network Analysis (WGCNA) [52] was used to construct coexpression networks for all seven plant taxa. Briefly, WGCNA calculates a Pearson’s correlation matrix for all genes (95% confidence interval), and transforms this matrix by raising all values to a power β (soft thresholding). The β value for a given taxon is a nonlinear transformation, which can influence the correlation between any two genes [53], weighting those with higher connectivity over those with lower. This influences the shape of network modules and creates a scale-free topology. We estimated a β value for each taxon based on underlying expression values using the function pickSoftThreshold in the WGCNA package, resulting in power values of 2, 5, 13, 3, 9, 5, and 4 for A. thaliana, G. max, O. sativa, Populus spp., S. lycopersicum, Vitis spp., and Z. mays, respectively (S1 Fig). All remaining parameters were kept at the recommended default values as stated in the manual. Estimates of per gene connectivity were extracted from the output by tallying the number of genes connected to each unique gene in a network (S2 Appendix). Network characteristics for all seven taxa are summarized in S3 Appendix.
Once connectivity was established, pairwise rates of molecular evolution were estimated for each taxon using PAML’s yn00 model [54]. For comparing relative rates of molecular evolution between taxa, a monocot (O. sativa) or dicot (A. thaliana) outgroup was used for the dicot or monocot species, respectively. For all seven taxa, sequence CDS files were gathered or generated to match with array probes. For A. thaliana, O. sativa, and G. max, these CDS files were downloaded from Phytozome v10.1 (http://phytozome.jgi.doe.gov/pz/portal.html), while sequences for the other species were downloaded from GenBank by matching probe annotations to GenBank sequence accession numbers. Putative orthologs between each taxon and its outgroup were estimated using reciprocal best BLAST hits with an e-value threshold of <1E-08. Sequence pairs were aligned using MUSCLE v.3.8 [55], and in-frame stretches of aligned sequence (≥ 30 bp) were identified and concatenated into a single contiguous sequence of ≥ 300 bp prior to analyses in PAML, using custom perl scripts (mani-seq; https://github.com/bewickaj). All orthologous sequence pairs can be found in S2 Appendix. An estimate of per gene connectivity was determined for each taxon (range of all taxon-wide averages: 228 to 4682) using the coexpression networks generated.
Rates of nonsynonymous substitutions per nonsynonymous site (dN), synonymous substitutions per synonymous site (dS), and estimates of adaptive evolution (ω = dN/dS) were visualized via linear regression against our estimates of gene connectivity (Fig 2). Significance of the correlations, estimated as Kendall's tau (τ) to address tied correlation ranks, was assessed via randomization tests. Briefly, this involved randomizing the parameter of interest vs. connectivity for each comparison in Fig 2 and recalculating the correlation. This procedure was repeated 10,000 times to generate a null distribution against which the observed values were compared. To account for multiple comparisons across taxa, we applied a sequential Bonferroni correction at α = 0.05 [56, 57]. All statistical tests were performed in R [58].
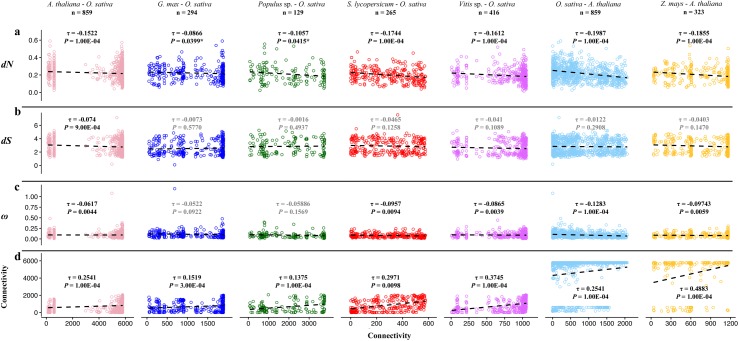
Fig 2. Linear regression of gene connectivity of seven taxa analyzed.
Taxa: A. thaliana, G. max, Populus spp., S. lycopersicum, Vitis spp., O. sativa, and Z. mays, against (a): non-synonymous substitutions (dN), (b): synonymous substitutions (dS), (c): estimates of adaptive evolution (ω = dN/dS) and (d): number of connections in ortholog comparison. Circles represent genes, while the regression coefficient, represented as Kendall's tau (τ) coefficient, is the dashed line. Significance is indicated by bold text. Note that all significant results except the two marked with an asterisk (*) remained significant after correcting for multiple comparisons (see text for details).
Results
In terms of the relationship between connectedness and evolutionary constraint, our results align with what has previously been found in biochemical pathways and protein-protein interaction networks. Our analyses (based on n = 859, 294, 139, 265, 416, 859, 323 orthologous sequence pairs for A. thaliana—O.sativa, G. max—O. sativa, Populus spp—O. sativa, S. lycopersicum—O. sativa, Vitis spp.—O. sativa, O. sativa—A. thaliana, Z. mays—A. thaliana, respectively) revealed that the nonsynonymous substitution rate (dN) was significantly negatively correlated with connectivity in the majority of taxa investigated (the results for G. max and Populus spp. were nominally significant based on our randomization tests, but not significant after controlling for multiple comparisons; Fig 2A). The same overall pattern was evident for our estimates of adaptive evolution (ω = dN/dS) (Fig 2C), though the correlations between ω and connectedness were generally weaker than those between dN and connectedness. While the correlations between dN or ω and connectedness were not significant for two of the seven taxa (G. max and Populus spp.), they did exhibit the same overall trend (i.e., a negative relationship) as was observed in the other five taxa. When combining our results across all seven taxa, we found that the overall pattern–i.e., more highly interconnected hub genes exhibited stronger evolutionary constraint–was highly significant (Fisher’s combined probability test, P < 1E-08 for both dN and ω [using CombinePValue; https://CRAN.R-project.org/package=CombinePValue]; S2 Appendix). The synonymous substitution rate (dS) was only significantly correlated with connectivity in the A. thaliana—O. sativa comparison (Fig 2B). While all dS-related correlations had the same (negative) sign, resulting in a combined probability of P < 1E-02, it is noteworthy that this result is largely attributable to the A. thaliana—O. sativa comparison (P = 0.0009, with the other six P-values ranging from 0.11–0.58). Despite overall differences in the numbers of connections amongst genes within a species, the connectivity of orthologous gene pairs between each taxon and its outgroup (i.e., A. thaliana for monocots and O. sativa for dicots) was positively correlated (all P < 0.05, Fig 2D; S4 Appendix).
Discussion
Taken together, these findings indicate that more centrally located and highly interconnected (i.e., hub) genes exhibit reduced nonsynonymous substitution rates and rates of adaptive evolution. This observation is consistent with our hypothesis that such genes are subject to greater functional constraint than less connected genes that can be found on the periphery of coexpression networks. The consistency of this result across disparate taxa suggests that it holds for flowering plants in general, as opposed to being a species-specific phenomenon. Though our results align with the findings of previous work done in both protein-protein interaction networks [33–36] and biochemical pathways (e.g., [43–47]), it is important to note that patterns of coexpression do not necessarily translate into direct molecular interactions or biochemical relationships. So why do we see this negative correlation between connectivity and rates of molecular evolution, both here and in other recent studies in plants (i.e., [49,50])? While coexpression patterns are not necessarily indicative of direct molecular interactions, it seems likely that more centrally located (i.e., hub) genes will tend to influence more aspects of organismal biology than peripheral genes, and might thus be expected to experience more antagonistic pleiotropy. That is, alterations of the amino acid sequence of a hub gene could have a multitude of associated effects, some of which may be deleterious, whereas tinkering at the periphery of a network might result in variants with fewer negative consequences.
While two recent studies have documented a significant, negative correlation between dS and connectivity in plants [49,50], we saw little evidence of such a correlation in our study. Indeed, only one of the comparisons (A. thaliana—O. sativa) revealed a highly significant (negative) correlation between dS and connectivity. While the remaining six comparisons resulted in negative τ values, none of them individually approached significance. Nonetheless, the combined probability of these results was significant (P < 1E-02), and the likelihood of observing seven negative correlations by chance is extremely low in the absence of a true relationship between dS and connectivity (two-tailed sign test: P < 0.05). Such a relationship could be a byproduct of mutation rate variation, with more highly connected genes experiencing fewer mutations, or it could be due to variation in selective constraint on synonymous sites (e.g., due to codon usage bias; [49]) as a function of connectedness. As noted by Josephs et al. (2017), heterogeneity in dS due to variation in synonymous constraint could explain our observation of a weaker correlation between ω and connectedness as compared to dN and connectedness.
Interestingly, a general tendency toward reduced expression variation in hub genes, evidenced in part by a paucity of “local” eQTLs associated with such genes, has also been observed [49,50]. This observation suggests that expression level variation may be subject to stabilizing selection in more highly connected genes as compared to those with fewer connections. As such, it may be that constraint on both sequence and expression changes is relaxed for genes on the periphery of coexpression networks as compared to more centrally located genes. Unfortunately, the data analyzed herein do not allow us to perform equivalent analyses, and so a more complete understanding of the relationship between expression variation and network position across flowering plants awaits further study. Likewise, a more holistic understanding of the influence of network topology on patterns of both molecular evolution and expression variation across the entirety of the plant kingdom awaits a more complete taxonomic sampling.
Supporting information
(XLS)
(XLS)
(XLS)
(XLS)
(PDF)
Acknowledgments
We thank members of the Burke lab for comments on an earlier version of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was funded by a grant from the National Science Foundation Plant Genome Research Program (DBI-1444522; https://nsf.gov/funding/pgm_summ.jsp?pims_id=5338) received by John M. Burke. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schulze a, Downward J. Navigating gene expression using microarrays—a technology review. Nat Cell Biol. 2001;3(8):E190–5. doi: 10.1038/35087138 [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet [Internet]. 2009;10(1):57–63. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2949280&tool=pmcentrez&rendertype=abstract doi: 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc [Internet]. 2012;7(3):562–78. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3334321&tool=pmcentrez&rendertype=abstract doi: 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lovén J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, et al. Revisiting global gene expression analysis. Cell. 2012;151(3):476–82. doi: 10.1016/j.cell.2012.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barabási A, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5(2):101–13. doi: 10.1038/nrg1272 [DOI] [PubMed] [Google Scholar]
- 6.Cork JM, Purugganan MD. The evolution of molecular genetic pathways and networks. Vol. 26, BioEssays. 2004. p. 479–84. doi: 10.1002/bies.20026 [DOI] [PubMed] [Google Scholar]
- 7.Proulx SR, Promislow DEL, Phillips PC. Network thinking in ecology and evolution. Vol. 20, Trends in Ecology and Evolution. 2005. p. 345–53. doi: 10.1016/j.tree.2005.04.004 [DOI] [PubMed] [Google Scholar]
- 8.Pigliucci M. An extended synthesis for evolutionary biology. Vol. 1168, Annals of the New York Academy of Sciences. 2009. p. 218–28. doi: 10.1111/j.1749-6632.2009.04578.x [DOI] [PubMed] [Google Scholar]
- 9.Ficklin SP, Luo F, Feltus FA. The association of multiple interacting genes with specific phenotypes in rice using gene coexpression networks. Plant Physiol. 2010;154(1):13–24. doi: 10.1104/pp.110.159459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerin C, Joët T, Serret J, Lashermes P, Vaissayre V, Agbessi MDT, et al. Gene coexpression network analysis of oil biosynthesis in an interspecific backcross of oil palm. Plant J [Internet]. 2016;1–19. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27145323 [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Zheng H, Li Y, Li H, Liu X, Qin H, et al. Coexpression network analysis of the genes regulated by two types of resistance responses to powdery mildew in wheat. Sci Rep [Internet]. 2016;6(April):23805 Available from: http://www.nature.com/srep/2016/160401/srep23805/full/srep23805.html%5Cnhttp://www.nature.com/articles/srep23805 doi: 10.1038/srep23805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver S. Guilt-by-association goes global. Nature. 2000;403(6770):601–3. doi: 10.1038/35001165 [DOI] [PubMed] [Google Scholar]
- 13.Saito K, Hirai MY, Yonekura-Sakakibara K. Decoding genes with coexpression networks and metabolomics—“majority report by precogs.” Vol. 13, Trends in Plant Science. 2008. p. 36–43. doi: 10.1016/j.tplants.2007.10.006 [DOI] [PubMed] [Google Scholar]
- 14.Lee I, Ambaru B, Thakkar P, Marcotte EM, Rhee SY. Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat Biotechnol [Internet]. 2010;28(2):149–56. Available from: http://dx.doi.org/10.1038/nbt.1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BMC Bioinformatics [Internet]. 2005;6(1):227 Available from: http://link.springer.com/article/10.1186/1471-2105-6-227/fulltext.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aoki K, Ogata Y, Shibata D. Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 2007;48(3):381–90. doi: 10.1093/pcp/pcm013 [DOI] [PubMed] [Google Scholar]
- 17.Usadel B, Obayashi T, Mutwil M, Giorgi FM, Bassel GW, Tanimoto M, et al. Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant, Cell Environ. 2009;32(12):1633–51. [DOI] [PubMed] [Google Scholar]
- 18.Wong DCJ, Sweetman C, Drew DP, Ford CM. VTCdb: a gene co-expression database for the crop species Vitis vinifera (grapevine). BMC Genomics [Internet]. 2013;14(1):882 Available from: http://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-14-882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Persson S, Wei H, Milne J, Page GP, Somerville CR. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci U S A [Internet]. 2005;102(24):8633–8. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1142401&tool=pmcentrez&rendertype=abstract doi: 10.1073/pnas.0503392102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atias O, Chor B, Chamovitz D a. Large-scale analysis of Arabidopsis transcription reveals a basal co-regulation network. BMC Syst Biol. 2009;3:86 doi: 10.1186/1752-0509-3-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao L, Van Hemert JL, Dash S, Dickerson JA. Arabidopsis gene co-expression network and its functional modules. BMC Bioinformatics. 2009;10:346 doi: 10.1186/1471-2105-10-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Hu Z, Yang Y, Chen X, Chen G. Function annotation of an SBP-box gene in arabidopsis based on analysis of co-expression networks and promoters. Int J Mol Sci. 2009;10(1):116–32. doi: 10.3390/ijms10010116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mutwil M, Usadel B, Schütte M, Loraine A, Ebenhöh O, Persson S. Assembly of an interactive correlation network for the Arabidopsis genome using a novel heuristic clustering algorithm. Plant Physiol. 2010;152(1):29–43. doi: 10.1104/pp.109.145318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res [Internet]. 2004;14(6):1085–94. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15173114%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC419787 doi: 10.1101/gr.1910904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacLennan NK, Dong J, Aten JE, Horvath S, Rahib L, Ornelas L, et al. Weighted gene co-expression network analysis identifies biomarkers in glycerol kinase deficient mice. Mol Genet Metab. 2009;98(1–2):203–14. doi: 10.1016/j.ymgme.2009.05.004 [DOI] [PubMed] [Google Scholar]
- 26.Lee T-H, Kim Y-K, Pham TTM, Song SI, Kim J-K, Kang KY, et al. RiceArrayNet: a database for correlating gene expression from transcriptome profiling, and its application to the analysis of coexpressed genes in rice. Plant Physiol. 2009;151(1):16–33. doi: 10.1104/pp.109.139030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ficklin SP, Feltus FA. Gene coexpression network alignment and conservation of gene modules between two grass species: maize and rice. Plant Physiol. 2011;156(3):1244–56. doi: 10.1104/pp.111.173047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stuart JM, Segal E, Koller D, Kim SK. A Gene-Coexpression Network for Global Discovery of Conserved Genetic Modules. Science (80-) [Internet]. 2003;302(5643):249–55. Available from: http://www.sciencemag.org/cgi/content/abstract/302/5643/249%5Cnpapers://d2952c50-9509-4ba2-9a03-22fbc04267d4/Paper/p908 doi: 10.1126/science.1087447 [DOI] [PubMed] [Google Scholar]
- 29.Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann S a. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol. 2004;14(3):283–91. doi: 10.1016/j.sbi.2004.05.004 [DOI] [PubMed] [Google Scholar]
- 30.Movahedi S, Van de Peer Y, Vandepoele K. Comparative Network Analysis Reveals That Tissue Specificity and Gene Function Are Important Factors Influencing the Mode of Expression Evolution in Arabidopsis and Rice. Plant Physiol [Internet]. 2011;156(3):1316–30. Available from: http://www.plantphysiol.org/content/156/3/1316.abstract doi: 10.1104/pp.111.177865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proost S, Mutwil M. Tools of the trade: Studying molecular networks in plants. Vol. 30, Current Opinion in Plant Biology. 2016. p. 130–40. [DOI] [PubMed] [Google Scholar]
- 32.Ruprecht C, Proost S, Hernandez-Coronado M, Ortiz-Ramirez C, Lang D, Rensing SA, et al. Phylogenomic analysis of gene co-expression networks reveals the evolution of functional modules. Plant J [Internet]. 2017. May [cited 2017 Apr 19];90(3):447–65. Available from: http://doi.wiley.com/10.1111/tpj.13502 [DOI] [PubMed] [Google Scholar]
- 33.Jeong H, Mason SP, Barabási a L, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–2. doi: 10.1038/35075138 [DOI] [PubMed] [Google Scholar]
- 34.Fraser HB, Hirsh a E, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science (80-). 2002;296(April):750–2. [DOI] [PubMed] [Google Scholar]
- 35.Fraser HB, Wall DP, Hirsh AE. A simple dependence between protein evolution rate and the number of protein-protein interactions. BMC Evol Biol [Internet]. 2003;3(1):11 Available from: http://bmcevolbiol.biomedcentral.com/articles/10.1186/1471-2148-3-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hahn MW, Kern AD. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol Biol Evol. 2005;22(4):803–6. doi: 10.1093/molbev/msi072 [DOI] [PubMed] [Google Scholar]
- 37.Vitkup D, Kharchenko P, Wagner A. Influence of metabolic network structure and function on enzyme evolution. Genome Biol [Internet]. 2006;7(5):R39 Available from: http://genomebiology.com/2006/7/5/R39 doi: 10.1186/gb-2006-7-5-r39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim PM, Korbel JO, Gerstein MB. Positive selection at the protein network periphery: evaluation in terms of structural constraints and cellular context. Proc Natl Acad Sci U S A. 2007;104(51):20274–9. doi: 10.1073/pnas.0710183104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alvarez-Ponce D, Fares MA. Evolutionary rate and duplicability in the Arabidopsis thaliana protein-protein interaction network. Genome Biol Evol. 2012;4(12):1263–74. doi: 10.1093/gbe/evs101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luisi P, Alvarez-Ponce D, Pybus M, Fares MA, Bertranpetit J, Laayouni H. Recent positive selection has acted on genes encoding proteins with more interactions within the whole human interactome. Genome Biol Evol. 2015;7(4):1141–54. doi: 10.1093/gbe/evv055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Promislow DEL. Protein networks, pleiotropy and the evolution of senescence. Proc Biol Sci [Internet]. 2004;271(1545):1225–34. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1691725&tool=pmcentrez&rendertype=abstract doi: 10.1098/rspb.2004.2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahn MW, Conant GC, Wagner A. Molecular Evolution in Large Genetic Networks: Does Connectivity Equal Constraint? J Mol Evol. 2004;58(2):203–11. doi: 10.1007/s00239-003-2544-0 [DOI] [PubMed] [Google Scholar]
- 43.Rausher MD, Miller RE, Tiffin P. Patterns of evolutionary rate variation among genes of the anthocyanin biosynthetic pathway. Mol Biol Evol. 1999;16(2):266–74. [DOI] [PubMed] [Google Scholar]
- 44.Lu Y, Rausher MD. Evolutionary Rate Variation in Anthocyanin Pathway Genes. Mol Biol Evol. 2003;20(11):1844–53. doi: 10.1093/molbev/msg197 [DOI] [PubMed] [Google Scholar]
- 45.Rausher MD, Lu Y, Meyer K. Variation in constraint versus positive selection as an explanation for evolutionary rate variation among anthocyanin genes. J Mol Evol. 2008;67(2):137–44. doi: 10.1007/s00239-008-9105-5 [DOI] [PubMed] [Google Scholar]
- 46.Livingstone K, Anderson S. Patterns of variation in the evolution of carotenoid biosynthetic pathway enzymes of higher plants. J Hered. 2009;100(6):754–61. doi: 10.1093/jhered/esp026 [DOI] [PubMed] [Google Scholar]
- 47.Ramsay H, Rieseberg LH, Ritland K. The correlation of evolutionary rate with pathway position in plant terpenoid biosynthesis. Mol Biol Evol. 2009;26(5):1045–53. doi: 10.1093/molbev/msp021 [DOI] [PubMed] [Google Scholar]
- 48.Alvarez-Ponce D, Aguadé M, Rozas J. Comparative genomics of the vertebrate insulin/TOR signal transduction pathway: A network-level analysis of selective pressures. Genome Biol Evol. 2011;3(1):87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Josephs EB, Wright SI, Stinchcombe JR, Schoen DJ. The relationship between selection, network connectivity, and regulatory variation within a population of Capsella grandiora. Genome Biol Evol [Internet]. 2017. April 8; Available from: https://academic.oup.com/gbe/article-lookup/doi/10.1093/gbe/evx068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mähler N, Wang J, Terebieniec BK, Ingvarsson PK, Street NR, Hvidsten TR. Gene co-expression network connectivity is an important determinant of selective constraint. bioRxiv [Internet]. 2017 Jan 30; Available from: http://biorxiv.org/content/early/2017/01/30/078188.1.abstract [DOI] [PMC free article] [PubMed]
- 51.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–15. doi: 10.1093/bioinformatics/btg405 [DOI] [PubMed] [Google Scholar]
- 52.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559 doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol [Internet]. 2005;4:Article17. Available from: http://www.degruyter.com/view/j/sagmb.2005.4.issue-1/sagmb.2005.4.1.1128/sagmb.2005.4.1.1128.xml [DOI] [PubMed] [Google Scholar]
- 54.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci [Internet]. 1997;13(5):555–6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9367129 [DOI] [PubMed] [Google Scholar]
- 55.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scand J Stat [Internet]. 1979;6(2):65–70. Available from: http://www.jstor.org/stable/10.2307/4615733 [Google Scholar]
- 57.Rice WR. Analyzing Tables of Statistical Tests. Evolution (N Y) [Internet]. 1989;43(1):223 Available from: http://www.jstor.org/stable/2409177?origin=crossref [DOI] [PubMed] [Google Scholar]
- 58.R Core team. R Core Team [Internet]. Vol. 55, R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: ISBN 3-900051-07-0, URL http://www.R-project.org/. 2015. p. 275–86. Available from: http://www.mendeley.com/research/r-language-environment-statistical-computing-96/%5Cnpapers2://publication/uuid/A1207DAB-22D3-4A04-82FB-D4DD5AD57C28 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
(XLS)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.