Abstract
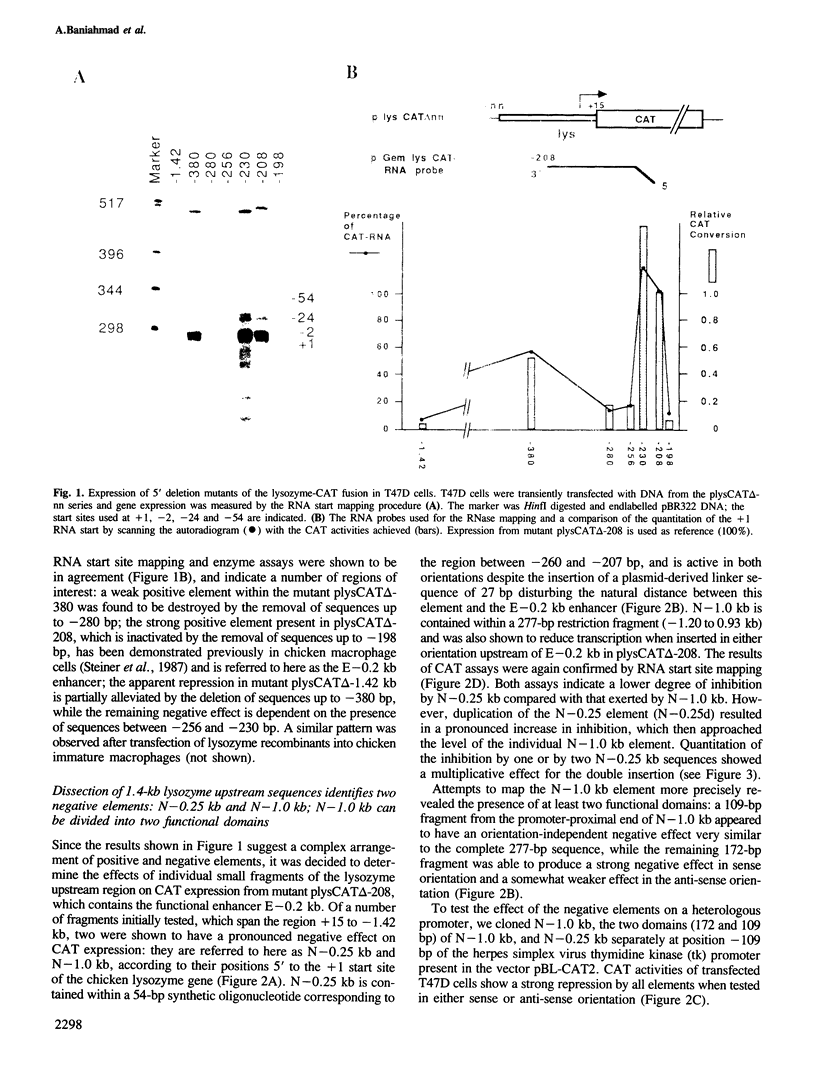
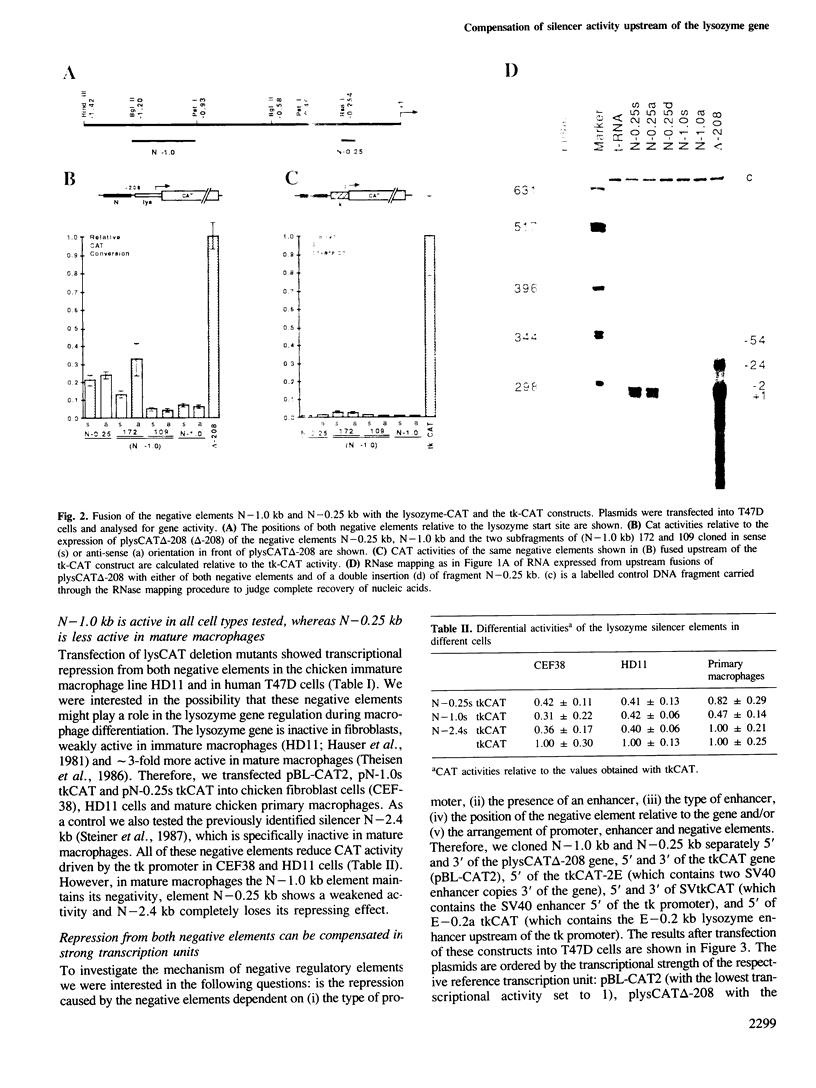
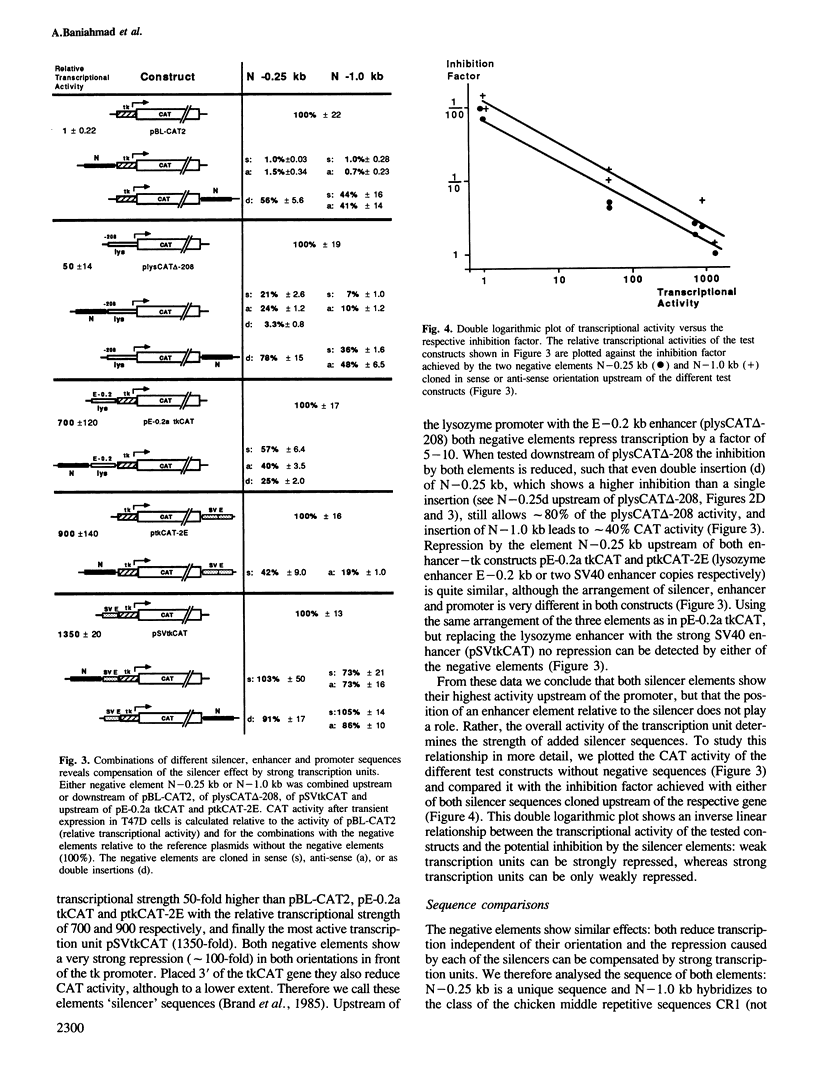
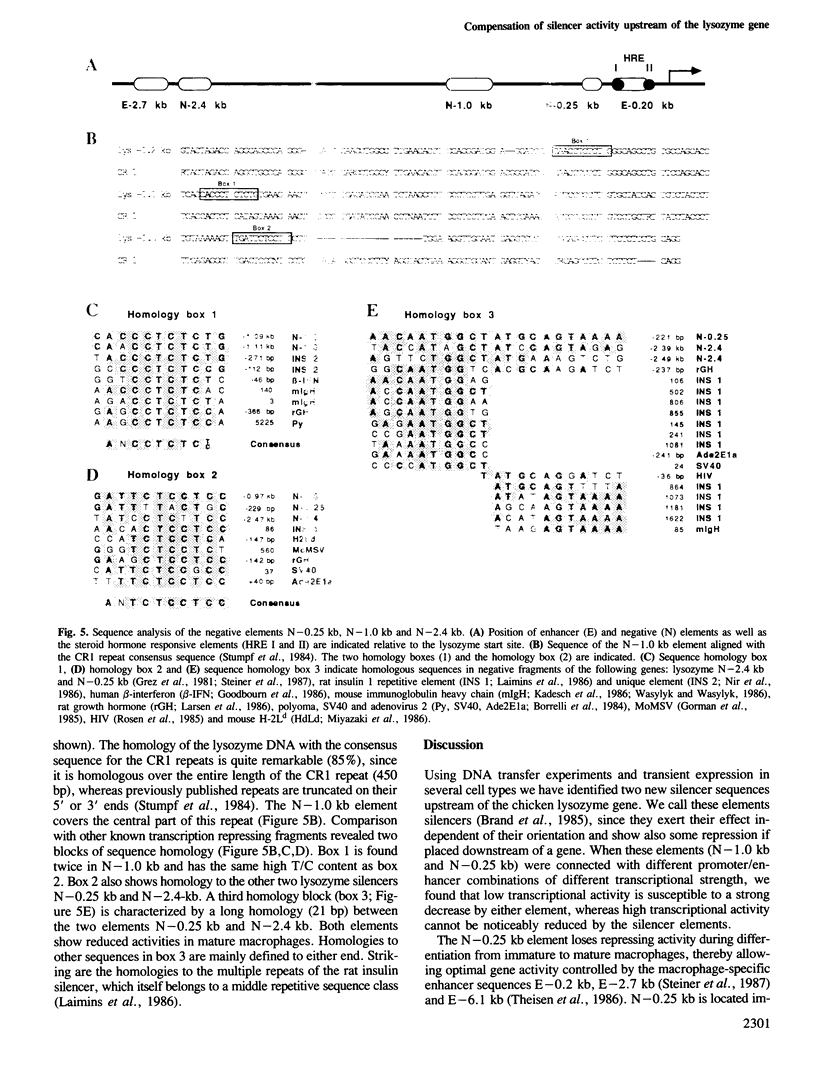
The chicken lysozyme gene is constitutively expressed in macrophages. Transfection of recombinant genes containing different portions of the lysozyme 5' upstream region revealed the existence of two negative transcriptional elements within 1 kb upstream of the start sites. Both elements placed upstream or downstream of a heterologous promoter-gene unit repress transcription independent of their orientation and are therefore called silencer elements, although their repressing activities 3' of the gene are reduced. One silencer (N-1.0 kb) at position -1 kb consists of the central region of the chicken middle repetitive sequence element CR1 and can be divided into two functional domains. N-1.0 kb is active in all cell types tested. The other silencer (N-0.25 kb) at position -0.25 kb shows reduced activity in primary macrophages. Despite their different specificities, the activity of both silencer elements can be influenced similarly. An inverse linear relationship between the transcriptional activity of the tested constructs and the potential inhibition by the silencer elements was found: weak transcription units can be strongly repressed, whereas strong transcription units can be only weakly repressed. Such a mechanism may help to turn off completely a particular gene in situations or tissues where strong positive regulators are inactive.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beug H., von Kirchbach A., Döderlein G., Conscience J. F., Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell. 1979 Oct;18(2):375–390. doi: 10.1016/0092-8674(79)90057-6. [DOI] [PubMed] [Google Scholar]
- Borrelli E., Hen R., Chambon P. Adenovirus-2 E1A products repress enhancer-induced stimulation of transcription. Nature. 1984 Dec 13;312(5995):608–612. doi: 10.1038/312608a0. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Breeden L., Abraham J., Sternglanz R., Nasmyth K. Characterization of a "silencer" in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985 May;41(1):41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- Cato A. C., Miksicek R., Schütz G., Arnemann J., Beato M. The hormone regulatory element of mouse mammary tumour virus mediates progesterone induction. EMBO J. 1986 Sep;5(9):2237–2240. doi: 10.1002/j.1460-2075.1986.tb04490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron J., Drouin J. Glucocorticoid inhibition of transcription from episomal proopiomelanocortin gene promoter. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8903–8907. doi: 10.1073/pnas.83.23.8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Colantuoni V., Pirozzi A., Blance C., Cortese R. Negative control of liver-specific gene expression: cloned human retinol-binding protein gene is repressed in HeLa cells. EMBO J. 1987 Mar;6(3):631–636. doi: 10.1002/j.1460-2075.1987.tb04801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodbourn S., Burstein H., Maniatis T. The human beta-interferon gene enhancer is under negative control. Cell. 1986 May 23;45(4):601–610. doi: 10.1016/0092-8674(86)90292-8. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Rigby P. W., Lane D. P. Negative regulation of viral enhancers in undifferentiated embryonic stem cells. Cell. 1985 Sep;42(2):519–526. doi: 10.1016/0092-8674(85)90109-6. [DOI] [PubMed] [Google Scholar]
- Grez M., Land H., Giesecke K., Schütz G., Jung A., Sippel A. E. Multiple mRNAs are generated from the chicken lysozyme gene. Cell. 1981 Sep;25(3):743–752. doi: 10.1016/0092-8674(81)90182-3. [DOI] [PubMed] [Google Scholar]
- Hauser H., Graf T., Beug H., Greiser-Wilke I., Lindenmaier W., Grez M., Land H., Giesecke K., Schütz G. Structure of the lysozyme gene and expression in the oviduct and macrophages. Haematol Blood Transfus. 1981;26:175–178. doi: 10.1007/978-3-642-67984-1_29. [DOI] [PubMed] [Google Scholar]
- Kaaden O. R., Lange S., Stiburek B. Establishment and characterization of chicken embryo fibroblast clone LSCC-H32. In Vitro. 1982 Oct;18(10):827–834. doi: 10.1007/BF02796323. [DOI] [PubMed] [Google Scholar]
- Kadesch T., Zervos P., Ruezinsky D. Functional analysis of the murine IgH enhancer: evidence for negative control of cell-type specificity. Nucleic Acids Res. 1986 Oct 24;14(20):8209–8221. doi: 10.1093/nar/14.20.8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L., Holmgren-König M., Khoury G. Transcriptional "silencer" element in rat repetitive sequences associated with the rat insulin 1 gene locus. Proc Natl Acad Sci U S A. 1986 May;83(10):3151–3155. doi: 10.1073/pnas.83.10.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P. R., Harney J. W., Moore D. D. Repression mediates cell-type-specific expression of the rat growth hormone gene. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8283–8287. doi: 10.1073/pnas.83.21.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenmaier W., Nguyen-Huu M. C., Lurz R., Stratmann M., Blin N., Wurtz T., Hauser H. J., Sippel A. E., Schütz G. Arrangement of coding and intervening sequences of chicken lysozyme gene. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6196–6200. doi: 10.1073/pnas.76.12.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miksicek R., Heber A., Schmid W., Danesch U., Posseckert G., Beato M., Schütz G. Glucocorticoid responsiveness of the transcriptional enhancer of Moloney murine sarcoma virus. Cell. 1986 Jul 18;46(2):283–290. doi: 10.1016/0092-8674(86)90745-2. [DOI] [PubMed] [Google Scholar]
- Muglia L., Rothman-Denes L. B. Cell type-specific negative regulatory element in the control region of the rat alpha-fetoprotein gene. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7653–7657. doi: 10.1073/pnas.83.20.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muglia L., Rothman-Denes L. B. Cell type-specific negative regulatory element in the control region of the rat alpha-fetoprotein gene. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7653–7657. doi: 10.1073/pnas.83.20.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir U., Walker M. D., Rutter W. J. Regulation of rat insulin 1 gene expression: evidence for negative regulation in nonpancreatic cells. Proc Natl Acad Sci U S A. 1986 May;83(10):3180–3184. doi: 10.1073/pnas.83.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne T. F., Goldstein J. L., Brown M. S. 5' end of HMG CoA reductase gene contains sequences responsible for cholesterol-mediated inhibition of transcription. Cell. 1985 Aug;42(1):203–212. doi: 10.1016/s0092-8674(85)80116-1. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D. Regulation of protein synthesis in chick oviduct. I. Independent regulation of ovalbumin, conalbumin, ovomucoid, and lysozyme induction. J Biol Chem. 1972 Oct 25;247(20):6450–6461. [PubMed] [Google Scholar]
- Remmers E. F., Yang J. Q., Marcu K. B. A negative transcriptional control element located upstream of the murine c-myc gene. EMBO J. 1986 May;5(5):899–904. doi: 10.1002/j.1460-2075.1986.tb04301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz R., Schütz G., von der Ahe D., Beato M. Sequences in the promoter region of the chicken lysozyme gene required for steroid regulation and receptor binding. Cell. 1984 Jun;37(2):503–510. doi: 10.1016/0092-8674(84)90380-5. [DOI] [PubMed] [Google Scholar]
- Rosen C. A., Sodroski J. G., Haseltine W. A. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell. 1985 Jul;41(3):813–823. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- Steiner C., Muller M., Baniahmad A., Renkawitz R. Lysozyme gene activity in chicken macrophages is controlled by positive and negative regulatory elements. Nucleic Acids Res. 1987 May 26;15(10):4163–4178. doi: 10.1093/nar/15.10.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumph W. E., Hodgson C. P., Tsai M. J., O'Malley B. W. Genomic structure and possible retroviral origin of the chicken CR1 repetitive DNA sequence family. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6667–6671. doi: 10.1073/pnas.81.21.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumph W. E., Kristo P., Tsai M. J., O'Malley B. W. A chicken middle-repetitive DNA sequence which shares homology with mammalian ubiquitous repeats. Nucleic Acids Res. 1981 Oct 24;9(20):5383–5397. doi: 10.1093/nar/9.20.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen M., Stief A., Sippel A. E. The lysozyme enhancer: cell-specific activation of the chicken lysozyme gene by a far-upstream DNA element. EMBO J. 1986 Apr;5(4):719–724. doi: 10.1002/j.1460-2075.1986.tb04273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk C., Wasylyk B. The immunoglobulin heavy-chain B-lymphocyte enhancer efficiently stimulates transcription in non-lymphoid cells. EMBO J. 1986 Mar;5(3):553–560. doi: 10.1002/j.1460-2075.1986.tb04246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell. 1978 Jul;14(3):725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]
- Zinn K., DiMaio D., Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983 Oct;34(3):865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]
- von der Ahe D., Janich S., Scheidereit C., Renkawitz R., Schütz G., Beato M. Glucocorticoid and progesterone receptors bind to the same sites in two hormonally regulated promoters. Nature. 1985 Feb 21;313(6004):706–709. doi: 10.1038/313706a0. [DOI] [PubMed] [Google Scholar]






