Summary
Chromosomes in multicellular animals are subdivided into a series of looped domains. In addition to being the underlying principle for organizing the chromatin fiber, looping is critical for processes ranging from gene regulation to recombination and repair. The subdivision of chromosomes into looped domains depends upon a special class of architectural elements called boundaries or insulators. These elements are distributed throughout the genome and are ubiquitous building blocks of chromosomes. In this review, we focus on features of boundaries that are critical in determining the topology of the looped domains and their genetic properties. We highlight the properties of fly boundaries that are likely to have an important bearing on the organization of looped domains in vertebrates, and discuss the functional consequences of the observed similarities and differences.
Keywords: chromosome architecture, gene regulation, insulators, looped domain, loop topology, TADs
Introduction
The 3D organization of the genome in multicellular eukaryotes is thought to play a crucial role in regulation, recombination and repair [1-5].
The first evidence that the chromatin fiber in chromosomes of multicellular eukaryotes has a regular, stereotypic 3D organization came from cytological studies dating back more than 100 years. One classic example are the polytene chromosomes in Diptera salivary glands, fat bodies and nurse cells [6]. The euchromatic regions of these polytene chromosomes are replicated tens to hundreds of times, and the copies are aligned in precise register. The homologs are also paired, again in register. The resulting chromosomes have banding patterns that reflect the relative degree of compaction in each chromosomal segment. This pattern (bands and interbands) is highly reproducible from one cell to the next and between individuals. Moreover, analysis of deletion and inversion mutants shows that the banding pattern is dictated bythe underlying DNA sequence.
Another classic example found in both invertebrates and vertebrates are the lampbrush chromosomes of oocytes arrested at the diplotene stage of meiosis I [7-9]. At this stage, the two sister chromatids are paired in register. Emanating from the condensed chromatin that forms the main axis of the chromosome are loops that contain actively transcribed genes. The loops are arranged in pairs, one from each chromatid. Like polytenes, the looping pattern is dictated by the underlying DNA sequence.
These examples gave rise to the idea that chromosomes in somatic cells are subdivided into a series of looped domains. This model was supported by biochemical studies of Benyajati and Worcel which suggested that Drosophila chromosomes are organized into loops of about 80 kb [10]. Further support came from EM micrographs of metaphase chromosomes by Laemmli and colleagues [11-13], which showed thousands of large loops anchored at their base by a scaffold-like structure.
If chromosomes are subdivided into a series of precisely defined loops, a plausible inference is that there are special cis-acting elements that delimit the ends or boundaries of each loop. The first such elements (called boundary elements or insulators) were discovered in Drosophila several decades ago. Three were identified genetically. One, facetstrawberry is an ∼900 bp deletion upstream of the promoter of the Notch gene[14,15]. It alters the banding pattern in polytene chromosomes, fusing the band that contains Notch with the adjacent band, and causing a chromosomal position effect that reduces Notch expression. The second boundary, Fab-7, is located between two regulatory domains in the Bithorax complex (BX-C), iab-6 and iab-7, which direct the expression of the Abd-B gene in parasegments 11(PS11/abdominal segment A6) and 12 (PS12/A7), respectively. One function of this boundary is to block adventitious interactions between initiators, enhancers and Polycomb-dependent silencers in the two regulatory domains. In Fab-7 mutants, the two domains fuse, and as a consequence, cells in PS11 (A6) assume either a PS12 (A7) or a PS10 (A5) identity [16-19]. In the former case, the iab-6 initiator activates iab-7 inappropriately in PS11, while in the latter case, the iab-7 Polycomb Response Element (PRE) silences iab-6 [19]. The third boundary, su(Hw), is in the gypsy retrotransposon [20-22]. A large percentage of spontaneous mutations in flies are gypsy insertions that block normal enhancer-promoter interactions. These mutagenic effects can be suppressed by mutations in the gene encoding the Su(Hw) protein[23]. This protein binds to multiple sites in su(Hw) and is responsible for its blocking activity [22, 24, 25]. Two other boundaries, scs and scs', were identified molecularly. They flank the 87A7 heat shock locus and are located close to the borders of the “puff” of decondensed chromatin that forms when the hsp70 genes are induced [26, 27].
Many other boundaries have subsequently been discovered in organisms ranging from yeast (S. pombe) to man [28-31]. While fewer than 100 or so have actually been characterized, ChIP experiments suggest that there are thousands in chromosomes of multicellular eukaryotes. For example, human chromosomes have over 15,000 cell type-independent sites for the CTCF boundary protein,, as well as many cell type-specific sites [32].
Boundaries are architectural elements that define topologically independent domains
As initially envisioned, the primary function of boundaries is architectural; they subdivide the chromosome into a series of topologically independent looped domains. The first findings supporting an architectural function came from the discovery that su(Hw) and a BX-C boundary, Mcp [18],mediate long-distance regulatory interactions [33-36].For example, a white enhancer in an Mcp transgene can activate a white reporter in a second Mcp transgene inserted several thousand kb away. Similarly, PRE-dependent silencing interactions were observed over even larger distances, including between Mcp inserts located on different chromosomes[34,36]. Subsequent studies showed that these long-distance regulatory interactions involve direct physical contacts between the boundaries [36-39]. Moreover, regulatory interactions and physical contacts are eliminated when one of the boundaries is excised [37, 38].
The architectural functions of boundaries have been most clearly demonstrated by chromosome conformation capture (3C) experiments [40, 41]. 3C experiments on the mouse β-globin and the 87A7 heat shock loci demonstrated that the boundaries flanking each locus contact each other, forming a loop containing the locus [42]. Moreover, in the β-globin locus, there are also contacts connecting the boundaries to enhancers and the active β-globin gene [42].
The genome-wide 3C experiments have shown that chromosomes in mammals and flies are subdivided into rather large “topologically associating domains” (TADs). The TADs initially detected had an average length of nearly 1 Mb in mammals [43-46] and 100 kb in flies [47, 48]. Although these studies showed that CTCF binding sites often marked the end points of TADs, multiple CTCF sites were also found buried within them, suggesting that in many cases, CTCF-dependent boundaries don't delimit TAD endpoints [43, 45, 47-49]. However, in more recent, higher resolution 3C experiments, the length of TADs in human cultured cells averaged only 180 kb [50]. About 40% of these smaller TADs are connected at their base, forming loops like those described above. Moreover, for 90% of the loops that are anchored by a pair of CTCF sites, the two CTCF sites are in a convergent orientation (more on this later).
Boundaries have genetic activities
In additional to their architectural functions, boundaries have regulatory activities. The first functional assays used transgenes to show that boundaries could insulate reporters against chromosomal position effects, and block enhancers from activating reporters [24, 51-54].
Both of these activities, as well as blocking silencers from repressing transcription, are characteristic features of boundaries from flies to man [55,56]. Insulating activity requires two boundaries, one on each side of the reporter (Fig. 1A). In the blocking assay, the boundary is placed between an enhancer/silencer and a reporter (Fig. 1B). A boundary only blocks regulatory interactions in this proximal position, while it has no effect when placed distal to the enhancer/silencer.
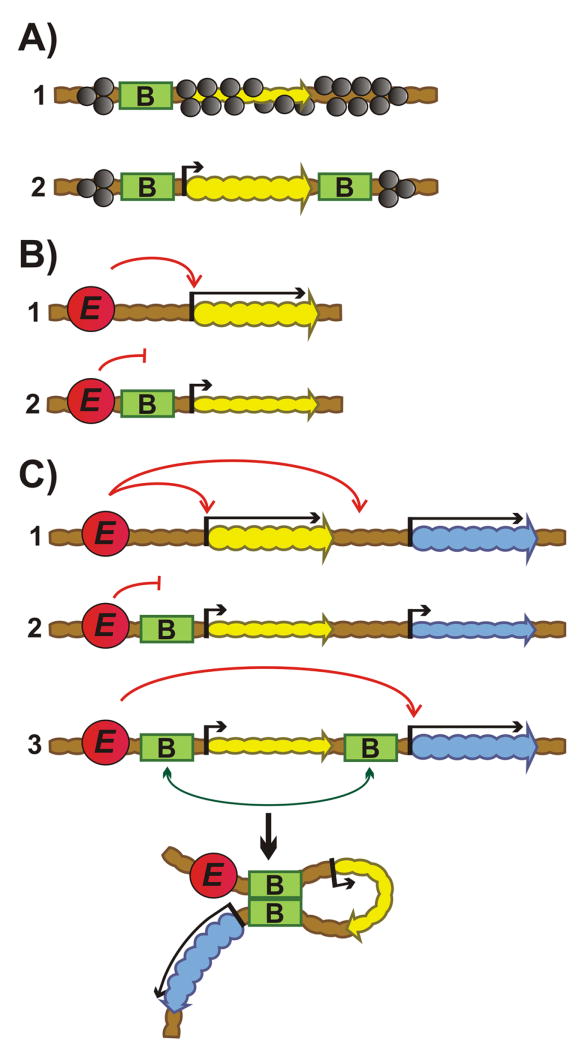
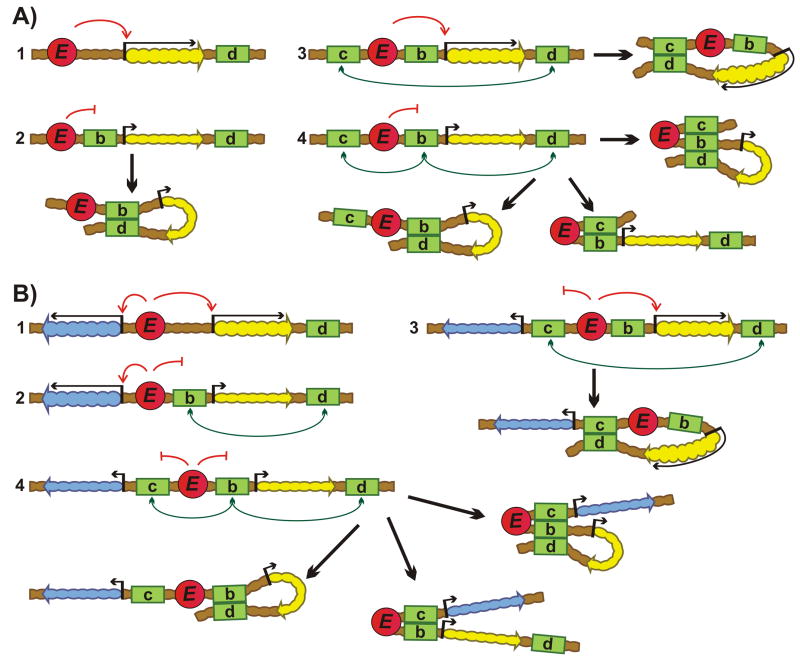
Figure 1.
Transgene assays for boundaries. A: Insulator assay. (1) One boundary is not effective in protecting repaorter transcription from heterochromatic/Polycomb repression. (2) Two boundaries flanking reporter on both sides protect against silencing. Green rectangles “B” – boundaries. The yellow and blue wavy arrows indicate genes; upstream arrow indicates promoter: short – isolated from enhancer; long – activated by enhancer. Absence of an arrow upstream of the gene in (1) indicates gene repression by heterochromatin/Polycomb. Heterochromatin/Polycomb factors– gray ovals. B: Enhancer-blocking assay. (1) Enhancer activates promoter. (2) Boundary inserted between enhancer and promoter blocks enhancer action. Red circle “E” – enhancer. C: The bypass assay. (1) Two reporters are regulated by an upstream enhancer. (2) Placing a boundary between the upstream enhancer and the promoter for the proximal reporter blocks activation of both reporters. (3) Addition of second boundary in between the two reporters leads to bypass. The two boundaries pair with each other and this brings the enhancers in close proximity to the distal reporter leading to its activation.
Architectural and genetic activities are non-autonomous and depend upon boundary-boundary interactions
Because the transgenes used in the first blocking assays contained only a single boundary, it was assumed that their activities were autonomous, and many of the models proposed for their mechanism were based on this assumption. However, architectural functions require interactions either with other boundaries or with some other structure such as the nuclear matrix, and for this reason are non-autonomous. A critical question is whether the genetic activities of these elements depend upon their architectural functions. If so, blocking and also insulation must be non-autonomous.
Bypass experiments
The first compelling evidence that this is the case came from “insulator (boundary) bypass” assays [57, 58]. In one version of this assay (see Fig. 1C), two reporters in tandem are regulated by upstream enhancers. Introduction of a boundary between the enhancers and the proximal reporter blocks activation of both reporters. However, when a second boundary is placed between the two reporters, the enhancers activate the distal but not the proximal reporter. Similar results are observed when the enhancers are replaced by a PRE silencer [59] (arguing against an autonomous “barrier” model for blocking Polycomb silencing).
Were boundary activity strictly autonomous, any combination of elements in the upstream and downstream position would support blocking and bypass in this assay. However, this is not the case. While all boundaries tested support bypass when paired with themselves, some heterologous combinations do, while others do not [37, 60-63].
Though the basis for this specificity is not yet understood, it clearly depends upon the proteins that are associated with the two boundaries. This is illustrated in bypass experiments with multimerized binding sites for different architectural proteins [61]. For example, multimerized sites for either dCTCF or Su(Hw) support bypass when paired with themselves, but not when paired with each other[61]. Evidence for non-autonomy also comes from boundary competition and domain definition experiments [64] (see Box 1).
Box 1. Boundary competition experiments.
As illustrated for the boundary competition experiment in Fig. 2A, one boundary is placed downstream of a reporter (d) while a second boundary is placed in the blocking position (b) between the reporter and an enhancer. The boundary in the blocking position is then challenged by a third boundary (c) placed upstream of the enhancer. Depending upon which pair of boundaries defines the looped domain, blocking activity will be retained or lost. If, for example, the boundary in the blocking position pairs with the downstream boundary (d), blocking activity will be retained. In contrast, if the upstream boundary (c) is a better match, a loop will form between (c) and (d), leaving the “blocking” boundary unpaired, and the enhancer will activate the reporter. The domain definition assay also uses competition to measure boundary matching, but with two reporters (Fig. 2B). Interestingly, in the domain definition assay, pairing preferences depend not only upon the boundary combination, but also on chromosomal insertion site, and stage/tissue. Stage/tissue specificity for blocking activity has also been observed in experiments in which su(Hw )was used to replace Fab-7 in BX-C [64]. .
Boundary:boundary interactions are orientation-dependent
Another property of fly boundaries uncovered in bypass assays is orientation dependence. As illustrated in Fig. 3A, self-pairing interactions are generally head-to-head. This means that the topology of the loop formed between two identical boundaries in cis will depend on their relative orientation. If both are oriented in the “forward” direction, head-to-head pairing will generate a circle-loop. In contrast, if one boundary is in the forward direction and the other is “reversed”, head-to-head pairing generates a stem-loop. In this latter case, the paired boundaries bring enhancers upstream of the stem-loop in proximity with the reporter downstream of the loop, resulting in activation. In the former configuration, the enhancer and reporter are on opposite sides of the paired boundary, and this blocks activation [37, 61-63].
Figure 3.
Self-pairing interactions are generally head-to-head. A: The bypass assay. The topology of the loop formed by paired boundaries depends upon their relative orientation and whether they pair head-to-head or head-to-tail.(1) If the two boundaries pair head-to-head, and they are arranged in the same orientation, a circle- loop will be formed. This topology positions the enhancer and promoter on opposite sides of the paired boundaries, leading to isolation of the reporter. (2) When the boundaries are arranged in opposite orientation, head-to-head pairing generates a stem-loop. This brings the enhancer and promoter into close proximity and leads to activation of transcription. Green arrows with straight sides– boundaries, arrowheads indicate orientation. B: Interactions between boundaries over large (2Mb) distances. (1) In this configuration, the enhancer and the reporter are “downstream” of the boundary. Here, head-to-head self-pairing between the two boundaries brings the enhancer and promoter into close proximity, activating transcription. (2) In this configuration, the enhancer is “downstream” of the boundary, while the reporter is in the “upstream” position. When the two boundaries pair with each other head-to-head, the enhancer and the reporter are on opposite sides of the paired boundary, blocking regulatory interactions. C: Self-pairing Interactions between boundaries on each homolog aligns the homologs in register and maintains homolog pairing.(1) Head-to-head self-pairing interactions align homologs in register. Different colored arrows – boundaries. (2) Head-to-tail self-pairing interactions would disrupt chromosome alignment and interfere with transvection.
Orientation-dependent pairing interactions are not limited in scale to the 10-15 kb DNA segments in typical transgenes, nor are they restricted to interactions in cis. For the two boundaries flanking the even-skipped (eve) locus, homie and nhomie, Fujioka et al. [65] observed orientation-dependent self-pairing interactions in trans between attP-mediated insertions more than 1 Mb apart. When head-to-head self-pairing places the enhancer and the reporter on the same side of the paired boundaries (in trans), the enhancer activates expression. In contrast, when the enhancer and reporter are on opposite sides of the self-paired boundaries, regulatory interactions are blocked (Fig.3B).
Results from these orientation-dependent assays give us a hint about how boundary pairing can facilitate or block regulatory interactions that can occur by looping. if the regulatory elements and gene end up on the same side of the paired boundaries, they can be brought together in 3D space and interact; however, if they are on opposite sides of the boundary pair, interactions are disfavored. This is true irrespective of whether the interaction is between elements that are in the same topological loop or in different loops.
Self-pairing boundary interactions align homologs in register
What is the function of boundary self-pairing in flies? Unlike many eukaryotes, homologs in Drosophila somatic cells are paired throughout much of development. This pairing is required for transvection, a phenomenon in which regulatory elements on one homolog control gene activity on the other [66, 67]. Trans regulation of this type occurs throughout the fly genome, augmenting gene activity [33, 68-70]. Transvection between homologous loci is disrupted by chromosomal rearrangements that interfere with pairing of the homologs in their immediate vicinity. Recent studies using attP technology showed that boundary self-paring substantially enhances trans-regulation between a reporter on one homolog and enhancers at the same site on the other homolog. However, in order for trans-regulation to occur, the two boundaries on each homolog have to be in the same orientation. Additionally, the reporter and enhancer must be on same side of the paired boundaries [65].
The ability of fly boundaries to self-pair with high specificity, taken together with the fact that there are hundreds distributed along the chromosome, provides a compelling argument that boundaries are responsible for aligning and pairing homologs in precise register [65]. This mechanism for homolog alignment fits with the fact that almost all of the characterized self-pairing interactions are head-to-head [37,61-63](the exceptions are bi-directional). As illustrated in Fig. 3C, head-to-head self-pairing facilitates the alignment of DNA sequences located between boundaries, while head-to-tail self-pairing would generate unpaired DNA sequences to either side of the paired boundary.
While homolog pairing is a characteristic feature of chromosomes in the somatic cells of dipteran species like D. melanogaster, it doesn't occur in the soma of most other animals. However, as mentioned above, pairing is observed for the lampbrush chromosomes that are present in many invertebrate and vertebrate oocytes [7-9].A plausible speculation is that a similar mechanism—the head-to-head self-pairing of boundaries--is deployed in aligning homologs in oocyte nuclei. Further, in all cells in G2, there are two copies of each chromosome, and these sister chromatids remain intertwined along their length until they condense during mitosis [71,72]. Paired boundaries may function within this configuration to keep PREs and other epigenetic elements in close proximity, helping to template epigenetic marks from one cellular generation to the next.
Boundary interactions in cis
To subdivide chromosomes into a series of looped domains, boundaries have to interact either with a nuclear substructure, such as the nuclear matrix, or with other boundaries in cis. In flies, it is now clear that boundary-boundary interactions serve this function. Several lines of evidence indicate that heterologous interactions are specific; that is, boundaries have a hierarchy of preferred partners. For example, scs and scs' prefer to pair with each other rather than with Fab-7, Fab-8, or su(Hw) [64]. Similarly, su(Hw) is a better match for scs' than for Fab-7, while Fab-8 is a better match for Fab-7 than scs'[64]. While there have not yet been systematic studies to assess the partner preferences for boundaries located throughout a neighborhood, the available evidence suggests that boundaries are usually matched with their immediate or other nearby neighbors (for example [65, 73]).
On the other hand, because fly boundaries utilize a combination of many different architectural proteins, they can pair with boundaries that are not ideal matches, but nevertheless share matching factors/co-factors.
Orientation dependence and loop topology
In addition to partner preference, the interactions between heterologous boundaries are with a few exceptions orientation-dependent. Unlike self-interactions, which are head-to-head (or have no preference), both head-to-head and head-to-tail pairing have been observed for heterologous combinations. Since the chromosome architectures generated by head-to-tail pairing are less complicated, these will be considered first.
Heterologous head-to-tail pairing generates stem-loops
Among the most thoroughly studied heterologous partners are the eve boundaries homie and nhomie [65, 74, 75], which flank the ∼17kb eve locus. nhomie is located ∼7 kb upstream of eve, while homie is ∼10kb downstream (Fig. 4A).Like self-pairing, heterologous nhomie ←→homie pairing can mediate trans-regulatory interactions over distances in excess of 1 Mb. However, while self-pairing is head-to-head, heterologous pairing is head-to-tail, which generates a stem-loop containing the entire eve locus [65].
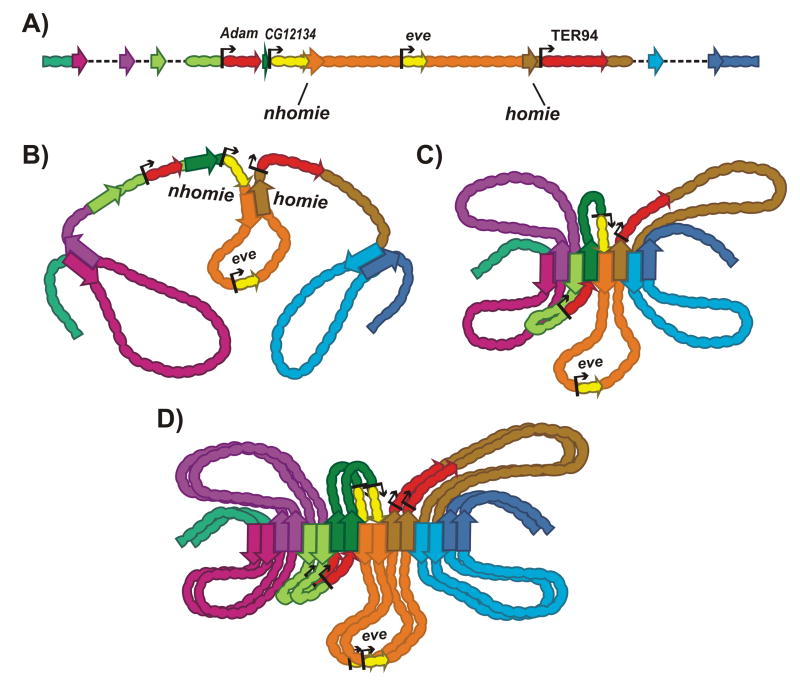
Figure 4.
Heterologous head-to-tail pairing between boundaries in cis. A: Schematic of the eve regulatory region. Boundaries: nhomie (orange), homie (brown), others are not specified. B: Interaction between nhomie-homie and other flanking boundaries generates independent stem-loops that are separated by unanchored segments. C: The nhomie-homie pair interacts with their neighbors. D: The interactions between boundaries are further stabilized by homolog pairing.
The organization of the looped domains surrounding eve will depend upon the pattern of interactions between nhomie, homie and other boundaries in the neighborhood. If nhomie and homie can pair with each other, but don't interact with the flanking boundaries, there will be unanchored segments on both sides of eve. In the configuration shown in Fig. 4B, the flanking boundaries pair head-to-tail with their more distal neighbors, generating a set of anchored stem-loops that are connected by an unanchored segment. In this case, the main axis of the chromosome in the region around eve would be defined by the unanchored segments containing CG12134 upstream and TER94 downstream of eve. However, a more likely scenario is that nhomie and homie simultaneously pair with each other and with flanking boundaries. In this case, this region of the chromosome will be organized into a series of stem-loops. As illustrated in Fig. 4C, the loops would emanate from the main axis of the chromosome, which would be defined by an array of paired boundaries(Fig. 4C).
Since pairing interactions have finite half-lives, the configuration shown in Fig. 4C will not be static, and the local architecture will oscillate between configurations in Fig. 4C and Fig. 4B, and variants thereof. (The same would be true for the circle-loops discussed below). A number of factors could favor one arrangement over the other. For example, if heterologous interactions are maintained when boundaries self-pair, homolog pairing could stabilize pairing between flanking boundaries and support a configuration in which boundaries define the main axis of both homologs (Fig. 4D). Note that in the stem-loop configuration, the CG12134, eve and TER94 loops on one homolog are juxtaposed with their counterparts on the other homolog in a manner that would facilitate trans-regulatory interactions. There are also mechanisms that would favor the formation of unanchored segments. For example, an unanchored segment would be formed if a boundary immediately upstream of eve were unable to pair with nhomie, and instead paired exclusively with its distal neighbor. This unanchored segment would correspond to a transition point, separating domains that are topologically connected by boundaries at their base.
Heterologous head-to-head pairing generates circle loops
A different loop topology, circle-loops, is generated by head-to-head pairing of heterologous boundaries in cis. For BX-C boundaries in the abd-A/Abd-B region (Fig. 5A, [76]), transgene assays have shown that they can pair with each other head-to-head [62,63,77]. If this pairing pattern is maintained in the endogenous setting, then circle-loops should be formed.
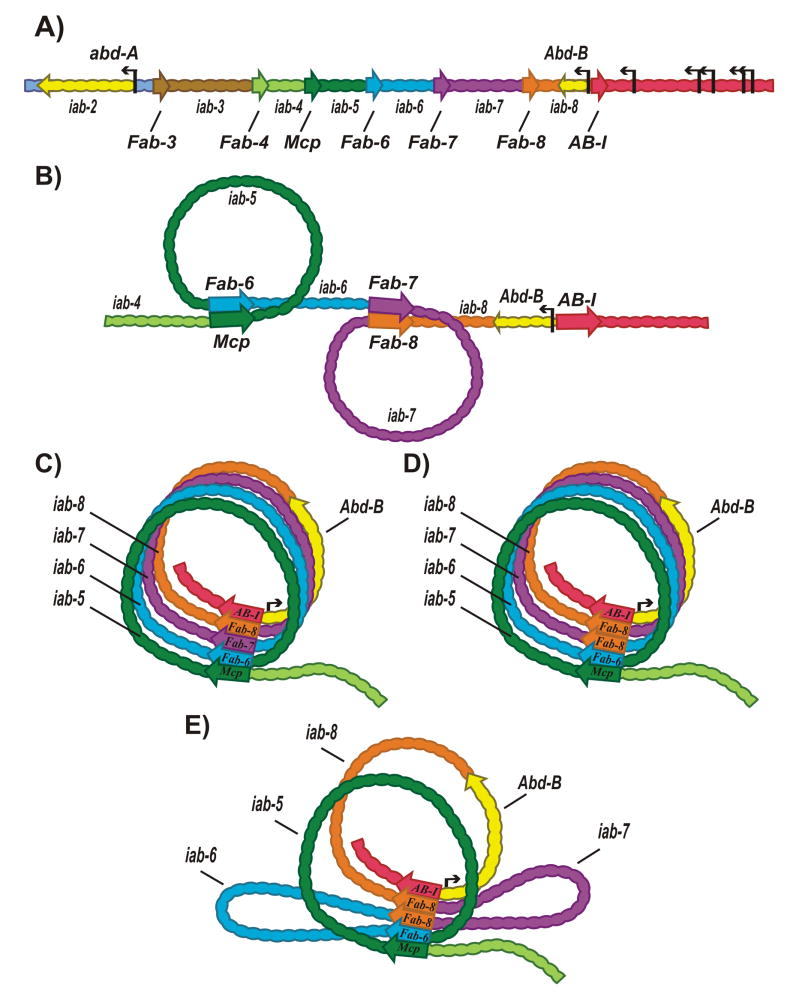
Figure 5.
Heterologous head-to-head pairing between boundaries in cis. A: Schematic of the abd-A and Abd-B gene regions. The regulatory domains, iab-2–iab-4 and iab-5 – iab8 of the abd-A and the Abd-B genes, respectively, are separated by boundaries (Fab-3, Fab-4, Mcp, Fab-6, Fab-7, Fab-8). AB-I is a boundary-like element located upstream of the Abd-B promoter. B: Head-to-head pairing between boundaries forms circle-loops that can be wound counter-clockwise (left) or clockwise (right). C: Series of circle-loops are connected by interacting boundaries. D, E: The Fab-7 replacement assay. D: A copy of Fab-8 copy is inserted in the “forward” orientation in place of Fab-7. In this orientation Fab-8 fully substitutes forFab-7 and would maintain the loop topology. E: Fab-8 is inserted in the “reverse” orientation. In this case head-to-head pairing interactions with neighboring boundaries would disrupt the circle-loop topology, introducing two stem-loops instead. In this orientation Fab-8 still blocks cross-talk between iab-6 and iab-7.However, it is no longer permissive for iab-6 (and to a lesser extent, iab-5) regulation of Abd-B.
Circle-loops can be wound clockwise (Fig. 5B right) or counter-clockwise (Fig. 5B left). In the configuration shown in Fig. 5B, the circle-loops are separated from each other by unanchored segments. As was the case for head-to-tail pairing, the unanchored segments separating the circle-loops define the main axis of the chromosome. Head-to-head pairing could also define the main axis of the chromosome. For example, the Abd-B boundaries (Fab-6, Fab-7, Fab-8 and AB-I) can pair with each other and (except for Fab-7and AB-I) with Mcp, which marks the border between the abd-A and Abd-B regulatory domains [62,63]. If these boundaries interact simultaneously with multiple partners, the Abd-B region would be assembled into a series of circle-loops connected at their base by boundaries. In the illustration in Fig. 5C, the Abd-B circle-loops are all wound in a clockwise direction, but winding could instead be all counter-clockwise, or be a mixture of clockwise and counter-clockwise winding.
Head-to-head pairing supports boundary bypass
Head-to-head pairing may be important in BX-C regulation. The reason is that boundaries in BX-C have two seemingly paradoxical activities. As described above, the first is to ensure functional autonomy by blocking adventitious interactions between regulatory elements in adjacent domains. Consistent with this blocking function, enhancer traps inserted into BX-C [78] and also transgene assays show that BX-C boundaries block enhancer/silencer<->promoter interactions like other fly boundaries. Seemingly inconsistent with this blocking activity, BX-C boundaries must also permit interactions between more distal regulatory domains and their target promoters. As shown for Abd-B in Fig. 5A, three (iab-5, iab-6, iab-7) of the four Abd-B regulatory domains have at least one boundary between them and Abd-B. Indeed, replacement experiments indicate that intervening boundaries can interfere with Abd-B regulation. For example, when Fab-7 is replaced by scs, it blocks iab-6<->iab-7 cross-talk just like Fab-7; however, it also interferes with the normal regulation of Abd-B by iab-5 and iab-6 [79]. Other heterologous boundary replacements give similar results[79,80].
Recent experiments in which the neighboring Fab-8 boundary replaced Fab-7 suggest a possible solution to this paradox [81]. Unlike scs, Fab-8 is fully functional when inserted in its normal orientation in place of Fab-7 (Fig.5D). However, its rescuing activity is orientation-dependent. When Fab-8 is inserted in the opposite orientation, it can still block cross-talk, but is no longer bypass-permissive. The inverted boundary prevents iab-6 from activating Abd-B, and weakens iab-5 regulation. As shown in Fig. 5E, flipping Fab-8 changes the loop configuration. Instead of a series of circle-loops, pairing of the inverted Fab-8 with its neighbors generates two stem-loops. The first stem-loop contains iab-6; the second contains iab-7. While the iab-7 stem-loop is on the same side of the boundary array as the Abd-B promoter, the iab-6 stem-loop is on the opposite side, precluding contact with the Abd-B promoter [81].
These findings suggest that the circle-loop topology of Abd-B regulatory domains generated by head-to-head pairing is important for boundary bypass. Supporting this idea is the fact that Fab-7 is fully functional in both orientations. In contrast to Fab-8, its pairing interactions are orientation-independent, so that inverting it should not perturb the circle-loop topology [63, 77, 80].
Boundaries and transcriptional regulation
The activation of Abd-B by the distal regulatory domains is expected to require contact between enhancers in the domains and the Abd-B promoter. Though not yet tested by mutations, pairing between Ab-I,upstream of the Abd-B promoter, and the downstream boundaries [62]may facilitate contact. In addition, the Abd-B promoter has a promoter-tethering element that stabilizes interactions with distant enhancers [82-84].
That boundary-like architectural elements are important in establishing and maintaining long-distance regulatory interactions is supported by the discovery of a developmentally regulated dCTCF element in the Ubx intron just upstream of the enhancers (abx/bx) that regulate Ubx in PS5 [85]. In PS4 and in more anterior parasegments, where Ubx is turned off by Polycomb repression, this dCTCF element is unoccupied. However, in parasegments where Ubx is active, dCTCF is bound. Regulated dCTCF binding correlates with chromosome topology. In parasegments where Ubx is off, contact between sequences around the dCTCF site and the Ubx promoter is absent. In contrast, when Ubx is active, physical contact is observed.
Stem-loops versus circle-loops
While the topology of looped domains, stem-loop or circle-loop, and the resulting regulatory readouts will be dictated by the orientation-dependence of boundary pairs, there are many questions that remain unresolved. For the Abd-B region, circle-loops appear to provide a mechanism for boundary bypass, enabling distal regulatory domains to “jump over” intervening boundaries and contact the Abd-B promoter. However, it isn't clear why circle-loops aren't also permissive for cross-talk between neighboring regulatory domains. Winding successive circle-loops in the opposite direction (clockwise, and then counter-clockwise) would place them on opposites sides of the paired boundaries, and this could block regulatory interactions between adjacent domains. However, it would also preclude Abd-B promoter interactions with iab-7 and iab-5. Thus, this solution requires a mechanism(s) for regulating the direction of winding during development. Another puzzle posed by the circle-loop configuration is homolog pairing and transvection. Here again, only circle-loops wound in the same direction on both homologs (and intercalated) would allow transvection.
There are also issues with stem-loops. For example, in regions where boundaries define the main chromosomal axis, genes and their regulatory elements in adjacent loops would be on opposite sides of a boundary pair, and therefore insulated from each other. However, every other loop would be located on the same side of the boundary array, which would seemingly be permissive for regulatory interactions.
One mechanism that might suppress cross-loop regulatory interactions would be a radial array of loops emanating from the main axis, like that proposed for metaphase chromosomes [12]. Alternatively, it is possible that there are special mechanisms that specifically promote and sustain contacts between enhancers/silencers and target genes within the same loop. One mechanism would involve the deployment of internal architectural elements like the developmentally regulated dCTCF site in the Ubx intron [85]. Another would be facilitated tracking in which molecular motors bring the enhancer and promoter together. By contrast, contacts between enhancers and promoters in different loops would depend upon thermal motion and be fleeting.
Yet another question is which of the possible loop configurations—unanchored versus anchored, stem-loops versus circle-loops—are more prevalent. If boundaries can simultaneously engage in pairing interactions with two heterologous partners, a mostly anchored configuration of loops would be expected. That this is possible is suggested by transgene experiments, which have shown that fly boundaries can (to greater or lesser extent) function in many different chromosomal contexts. This promiscuity likely arises because multiple DNA-bound proteins contribute to fly boundary activity. This means that in most instances the boundaries near the insertion site will utilize factors that can facilitate heterologous pairing. Nonetheless, for the BX-C, eve and 87A7 heat shock loci, neighboring boundaries seem to preferentially pair with each other, and to be orientation-specific.
It is also unclear whether stem-loops or circle-loops are more prevalent. A stem-loop is likely for the eve locus, but other boundaries in the neighborhood have not yet been studied. The only region of the fly genome that contains a series of well-characterized boundaries is the Abd-B region of BX-C. They interact head-to-head[62,63,77], and are thus expected to generate circle-loops. However, because BX-C regulation is special in requiring both domain insulation and bypass, deployment of the circle-loop topology there may be an unusual case. A systematic analysis of the boundary interaction pattern in other chromosomal regions will be needed to answer this question.
Mammalian boundaries
Though much less is known about the properties of mammalian boundaries, it is clear that they have architectural and regulatory functions similar to those in flies. For example, CTCF-dependent looping plays a key role in immunoglobulin gene rearrangement and regulation to generate antibody diversity. Here, CTCF binding elements influence looping configurations and rearrangement choice, along with a variety of other factors [86]. In one specific example, at the TCRβ locus, regulatory switching between enhancer blocking activity and facilitation of long-range interactions by one CTCF element is influenced by the presence of another such element [87], suggesting that partner choice may underlie CTCF functions in this process, and that other factors may help distinguish one CTCF site from another.
While a number of direct DNA binding proteins contribute to boundary function in flies, only one, CTCF, has been strongly correlated with boundaries so far in mammals [29, 31]. Not surprisingly, associated proteins (including the cohesin complex in mammals [88-90] mediate architectural functions, while other sequence-specific factors, notably ZNF143 [91-93], may help target CTCF to a subset of its genomic sites. In addition, tRNA genes and their associated factors (which include TFIIIC and CTCF) act as boundaries and affect chromosome organization [94]. Boundary factors in flies include both C2H2 zinc finger and BEN domain proteins[29,95,96]. Each of these families is represented in mammalian genomes, and consequently could function in chromosome architecture. If it turns out that mammals do indeed have multiple DNA binding architectural factors in addition to CTCF, then mammalian boundaries would likely resemble fly boundaries, and not only be non-autonomous, but also display pairing preferences. Pairing partner preference would then dictate how looped domains are assembled along the chromosome, including loop topology.
Consistent with directional pairing, high resolution 3C experiments have shown that the large majority of the anchored loops in human chromosomes are associated with a pair of convergently oriented CTCF sites [50, 97]. While fly dCTCF appears to form tetrameric complexes [98], only dimeric complexes have been reported for mammalian CTCF [99, 100]. Thus the predicted topology of loops anchored by convergent CTCF sites would be a stem-loop, like that containing the eve locus in flies [65]). The reason for the apparent prevalence of stem-loops is unclear, although this topology may provide for both a simpler chromosome organization and more predictable effects on gene regulation (discussed in [65]). Intriguingly, the convergent orientation of CTCF site pairs may be a major determinant not only of the topology of the loops that form, but also of partner choice, as reversing the orientation of a CTCF-anchored boundary can change not only nearby gene expression patterns but also the prevailing loop arrangement [101]. A subset of stem-loops appear to be adjacent to unanchored segments, as illustrated in Fig. 4B, and the unanchored segments in such regions would correspond to the main chromosomal axis [50,97]. Other stem-loops are adjacent to another anchored loop (Fig. 4C), and here, the boundaries that anchor the loops would form the main axis.
As in flies, the endpoints of each loop are defined by boundaries. A puzzle arises if CTCF is the only architectural DNA binding protein, namely, how are pairing partners chosen? In flies, the choice of pairing partners depends on two factors, boundary matching and proximity. For example, homie and nhomiecan interact with each other over distances of several Mb, “skipping over” many intervening boundaries [65,75] Here, pairing preference plays the key role in partner choice. However, if the only sequence-specific DNA binding protein responsible for boundary pairing is CTCF, the basis for pairing preference is not obvious, and the choice of partners might be dictated by proximity alone. But if this is the case, why do convergent sites typically anchor loops? Presumably there are many instances in which a non-convergent pair of CTCF sites are in closer proximity to each other than their nearest convergent neighbors. Is there a bias against the pairing of non-convergent CTCF sites? In flies, pairing between compatible boundaries can occur whether they are in the same orientation, divergent or convergent. The relative orientation and pairing specificity (head-to-head or head-to-tail) determines the topology of the loop that is formed, and this in turn influences the regulatory consequences. Whether these considerations are central to understanding mammalian boundaries is still unclear.
Conclusions and outlook
In contrast to the view that prevailed until recently, boundaries do not function autonomously, but require pairing with other boundaries or nuclear substructures to organize chromosomal loops. The effects of boundaries on gene regulation are critically dependent on this ability to organize loops, and on the details of the resulting topology. Presumably, similar considerations are central to their roles in other nuclear processes, such as recombination and DNA repair, as well as chromatin compaction during metaphase. Genetic analysis of loop formation in flies has given us a clearer picture of the pairing properties of boundaries. We now know that not only the choice of pairing partner, but also the orientation of the resulting boundary pair, dictates chromosome organization and regulatory readout. For example, whether a boundary pair organizes the intervening region into a stem-loop or a circle-loop determines which enhancer-promoter interactions are facilitated and which are blocked, and also whether a block can be bypassed in a regulated manner. Compelling new evidence suggests that similar considerations apply to vertebrate boundaries: most CTCF binding sites that anchor loops are oppositely oriented in the chromosome, suggesting that the intervening DNA is organized into a stem-loop. It is so far unclear what evolutionary forces have caused this bias, but the lessons from flies are that the implications for gene regulation are likely to be a major part of the story. Surely, much remains to be learned about boundary functions and mechanisms in mammals, and how closely they parallel those in Drosophila. Nonetheless, it seems increasingly clear that there are fundamentally important parallels yet to be fully charted and explored.
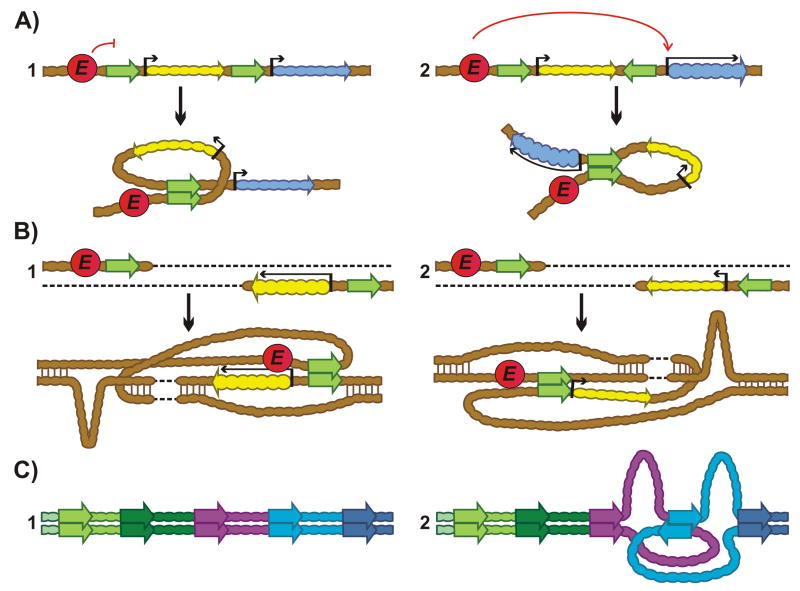
Figure 2.
Boundary competition and domain definition assays. A: Competition assay. One reporter and three boundaries (d, b, c) are used in this assay. (1) Downstream (d) boundary doesn't influence enhancer activity. (2) Insertion of second boundary (b) between the enhancer and promoter blocks activation. A loop isolating reporter is formed between the paired boundaries. (3-4) Insertion of a third boundary (c) upstream of enhancer can lead to activation (3) or isolation (4) of the reporter depending on their partner preference. B: Domain definition assay. Second reporter is inserted upstream of enhancer (red circle).(1) Both reporters are active. (2) Interaction between (b) and (d) boundaries isolates “yellow” colored reporter, leaving “blue” reporter unaffected. (3) Interaction between (c) and (d) boundaries isolates “blue”, but not “yellow” reporter. (4) Both reporters are isolated from enhancers.
Acknowledgments
This work was supported by NIH Awards GM050231 and GM117458 and NSF Award MCB-0818118 to JBJ, and byGM043432 to PS. PS would also like to acknowledge support from a grant to the Gene Biology Institute by the Russian Federation Ministry ofEducation and Science (14.B25.31.0022), by Russian Science Foundation N 14-24-00166 to PG.
Abbreviation
- TAD
topologically associating domain
References
- 1.Bickmore WA. The spatial organization of the human genome. Annu Review Genomics Hum Genet. 2013;14:67–84. doi: 10.1146/annurev-genom-091212-153515. [DOI] [PubMed] [Google Scholar]
- 2.Ciabrelli F, Cavalli G. Chromatin-driven behavior of topologically associating domains. J Mol Biol. 2015;427:608–25. doi: 10.1016/j.jmb.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Cremer T, Cremer M, Hubner B, Strickfaden H, et al. The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 2015;589:2931–43. doi: 10.1016/j.febslet.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 4.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 5.Mizuguchi T, Barrowman J, Grewal SI. Chromosome domain architecture and dynamic organization of the fission yeast genome. FEBS letters. 2015;589:2975–86. doi: 10.1016/j.febslet.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhimulev IF, Belyaeva ES, Semeshin VF, Koryakov DE, et al. Polytene chromosomes: 70 years of genetic research. Int Rev Cytol. 2004;241:203–75. doi: 10.1016/S0074-7696(04)41004-3. [DOI] [PubMed] [Google Scholar]
- 7.Andraszek K, Smalec E. Structure and functions of lampbrush chromosomes. J Biotechnol Comput Biol Bionanotechnol. 2011;92:337–44. [Google Scholar]
- 8.Gaginskaya E, Kulikova T, Krasikova A. Avian lampbrush chromosomes: a powerful tool for exploration of genome expression. Cytogenet Genome Res. 2009;124:251–67. doi: 10.1159/000218130. [DOI] [PubMed] [Google Scholar]
- 9.Morgan GT. Lampbrush chromosomes and associated bodies: new insights into principles of nuclear structure and function. Chromosome Res. 2002;10:177–200. doi: 10.1023/a:1015227020652. [DOI] [PubMed] [Google Scholar]
- 10.Benyajati C, Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976;9:393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- 11.Earnshaw WC, Laemmli UK. Architecture of metaphase chromosomes and chromosome scaffolds. J Cell Biol. 1983;96:84–93. doi: 10.1083/jcb.96.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsden MP, Laemmli UK. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979;17:849–58. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- 13.Saitoh Y, Laemmli UK. Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell. 1994;76:609–22. doi: 10.1016/0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 14.Vazquez J, Schedl P. Deletion of an insulator element by the mutation facet-strawberry in Drosophila melanogaster. Genetics. 2000;155:1297–311. doi: 10.1093/genetics/155.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welshons WJ, Keppy DO. Intragenic deletions and salivary band relationships in Drosophila. Genetics. 1975;80:143–55. doi: 10.1093/genetics/80.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galloni M, Gyurkovics H, Schedl P, Karch F. The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 1993;12:1087–97. doi: 10.1002/j.1460-2075.1993.tb05750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gyurkovics H, Gausz J, Kummer J, Karch F. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 1990;9:2579–85. doi: 10.1002/j.1460-2075.1990.tb07439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karch F, Galloni M, Sipos L, Gausz J, et al. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 1994;22:3138–46. doi: 10.1093/nar/22.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mihaly J, Hogga I, Gausz J, Gyurkovics H, et al. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development. 1997;124:1809–20. doi: 10.1242/dev.124.9.1809. [DOI] [PubMed] [Google Scholar]
- 20.Geyer PK, Green MM, Corces VG. Mutant gene phenotypes mediated by a Drosophila melanogaster retrotransposon require sequences homologous to mammalian enhancers. Proc Natl Acad Sci USA. 1988;85:8593–7. doi: 10.1073/pnas.85.22.8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peifer M, Bender W. Sequences of the gypsy transposon of Drosophila necessary for its effects on adjacent genes. Proc Natl Acad Sci USA. 1988;85:9650–4. doi: 10.1073/pnas.85.24.9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spana C, Harrison DA, Corces VG. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 1988;2:1414–23. doi: 10.1101/gad.2.11.1414. [DOI] [PubMed] [Google Scholar]
- 23.Parkhurst SM, Harrison DA, Remington MP, Spana C, et al. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 1988;2:1205–15. doi: 10.1101/gad.2.10.1205. [DOI] [PubMed] [Google Scholar]
- 24.Holdridge C, Dorsett D. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol Cell Biol. 1991;11:1894–900. doi: 10.1128/mcb.11.4.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith PA, Corces VG. The suppressor of Hairy-wing binding region is required for gypsy mutagenesis. Mol Gen Genet. 1992;233:65–70. doi: 10.1007/BF00587562. [DOI] [PubMed] [Google Scholar]
- 26.Udvardy A, Schedl P. Chromatin organization of the 87A7 heat shock locus of Drosophila melanogaster. J Mol Biol. 1984;172:385–403. doi: 10.1016/s0022-2836(84)80013-3. [DOI] [PubMed] [Google Scholar]
- 27.Udvardy A, Schedl P. The dynamics of chromatin condensation: redistribution of topoisomerase II in the 87A7 heat shock locus during induction and recovery. Mol Cell Biol. 1993;13:7522–30. doi: 10.1128/mcb.13.12.7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali T, Renkawitz R, Bartkuhn M. Insulators and domains of gene expression. Curr Opin Genet Dev. 2016;37:17–26. doi: 10.1016/j.gde.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Chetverina D, Aoki T, Erokhin M, Georgiev P, et al. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. BioEssays. 2014;36:163–72. doi: 10.1002/bies.201300125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matharu NK, Ahanger SH. Chromatin Insulators and Topological Domains: Adding New Dimensions to 3D Genome Architecture. Genes. 2015;6:790–811. doi: 10.3390/genes6030790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogelmann J, Valeri A, Guillou E, Cuvier O, et al. Roles of chromatin insulator proteins in higher-order chromatin organization and transcription regulation. Nucleus. 2011;2:358–69. doi: 10.4161/nucl.2.5.17860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H, Tian Y, Shu W, Bo X, et al. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS One. 2012;7:e41374. doi: 10.1371/journal.pone.0041374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kravchenko E, Savitskaya E, Kravchuk O, Parshikov A, et al. Pairing between gypsy insulators facilitates the enhancer action in trans throughout the Drosophila genome. Mol Cell Biol. 2005;25:9283–91. doi: 10.1128/MCB.25.21.9283-9291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller M, Hagstrom K, Gyurkovics H, Pirrotta V, et al. The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics. 1999;153:1333–56. doi: 10.1093/genetics/153.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sigrist CJ, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–21. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vazquez J, Muller M, Pirrotta V, Sedat JW. The Mcp element mediates stable long-range chromosome-chromosome interactions in Drosophila. Mol Biol Cell. 2006;17:2158–65. doi: 10.1091/mbc.E06-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyrchanova O, Toshchakov S, Parshikov A, Georgiev P. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: effects of insulator pairing on enhancer-promoter communication. Mol Cell Biol. 2007;27:3035–43. doi: 10.1128/MCB.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li HB, Muller M, Bahechar IA, Kyrchanova O, et al. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol Cell Biol. 2011;31:616–25. doi: 10.1128/MCB.00849-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang J, Lacroix L, Gamot A, Cuddapah S, et al. Chromatin immunoprecipitation indirect peaks highlight long-range interactions of insulator proteins and Pol II pausing. Mol Cell. 2014;53:672–81. doi: 10.1016/j.molcel.2013.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dekker J, Misteli T. Long-Range Chromatin Interactions. Cold Spring Harb Perspect Biol. 2015;7:a019356. doi: 10.1101/cshperspect.a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 42.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, et al. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–65. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 43.Dixon JR, Selvaraj S, Yue F, Kim A, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin F, Li Y, Dixon JR, Selvaraj S, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–4. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sofueva S, Yaffe E, Chan WC, Georgopoulou D, et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–29. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48:471–84. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 49.Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–95. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rao SS, Huntley MH, Durand NC, Stamenova EK, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992;6:1865–73. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- 52.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–50. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 53.Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol. 1992;12:2424–31. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roseman RR, Pirrotta V, Geyer PK. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993;12:435–42. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–96. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 56.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–14. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 57.Cai HN, Shen P. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science. 2001;291:493–5. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- 58.Muravyova E, Golovnin A, Gracheva E, Parshikov A, et al. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science. 2001;291:495–8. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- 59.Comet I, Savitskaya E, Schuettengruber B, Negre N, et al. PRE-mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev Cell. 2006;11:117–24. doi: 10.1016/j.devcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 60.Chetverina D, Savitskaya E, Maksimenko O, Melnikova L, et al. Red flag on the white reporter: a versatile insulator abuts the white gene in Drosophila and is omnipresent in mini-white constructs. Nucleic Acids Res. 2008;36:929–37. doi: 10.1093/nar/gkm992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kyrchanova O, Chetverina D, Maksimenko O, Kullyev A, et al. Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements. Nucleic Acids Res. 2008;36:7019–28. doi: 10.1093/nar/gkn781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kyrchanova O, Ivlieva T, Toshchakov S, Parshikov A, et al. Selective interactions of boundaries with upstream region of Abd-B promoter in Drosophila bithorax complex and role of dCTCF in this process. Nucleic Acids Res. 2011;39:3042–52. doi: 10.1093/nar/gkq1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kyrchanova O, Toshchakov S, Podstreshnaya Y, Parshikov A, et al. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol Cell Biol. 2008;28:4188–95. doi: 10.1128/MCB.00229-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gohl D, Aoki T, Blanton J, Shanower G, et al. Mechanism of chromosomal boundary action: roadblock, sink, or loop? Genetics. 2011;187:731–48. doi: 10.1534/genetics.110.123752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fujioka M, Mistry H, Schedl P, Jaynes JB. Determinants of Chromosome Architecture: Insulator Pairing in cis and in trans. PLoS Genet. 2016;12:e1005889. doi: 10.1371/journal.pgen.1005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duncan IW. Transvection effects in Drosophila. Annu Rev Genet. 2002;36:521–56. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- 67.Kennison JA, Southworth JW. Transvection in Drosophila. Adv Genet. 2002;46:399–420. doi: 10.1016/s0065-2660(02)46014-2. [DOI] [PubMed] [Google Scholar]
- 68.Gohl D, Muller M, Pirrotta V, Affolter M, et al. Enhancer blocking and transvection at the Drosophila apterous locus. Genetics. 2008;178:127–43. doi: 10.1534/genetics.107.077768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mellert DJ, Truman JW. Transvection is common throughout the Drosophila genome. Genetics. 2012;191:1129–41. doi: 10.1534/genetics.112.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Savitsky M, Kahn T, Pomerantseva E, Georgiev P. Transvection at the end of the truncated chromosome in Drosophila melanogaster. Genetics. 2003;163:1375–87. doi: 10.1093/genetics/163.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goloborodko A, Imakaev MV, Marko JF, Mirny L. Compaction and segregation of sister chromatids via active loop extrusion. eLife. 2016;5:e14864. doi: 10.7554/eLife.14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- 73.Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 2003;17:664–75. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fujioka M, Sun G, Jaynes JB. The Drosophila eve insulator Homie promotes eve expression and protects the adjacent gene from repression by polycomb spreading. PLoS Genet. 2013;9:e1003883. doi: 10.1371/journal.pgen.1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fujioka M, Wu X, Jaynes JB. A chromatin insulator mediates transgene homing and very long-range enhancer-promoter communication. Development. 2009;136:3077–87. doi: 10.1242/dev.036467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kyrchanova O, Mogila V, Wolle D, Magbanua JP, et al. The boundary paradox in the Bithorax complex. Mech Dev. 2015;138(Pt 2):122–32. doi: 10.1016/j.mod.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodin S, Kyrchanova O, Pomerantseva E, Parshikov A, et al. New properties of Drosophila fab-7 insulator. Genetics. 2007;177:113–21. doi: 10.1534/genetics.107.075887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bender W, Hudson A. P element homing to the Drosophila bithorax complex. Development. 2000;127:3981–92. doi: 10.1242/dev.127.18.3981. [DOI] [PubMed] [Google Scholar]
- 79.Hogga I, Mihaly J, Barges S, Karch F. Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol Cell. 2001;8:1145–51. doi: 10.1016/s1097-2765(01)00377-x. [DOI] [PubMed] [Google Scholar]
- 80.Iampietro C, Cleard F, Gyurkovics H, Maeda RK, et al. Boundary swapping in the Drosophila Bithorax complex. Development. 2008;135:3983–7. doi: 10.1242/dev.025700. [DOI] [PubMed] [Google Scholar]
- 81.Kyrchanova O, Mogila V, Wolle D, Deshpande G, et al. Functional Dissection of the Blocking and Bypass Activities of the Fab-8 Boundary in the Drosophila Bithorax Complex. PLoS Genet. 2016;12:e1006188. doi: 10.1371/journal.pgen.1006188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Akbari OS, Bae E, Johnsen H, Villaluz A, et al. A novel promoter-tethering element regulates enhancer-driven gene expression at the bithorax complex in the Drosophila embryo. Development. 2008;135:123–31. doi: 10.1242/dev.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Akbari OS, Schiller BJ, Goetz SE, Ho MC, et al. The abdominal-B promoter tethering element mediates promoter-enhancer specificity at the Drosophila bithorax complex. Fly. 2007;1:337–9. doi: 10.4161/fly.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ho MC, Schiller BJ, Akbari OS, Bae E, et al. Disruption of the abdominal-B promoter tethering element results in a loss of long-range enhancer-directed Hox gene expression in Drosophila. PLoS One. 2011;6:e16283. doi: 10.1371/journal.pone.0016283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Magbanua JP, Runneburger E, Russell S, White R. A variably occupied CTCF binding site in the ultrabithorax gene in the Drosophila bithorax complex. Mol Cell Biol. 2015;35:318–30. doi: 10.1128/MCB.01061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ribeiro de Almeida C, Hendriks RW, Stadhouders R. Dynamic Control of Long-Range Genomic Interactions at the Immunoglobulin kappa Light-Chain Locus. Adv Immunol. 2015;128:183–271. doi: 10.1016/bs.ai.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 87.Majumder K, Koues OI, Chan EA, Kyle KE, et al. Lineage-specific compaction of Tcrb requires a chromatin barrier to protect the function of a long-range tethering element. J Exp Med. 2015;212:107–20. doi: 10.1084/jem.20141479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gause M, Schaaf CA, Dorsett D. Cohesin and CTCF: cooperating to control chromosome conformation? BioEssays. 2008;30:715–8. doi: 10.1002/bies.20787. [DOI] [PubMed] [Google Scholar]
- 89.Parelho V, Hadjur S, Spivakov M, Leleu M, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–33. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 90.Wendt KS, Yoshida K, Itoh T, Bando M, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 91.Bailey SD, Zhang X, Desai K, Aid M, et al. ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nat Commun. 2015;2:6186. doi: 10.1038/ncomms7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heidari N, Phanstiel DH, He C, Grubert F, et al. Genome-wide map of regulatory interactions in the human genome. Genome Res. 2014;24:1905–17. doi: 10.1101/gr.176586.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lundberg SM, Tu WB, Raught B, Penn LZ, et al. ChromNet: Learning the human chromatin network from all ENCODE ChIP-seq data. Genome Biol. 2016;17:82. doi: 10.1186/s13059-016-0925-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kirkland JG, Raab JR, Kamakaka RT. TFIIIC bound DNA elements in nuclear organization and insulation. Biochim Biophys Acta. 2013;1829:418–24. doi: 10.1016/j.bbagrm.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aoki T, Sarkeshik A, Yates J, Schedl P. Elba, a novel developmentally regulated chromatin boundary factor is a hetero-tripartite DNA binding complex. eLife. 2012;1:e00171. doi: 10.7554/eLife.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kyrchanova O, Georgiev P. Chromatin insulators and long-distance interactions in Drosophila. FEBS Lett. 2014;588:8–14. doi: 10.1016/j.febslet.2013.10.039. [DOI] [PubMed] [Google Scholar]
- 97.Vietri Rudan M, Barrington C, Henderson S, Ernst C, et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bonchuk A, Maksimenko O, Kyrchanova O, Ivlieva T, et al. Functional role of dimerization and CP190 interacting domains of CTCF protein in Drosophila melanogaster. BMC Biol. 2015;13:63. doi: 10.1186/s12915-015-0168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pant V, Kurukuti S, Pugacheva E, Shamsuddin S, et al. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol Cell Biol. 2004;24:3497–504. doi: 10.1128/MCB.24.8.3497-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–8. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 101.Guo Y, Xu Q, Canzio D, Shou J, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–10. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]