Abstract
Human induced pluripotent stem cell-derived neural progenitor cells (hiPSC-NPCs) are considered as a promising cell source for transplantation and have been used for organoid fabrication to recapitulate central nervous system (CNS) diseases in vitro. The establishment of three-dimensional (3D) in vitro model with hiPSC-NPCs and control of their differentiation is significantly critical for understanding biological processes and CNS disease and regeneration. Here we implemented 3D methacrylated hyaluronic acid (Me-HA) hydrogels with encapsulation of hiPSC-NPCs as in vitro culture models and further investigated the role of the hydrogel rigidity on the cell behavior of hiPSC-NPCs. We first encapsulated single dispersive hiPSC-NPCs within both soft and stiff Me-HA hydrogel and found that hiPSC-NPCs gradually self-assembled and aggregated to form 3D spheroids. Then, the hiPSC-NPCs were laden into Me-HA hydrogels in the form of spheroids to evaluate their spontaneous differentiation in response to hydrogel rigidity. The soft Me-HA hydrogel-encapsulated hiPSC-NPCs displayed robust neurite outgrowth and showed high levels of spontaneous neural differentiation. We further encapsulated Down Syndrome (DS) patient-specific hiPSC-derived NPCs (DS-NPCs) spheroids within our hydrogels. DS-NPCs remained excellent cell viability in both soft and stiff Me-HA hydrogels. Similarly, soft hydrogels promoted neural differentiation of DS-NPCs by significantly upregulating neural maturation markers. This study demonstrates that soft matrix promotes neural differentiation of hiPSC-NPCs and HA-based hydrogels with hiPSC-NPCs or DS-NPCs are effective 3D models for CNS disease study.
Graphical abstract
Stiffness tunable 3D HA-based hydrogel models were implemented to control the progenitor properties and neuronal differentiation of hiPSC-NPCs or DS-NPCs.
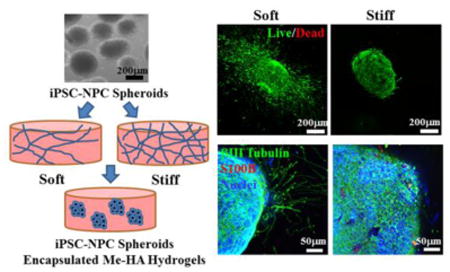
Introduction
Neurodegeneration-related injuries or diseases often make extensive neuronal death, subsequently resulting in permanent dysfunction due to the inferior regenerative capacity of the central nervous system (CNS).1-3 Cell replacement therapies hold great promise for neuronal regeneration and CNS functional recovery.4,5 Specifically, human induced pluripotent stem cells (hiPSCs) reprogrammed from somatic cells have been recently recognized as a promising cell source for personalized therapies by using the patient's autologous cells.6,7 Moreover, hiPSCs have been successfully differentiated into neural progenitor cells (NPCs) and then various CNS related cell phenotypes including neurons and glial cells, which are significantly invaluable for in vitro disease modeling, drug discovery, and developing regenerative therapies.8,9 However, the inferior survival rate of grafted cells and inappropriate cell scaffold platform limit the efficacy of hiPSC-based regenerative therapies.10-12 Therefore, development of physiologically relevant in vitro model with hiPSC-derived NPCs (hiPSC-NPCs) plays an important role in not only understanding neural network development, behavior and activities under physiological or pathological situations, but also identifying key characteristics of cell-matrix interactions to direct the appropriate design of functional grafts for CNS injuries or diseases. In addition, the cell fate of engrafted hiPSC-derived progenitor or precursor cells are typically poorly controlled, leading to the formation of teratomas, which is one of the major challenges in hiPSC-based cell therapies.13 It is thus of significant importance to delicately control the microenvironment and mediate the differentiation of hiPSC-NPCs to avoid tumor formation.
A promising in vitro model for the support of 3D hiPSC-NPC culture and differentiation involves the implementation of 3D biomimetic platforms that mimic the unique physiological microenvironment of extracellular matrix (ECM) in native CNS. Natural hydrogel-based scaffolds have been recognized as compelling materials for mimicking the native ECM recently.14-17 One example of the most commonly used hydrogel systems for neural stem cells and hiPSC based cells is Matrigel, a matrix extracting from the Englebreth-Holm-Swarm mouse sarcoma.18-20 Although Matrigel has been demonstrated as an effective CNS-related cell carrier, it cannot be employed for clinical usage due to its unknown growth factor composition and murine sarcoma origins.21-23 Therefore, there is an unmet need to develop other component-defined hydrogel systems to support hiPSC-NPCs and to improve the controllability. A number of attempts have been made to develop modified hyaluronic acid, agarose, alginate, or peptides-based hydrogel materials for hiPSC encapsulation.24-27 Promoting and controlling differentiation of the NPCs to neurons is a major aim in transplantation studies.28,29
Another important aspect for hiPSC-NPC in vitro model construction is to control the hydrogel matrix rigidity. In nature, ECM mechanics are one of key characteristics to adjust and support the differentiation and function of the corresponding cells.30-32 It has been found that the immature brain of rat and pig exhibited roughly twice as stiff as adult brain, demonstrating larger force requirement for brain development.33,34 Moreover, many studies have demonstrated that nerve-related cells could sense and respond to mechanical cues from the culture substrate.35-37 Softer substrates have been found to promote the neuronal maturation and differentiation,38,39 as well as neurite outgrowth and axon specification,40 whereas astrocytes spread less and adhered poorly to softer gels.41,42 Another study showed that neuronal cultures were more adherent to stiff gels and neurite length had no stiffness preference.43 However, information about the influence of the matrix rigidity on iPSC-NPC culture and differentiation is still lacking. Therefore, the development of ECM-mimicking hydrogel models with tailorable stiffness is an important target to extend the understanding of iPSC-NPC response to stiffness and provide material design parameters for future applications in regeneration of CNS system.
In response to the aforementioned challenges, we fabricated hydrogels based on methacrylated hyaluronic acid (Me-HA) as a 3D environment for the in vitro culture of hiPSC-NPCs. We hypothesize that the matrix stiffness of hyaluronic acid (HA, a major ECM component in the fetal mammalian brain) in CNS tissues directly affects NPC differentiation and that an HA-based 3D culture system could be employed to investigate and mimic these effects in vitro. We evaluated the effect of Me-HA hydrogel stiffness on the cell viability, migration, and spontaneous differentiation of hiPSC-NPCs. We further differentiated Down Syndrome (DS) patient-specific hiPSCs into NPCs (DS-NPCs) and investigated the viability and neural differentiation of DS-NPCs cultured in the same hydrogels.
Experimental
Cell culture and differentiation
The two healthy hiPSC lines and two DS hiPSC lines were reprogrammed from fibroblasts obtained from healthy and DS individuals, respectively, as described in our previous study.44 An embryoid body-based differentiation procedure was used for differentiation of these hiPSCs to NPCs.45 The hiPSC-NPCs and DS-NPCs were maintained in growth medium (GM) containing DMEM/F12 medium (Hyclone), 1×B27 (Thermo Fisher Scientific), 1×N2 (Thermo Fisher Scientific), basic fibroblast growth factor (20 ng/mL, bFGF, Peprotech), 1% penicillin/streptomycin (P/S, Thermo Fisher Scientific). The cells were seeded on 6-well polystyrene plates (Corning) coated with Matrigel (Corning) cultured at 37 °C with 5% CO2 using standard protocols.44 For spontaneous differentiation of these NPCs, cells were cultured in spontaneous differentiation medium (SDM) consisting of DMEM/F12 medium, 1×B27, 1×N2, and 1% P/S. For neuronal differentiation, cells were cultured in neuronal differentiation medium (NDM) consisted of neurobasal medium (Hyclone) and DMEM/F12 medium (1:1), 1×B27, 1×N2, 10 μM N6,2′-O-dibutyryladenosine 3′,5′-cyclic monophosphate sodium salt (cAMP, Sigma), 200 nM ascorbic acid, brain derived neurotrophic factor (10 ng/mL, BDNF, Peprotech), glial-derived neurotrophic factor (10 ng/mL, GDNF, Peprotech), laminin (1 μg/ml, Corning), and 1% P/S. All experiments conducted on hiPSCs were approved by the Stem Cell Research Oversight Committee at the University of Nebraska Medical Center.
Polymer modification and hydrogel preparation
Photocrosslinkable hyaluronic acid (HA, ∼1200 kDa, NovaMatrix) was synthesized as previously reported through the reaction of methacrylic anhydride (Sigma) with 0.5% HA in deionized water.46 A hydrogel precursor solution composed with Me-HA (0.75%w/v) was dissolved in cell culture medium with 0.05% w/v 2-hydroxy-1(4-(hydroxyethox)pheny)-2-methyl-1-propanone (Irgacure 2959, CIBA Chemicals). The gel precursor was transferred into silicone molds (8 mm in diameter, 1 mm in thickness, for single cell and spheroid encapsulation) and subsequently exposed to OmniCure S2000 UV lamp (Lumen Dynamics) for 30 s or 60 s at room temperature to generate stiffness-controlled hydrogel matrix. The distance between the silicone mold and UV lamp was maintained at 10 cm and the irradiance of the light was 320 μW/cm2 (Blak-Ray Ultraviolet meter, UVP Inc). For single cell encapsulation into hydrogels, the hiPSC-NPCs with a density of 1×106 cells/ml were utilized as shown in Fig. 1A. For cell spheroid encapsulation, hiPSC-NPCs or DS-NPCs spheroids were first prepared by the cell suspension culture as shown in Fig. 1B and C. Briefly, single dispersive cell resuspension was placed into low-attachment 6-well plate (Corning) with 1.5 ×105 cells per well. After 2 days of culture, the cells self-assembled as cell clusters in the shape of free floating spheroids due to spontaneous fusion among single cells. Then hiPSC-NPCs or DS-NPCs spheroids were resuspended within hydrogel precursor solutions before UV photocrosslinking. These cell-laden hydrogels scaffolds were maintained in SDM or NDM at 37 °C with 5% CO2 for 28 days, and the medium was replaced every 2 days.
Fig.1.
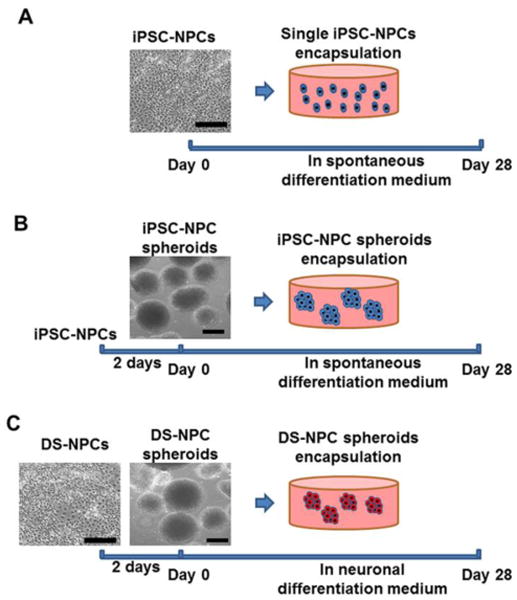
Schematic of experimental design. (A) Homogeneous hiPSC-NPC encapsulation within Me-HA hydrogels: single dispersive hiPSC-NPCs and hydrogel precursors were loaded into silicone molds and subsequently exposed to 365 nm UV light for 30 s or 60 s. The cell-hydrogel constructs were maintained in spontaneous differentiation medium (SDM) for 28 days. (B) hiPSC-NPC spheroids encapsulation within hydrogels: hiPSC-NPC cell spheroids were first fabricated by the cell self-assembly method after 2 days of culture and then encapsulated within the hydrogels. The spheroid laden constructs were conditioned in SDM for 28 days; (C) DS-NPC spheroids laden within hydrogels: DS-NPC spheroids were first manufactured, and then were incorporated into hydrogels. The hydrogel-encapsulated DS-NPC spheroids were conditioned in neural differentiation medium (NDM) for 28 days. Scale bars = 200 μm.
Physical characterization of the hydrogel scaffolds
Surface morphologies of Me-HA hydrogel scaffolds were characterized by utilizing scanning electron microscopy (SEM, FEI Quanta 200) after freeze-drying. The stiffness of Me-HA hydrogel scaffolds was conducted using a compression tester (Nanosute, Agilent Techologies) at room temperature.
Cell viability and morphology
An inverted optical microscope (Leica DMI1/MC120) with HD camera was employed to observe the morphology and migration of single cells or cell spheroids cultured in hydrogel-free medium or laden in hydrogel scaffolds. The viability of single cells or cell spheroids within the hydrogel scaffolds or hydrogel-free medium was determined after 28 days of culture by Live/Dead assay (Thermo Fisher Scientific) as previously described47 and fluorescence images were obtained using a confocal laser scanning microscopy (CLSM, LSM 710, Carl Zeiss, Germany).
Self-assembled hiPSC-NPC spheroid density and area
Based on Live/Dead assay images for encapsulated single hiPSC-NPCs within hydrogels, we calculated the self-assembled hiPSC-NPC spheroid density and area distribution in hydrogels with different stiffness. Spheroid density was quantified by counting the spheroid number via ImageJ and standardizing to image area. NIH Image J was used to measure the spheroid area and then the histograms were plotted. In total three samples for each condition were used and three images from each sample were analyzed.
Immunofluorescent staining
All samples were fixed in 4% paraformaldehyde, permeabilized in 0.2% Trion X-100 and then blocked with 1% bovine serum albumin (BSA) overnight at 4 °C. The samples were then treated with primary antibodies to βIII-Tubulin (1:500, Biolegend), and S100B (1:250, Sigma) overnight at 4 °C. Secondary fluorescent antibodies were incubated for 2 h and nuclear counterstaining (Draq 5, 1: 1000, Thermo Fisher Scientific) were performed for 30 minutes at room temperature. The stained samples were imaged with Zeiss 710 CLSM.
Length and extension speed of neurite outgrowth
For neurite outgrowth length and extension speed measurement, nine encapsulated hiPSC-NPC spheroids in three different hydrogel samples were randomly selected and imaged by using inverted microscope after 28-day culture. Ten longest and ten shortest neurites from one side of each spheroid were measured and averaged to generate the average neurite length for that particular side of the spheroids. The same analysis was then conducted on the other side. Then the average migration speed was calculated based the average length in both side and culture time. All measurements were conducted using NIH Image J software.
RNA isolation and qPCR
Total RNA was extracted from cell-encapsulated hydrogel constructs or cell spheroids cultured in hydrogel-free medium using QIA-Shredder and RNeasy mini-kits (QIAgen) according to the manufactures' instructions. Total RNA was synthesized into first strand cDNA in a 20 μL reaction using iScript cDNA synthesis kit (BioRad Laboratories). Real-time PCR analysis was performed in a StepOnePlus™ Real-Time PCR System (Thermo Scientific) using SsoAdvanced SYBR Green Supermix (Bio-Rad). cDNA samples were analyzed for the gene of interest and for the housekeeping gene 18S rRNA. The level of expression of each target gene was calculated using comparative Ct (2–ΔΔCt) method. All primers used in this study are listed in Supplementary Table S1.
Statistical analysis
All quantitative data were expressed as mean ± standard deviation (SD). Pairwise comparisons between groups were conducted using ANOVA with Scheffé post-hoc tests in statistical analysis. A value of p<0.05 was considered statistically significant.
Results
Me-HA Hydrogel Formation, Morphology and Mechanical Properties
Hydrogels with tunable stiffness were fabricated by photocrosslinking Me-HA precursor. SEM images showed that the microstructure morphologies of Me-HA hydrogels exhibited very irregular pore shapes and relatively open 3D network structure, which varied slightly between the two freeze-dried Me-HA hydrogels prepared from different crosslinking time (Fig. 2A, B). The varied UV crosslinking time (30 s or 60 s) was employed to control the stiffness of the resulting Me-HA hydrogel matrix. The compression modulus of the hydrogels was significantly raised by increasing the photocrosslinking time (Fig. 2C). Specifically, the stiffness of 60 s crosslinked Me-HA hydrogels increased to 1.41±0.27 kPa in comparison with 0.51±0.20 kPa for 30 s crosslinked counterparts. In this study, the softer hydrogel with stiffness of 0.51±0.20 kPa is denoted as “soft” hydrogel, and the stiffer one with the stiffness of 1.41±0.27 kPa is denoted as “stiff” hydrogel.
Fig.2.
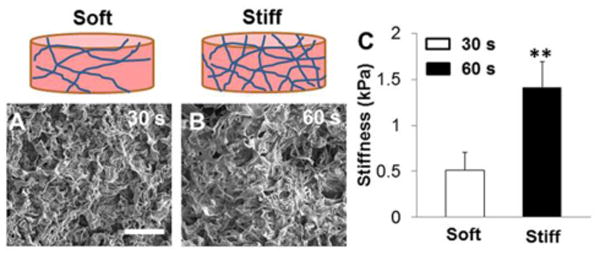
Physical properties comparison of Me-HA-based hydrogel constructs prepared at different crosslinking time. SEM images of freeze-dried Me-HA-based hydrogel constructs using varied crosslinking time: (A) 30 s and (B) 60 s. Scale bars = 100 μm. (C) Compressive modulus of these two different hydrogels performed at wet condition (**p<0.01).
3D In Vitro Culture of hiPSC-NPCs Single Cells Within Bioactive Me-HA Hydrogels
We first evaluated cell survival and morphology of hiPSC-NPCs encapsulated within our Me-HA hydrogels with the form of single cells (Fig. 3A). Although a few dead cells were observed in the Live/Dead assay, there were no significant differences in cell viability between two groups. The hiPSC-NPCs showed high viability in both soft and stiff hydrogels throughout 28-day culture. Importantly, single hiPSC-NPCs gradually self-assembled and aggregated to form 3D spheroids when they were encapsulated into both soft and stiff Me-HA hydrogels. The hiPSC-NPCs laden in soft hydrogels showed a similar spheroid density compared with those within stiff hydrogels (Fig. 3B). However, hiPSC-NPCs within soft hydrogels showed greater aggregating potency with significant larger spheroid diameters (Fig. 3C).
Fig.3.
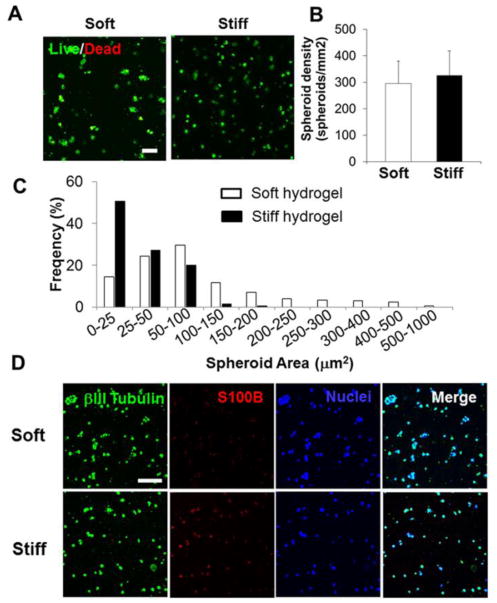
Soft Me-HA hydrogels promoted the spontaneous spheroid-shaped self-assembly and neuronal differentiation of single dispersive hiPSC-NPCs. (A) Representative fluorescent images of living cells (green) and dead cells (red) within single hiPSC-NPC laden hydrogels in SDM after 28-day culture. Scale bars = 100 μm. (B) Spheroid density and (C) spheroid area distribution analysis of single hiPSC-NPCs encapsulated into soft and stiff hydrogels. (D) Representative immunofluorescent staining for S100B (red), βIII-tubulin (green) and nuclei (blue) within single hiPSC-NPC laden hydrogels in SDM after 28-day culture.
The hiPSC-NPCs have the capacity to differentiate into different CNS related cell phenotypes, such as neurons (βIII-Tubulin+) and astrocytes (S100B+) depending on various cell substrates and culture conditions.12,48,49 We examined and compared the cell phenotype change of single cell dispersed hiPSC-NPCs in both soft and stiff Me-HA hydrogels in SDM for a period of 28 days. The encapsulated hiPSC-NPCs in the both hydrogel types expressed βIII-tubulin (green) and S100B (red) as shown after immunofluorescent staining, suggesting they spontaneously differentiated into mixed populations of cells (Fig. 3D). However, hiPSC-NPCs in the soft hydrogels appeared to express remarkably less S100B than βIII-Tubulin, indicating that soft hydrogel promoted spontaneous neuronal differentiation.
Soft Hydrogels Promoted iPSC-NPC Spheroid Migration, Neurite Outgrowth and Neural Differentiation
Encapsulated single hiPSC-NPCs have been demonstrated above to form spherical, multicellular aggregates in Me-HA hydrogels. In order to accommodate this cell self-assembly capacity and phenomenon, we first generated sphere-shaped cell aggregates with the diameter ranging between 50 and 300 μm by culturing the cells in suspension to form spheroids and then encapsulating these hiPSC-NPC spheroids into Me-HA hydrogels (Fig. 1B). The 3D organoids better mimic cell growth and differentiation observed in 3D CNS under physiological conditions, compared with 2D cultures.50 Cell behavioral differences for hiPSC-NPC spheroids within soft and stiff hydrogels were observed and evaluated by using phase contrast microscopy. After 28-day culture in SDM, the hiPSC-NPC spheroids within soft hydrogels showed robust neurite outgrowth, whereas limited neurites were observed in stiff hydrogels (Fig. 4A). Neurite morphometric analysis demonstrated that the mean value of neurite outgrowth length was significantly longer in soft hydrogels in comparison with the stiff counterparts (Fig. 4B). Interestingly, we observed that hiPSC-NPC spheroids migrated towards each other within soft hydrogel after 28 days of culture, indicating that the cell spheroids could remodel the hydrogels (Fig. 4C). We then performed Live/Dead assay to determine the effects of the hydrogel stiffness on viability of hiPSC-NPC spheroids at day 28 after conditioning in SDM. The hiPSC-NPC spheroids cultured in hydrogel-free SDM were utilized as a control group. The hiPSC-NPC spheroids within both soft and stiff hydrogels showed high viability, without obvious difference compared to hydrogel-free hiPSC-NPC spheroids (Fig. 5). In consistent with optical images, hiPSC-NPC spheroids showed extensive neurite outgrowth in soft hydrogels, whereas limited neurites were observed in stiff hydrogels and no neurites for hydrogel free spheroids.
Fig.4.
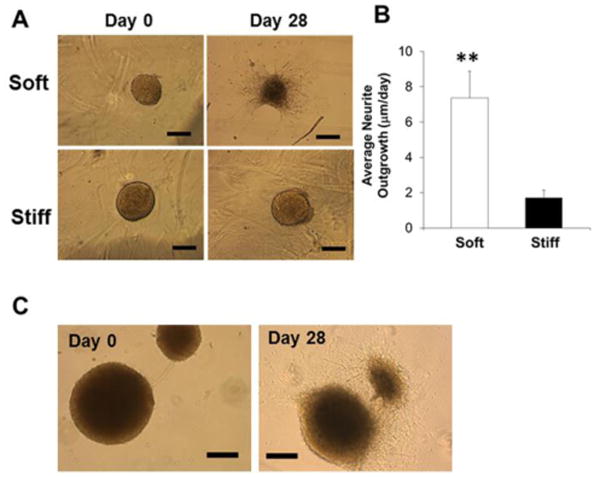
Soft hydrogels promoted spontaneous migration and neurite outgrowth of hiPSC-NPC spheroids. (A) Representative phase contrast images of hiPSC-NPC spheroids laden hydrogels in SDM after 28-day culture. Scale bars = 300 μm. (B) Average neurite outgrowth from hiPSC-NPC spheroids encapsulated into soft and stiff hydrogels (**p<0.01). (C) Representative migration images of hiPSC-NPC spheroids encapsulated into soft hydrogels in SDM after 28-day culture. Scale bars = 300 μm.
Fig.5.
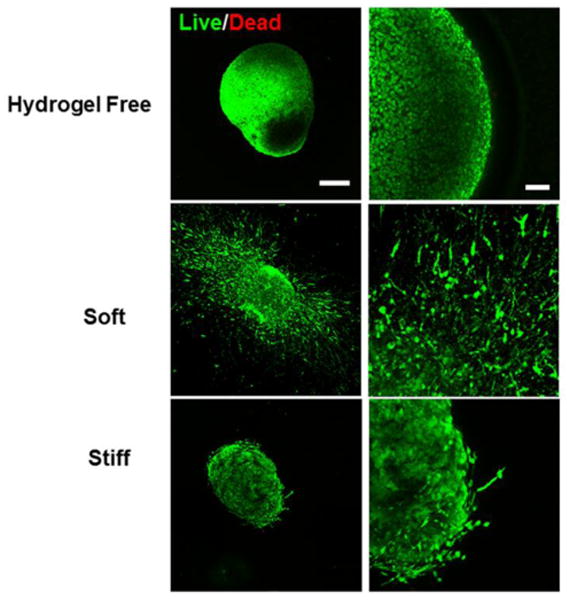
Both soft and stiff hydrogels supported high viability of hiPSC-NPC spheroids. Representative fluorescent images of living cells (green) and dead cells (red) of hiPSC-NPC spheroids cultured in hydrogel-free medium or laden in hydrogel scaffolds in SDM after 28-day culture. Scale bars on left panel= 200 μm, right panel= 50 μm.
We next evaluated the spontaneous differentiation potential of hiPSC-NPC spheroids in our Me-HA hydrogels and hydrogel-free SDM during 28 days of culture. Immunofluorescent staining showed that βIII-tubulin and S100B were expressed by hiPSC-NPC spheroids in all the three groups (Fig. 6A). However, lowest S100B expression was found in soft Me-HA hydrogel, which demonstrated that lower hydrogel stiffness suppressed the astrocyte differentiation capacity of hiPSC-NPC spheroids (Fig. 6A). Furthermore, βIII-tubulin was only observed within the spheroids in hydrogel-free group and stiff hydrogel group, while it was present in the cell extensions in soft hydrogel group, which confirmed that softer hydrogel noticeably promoted neurite outgrowth.
Fig.6.
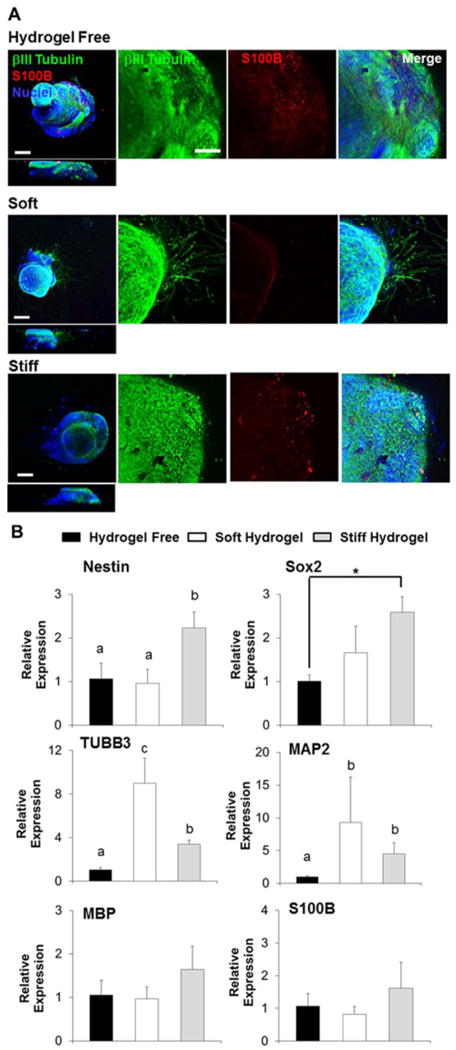
Soft hydrogels promoted spontaneous neurite outgrowth and neuronal differentiation of hiPSC-NPC spheroids, and stiff hydrogels were capable to better maintain the progenitor properties of hiPSC-NPCs. (A) Representative immunofluorescent staining for S100B (red), βIII-tubulin (green) and nuclei (blue) of hiPSC-NPC spheroids cultured in hydrogel-free medium or laden in hydrogel scaffolds in SDM after 28-day culture. Scale bars on left panel= 200 μm, right panel= 50 μm. (B) qPCR analysis of Nestin, Sox2, TUBB3, MAP2, MBP and S100B of hiPSC-NPC spheroids cultured in hydrogel-free medium or laden in hydrogel scaffolds in SDM after 28-day culture. Relative gene expression is presented as normalized to 18S and expressed relative to hiPSC-NPC spheroids cultured in hydrogel-free medium (n=3; bars that do not share letters are significantly different from each other (p<0.05); *p<0.05).
To further examine the expression levels of phenotypic markers, gene expression of the neural progenitor markers, Nestin and Sox2, the neuron and neuronal maturation markers, TUBB3 and MAP2, the astrocyte marker S100B, and the oligodendrocyte marker MBP were quantified by qPCR in soft and stiff hydrogel groups relative to the hydrogel-free group (Fig. 6B). The gene expression levels of Nestin and Sox2 were significantly higher in the stiff hydrogel group than the hydrogel-free group and soft hydrogel group, and the neuronal gene expression levels (i.e. TUBB3 and MAP2) in the soft hydrogel group were significantly upregulated comparing to the other two groups. The expression of the MBP and S100B was not significantly different among these three groups. These data demonstrated that softer hydrogel environment promoted the neuron differentiation and maturation. Moreover, stiffer hydrogel matrix may maintain the stemness of hiPSC-NPCs for a prolonged period.
Soft Hydrogels Promoted Neuronal Differentiation and Maturation of DS-NPC Spheroids
Since we have demonstrated that soft hydrogel promoted spontaneous neuronal differentiation of hiPSC-NPCs derived from normal fibroblasts, we then asked whether soft matrix could also facilitate neuronal differentiation of hiPSC-NPCs from patient with CNS disease. Here, we studied the NPCs derived from DS patient-specific hiPSCs, because previous studies in human brain tissues, transgenic mouse model and DS hiPSC models showed that DS-NPCs exhibited a gliocentric shift and impaired neurogenesis.44,51-53 We further generated DS-NPC spheroids and encapsulated the spheroids in our hydrogels with different stiffness and condition the constructs in NDM for 28 days. Live/dead assay showed that high survival ratio of DS-NPCs, but no discernible neurite outgrowth was found among all the three different groups (Fig. 7A). We did not find any neurite outgrowth from the spheroids. Immunofluorescent staining showed that neuron-related proteins, βIII-tubulin and NeuN, were expressed by DS-NPC spheroids in all the three groups, with an increased NeuN expression observed in soft Me-HA hydrogel (Fig. 7B). Results of qPCR also confirmed that DS-NPC spheroids showed highest gene expression level of neural differentiation and neuron maturation markers, including DLG4, Synapsin1, TUBB3 and NeuN, in soft Me-HA hydrogel, whereas stiffer hydrogel either had no effects or downregulated expression of these markers (Fig. 7C). Together, these results supported that lower hydrogel stiffness robustly enhanced the neural differentiation and maturation of DS-NPCs upon induction utilizing NDM.
Fig.7.
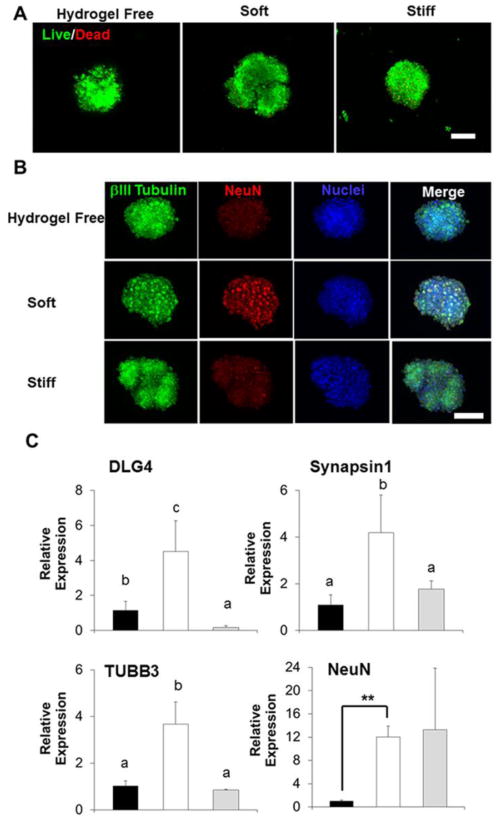
Soft hydrogels supported high viability of DS-NPC spheroids and promoted neuronal differentiation of DS-NPC spheroids. (A) Representative fluorescent images of living cells (green) and dead cells (red) of DS-NPC spheroids cultured in hydrogel-free medium or laden in hydrogel scaffolds in NDM after 28-day culture. Scale bars= 300 μm. (B) Representative immunofluorescent staining for NeuN (red), βIII-tubulin (green) and nuclei (blue) of DS-NPC spheroids cultured in hydrogel-free medium or laden in hydrogel scaffolds in NDM after 28-day culture. Scale bars= 300 μm, (C) qPCR analysis of DLG4, Synapsin1, TUBB3, and NeuN of DS-NPC spheroids cultured in hydrogel-free medium or laden in hydrogel scaffolds in NDM after 28-day culture. Relative gene expression is presented as normalized to 18S and expressed relative to DS-NPC spheroids cultured in hydrogel-free medium (n=3; bars that do not share letters are significantly different from each other (p<0.05); **p<0.01).
Discussion
Although several proposed cell types, including immortalized neural cell lines, and neural stem/progenitor cells, may be utilized for the CNS regeneration, hiPSC-derived NPCs hold great promise for patient-specific cell replacement therapies.54-56 Additionally, hiPSCs are highly expandable and are capable of generating large numbers of NPCs, which have also emerged as an unlimited cell source for disease modeling and drug screening.57,58 However, more detailed characterization of hiPSC-NPCs and better understanding of their cell-cell and cell-matrix interactions are needed, in order to maximize their therapeutic potential. Moreover, until now, biomaterial science still remains ongoing challenges for developing a well-controlled culture model for 3D hiPSC-NPC growth in vitro and a permissive environment for effectively manipulating cellular behavior of hiPSC-NPCs.59,60 For example, silk based scaffolds with collagen hydrogel has been used to support human induced neural stem cells and their co-culture to generate the engineered cortical brain tissue.61 The objective of this study is to determine whether the HA-based 3D hydrogel system could serve as an ECM-mimicking platform to improve the survival rate of hiPSC-NPCs and how the hydrogel rigidity control the phenotype change and functional maturation of hiPSC-NPCs in vitro. Our Me-HA hydrogel system possesses three advantages. First, HA is an essential organizational and structural component in the ECM of native tissues. It is particularly abundant in the fetal brain and surrounding immature neurons during differentiation in the spinal cord, indicative of its vital role in CNS development.62-64 Second, Second, the hydrogel formulation can be well controlled and defined with better reproducibility than Matrigel.24,65 Third, our Me-HA hydrogels can combine with other hydrogel systems (i.e. gelatin, collagen, and alginate) and also allow for the incorporation of specific neurotrophic factors or drugs to form a complex and advanced system for the application of neural tissue engineering.
Maintaining high cell survival of hiPSC-NPCs after encapsulation into hydrogel matrix is significantly important since this is the first step toward integration into 3D in-vitro culture model and realization of subsequent targeted differentiation. Low in vivo or in vitro survival rates and poor neurite outgrowth was observed when various NPCs were encapsulated within the hydrogels or injected into animal models.66,67 Locally injected cells generally had poor viability, often with as few as 1-20% of cells being able to survive after transplantation.35,68 Suspension culture usually lead to considerable cell agglomeration, cell death or uncontrolled differentiation.69,70 Moreover, low cell viability, limited cell growth, and uncontrolled differentiation were also observed in the hydrogel culture systems, including acrylate-modified HA hydrogels (60-90 kDa), agarose hydrogels and alginate hydrogels.27 In this study, HA with high molecular weight of ∼1200 kDa was utilized to fabricate hydrogel matrix. We demonstrated that when encapsulated into our Me-HA hydrogels in the form of single cells, the hiPSC-NPCs displayed robust survival rate. Previous studies also showed that HA with high molecular weight was an essential organizational and structural component in the native brain ECM and had neuroprotective effects.71,72 However, no discernible neurite outgrowth was generated in our Me-HA hydrogels. Instead, single hiPSC-NPCs spontaneously migrated and accumulated in dispersive sphere-like circular islands. Other studies also demonstrated poor neurite outgrowth capacity and spontaneous formation of spherical aggregates for single-cultured hiPSC-NPCs.26,62,73 This indicates that paracrine is important for survival and function of hiPSC-NPCs within matrix. This self-assembled cell behavior led us to pre-manufacturing hiPSC-NPC spheroids prior to the encapsulation into Me-HA hydrogels. The Me-HA hydrogel-encapsulated hiPSC-NPC spheroids exhibited robust survival and neurite outgrowth in vitro.
Mechanotransduction in soft tissues, particularly its influence on progenitor cell differentiation, has begun to be explored recently.74 Accumulating evidence suggests that the mechanical properties of the culture matrix can modulate important neuronal differentiation, maturation, and functions such as growth, extension, branching, and activity.40,75,76 Several studies have shown that the local elastic modulus of the brain is in the range of 0.1-1.5 kPa.42,77-79 Some other studies showed broader stiffness ranging from 0.1 to 10 kPa.40,62 The difference is probably due to the use of different brain anatomy and species, different age and different testing methods. In our study, the control of crosslinking time was utilized to directly manipulate the mechanical properties of the Me-HA hydrogel matrix for assessing the influence of the matrix stiffness on hiPSC-NPC responses. The stiffness of the soft hydrogel is in the lower native brain stiffness range, while the stiff one is in the middle or upper range. Notably, hiPSC-NPC single cells or spheroids showed more differentiation towards a neuronal phenotype in SDM in the soft Me-HA hydrogels over a period of 28 days of culture. In particular, neurites with extended length outgrowth from hiPSC-NPC spheroids positive for the neuronal marker βIII-tubulin were only found in soft hydrogel group, and significantly higher mature neuronal gene expressions were found in soft hydrogel group. In contrast, stiff Me-HA hydrogel was capable to restrict hiPSC-NPC spheroids spontaneous neural differentiation and better maintain the progenitor properties of hiPSC-NPC.
Down syndrome (DS) is the most common genetic cause of intellectual disability, and development of the DS brain is associated with decreased neuronal and abnormal differentiation.80,81 However, the cause of the neurodegenerative process in DS is unknown. In our present study, the DS-NPC-based spheroids were also encapsulated into Me-HA hydrogels with different stiffness. The DS-NPCs remained good cell viability after encapsulation. Although the expected neurite outgrowth was not obtained in both soft and stiff hydrogel group, our immunofluorescent staining assay and together with qPCR results showed that DS-NPCs laden into softer hydrogels presented robustly highest neuron-specific protein and gene expression in NDM, demonstrating the softer stiffness could promote the transformation of DS-NPCs into neuronal phenotype. This also indicates that only mechanical cue is probably not enough to promote functional neural differentiation of DS-NPCs. Other physical and biochemical cues, hydrogel recipes, or their combination may have more effects on DS-NPCs fate in 3D culture. To the best our knowledge, this is the first demonstration of using 3D hydrogel culture micro-environment to support high cell viability and enhanced neural differentiation of DS-NPCs in vitro.
Conclusions
In this study, we demonstrated the feasibility and benefit of using Me-HA based hydrogel matrix as 3D models for in vitro hiPSC-NPC culture and differentiation. Human iPSC-NPCs or DS-NPCs with the states of single-cell suspension, or multi-cell spheroids were encapsulated into Me-HA hydrogels with different matrix stiffness. We reported the ability of our stiffness-controlled Me-HA hydrogels to support cell viability. Lower hydrogel stiffness promoted the differentiation of hiPSC-NPCs or DS-NPCs towards a mature neuronal phenotype. Specially, robust neurite outgrowth was observed when hiPSC-NPC spheroids were laden into softer hydrogel matrix. However, higher hydrogel stiffness was capable to better maintain the progenitor properties of hiPSC-NPCs. Together, our results presented remarkable evidence, demonstrating that our Me-HA hydrogel matrix not only showed potential as model systems to mechanistically investigate hiPSC-NPC or DS-NPC differentiation and eventually direct their fate into matured neurons in vitro, but also provided meaningful information on scaffold construction for neural tissue engineering.
Supplementary Material
Acknowledgments
This work has been supported by Mary & Dick Holland Regenerative Medicine Program start-up grant and Nebraska Research Initiative funding to B.D., and supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the NIH under grant number P30GM110768, and Edna Ittner Pediatric Research Support Fund to P.J. We would like to thank Janice A. Taylor and James R. Talaska of the Advanced Microscopy Core Facility at the University of Nebraska Medical Center (UNMC) for providing assistance with confocal microscopy. Support for the UNMC Advanced Microscopy Core Facility was provided by the Nebraska Research Initiative, the Fred and Pamela Buffett Cancer Center Support Grant (P30CA036727), and an Institutional Development Award (IDeA) from the NIGMS of the NIH (P30GM106397). The authors declare no competing financial interest.
Footnotes
Electronic Supplementary Information (ESI) available: [qPCR primers]. See DOI: 10.1039/x0xx00000x
References
- 1.Menzies FM, Fleming A, Rubinsztein DC. Nature Reviews Neuroscience. 2015;16:345–357. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- 2.Kabu S, Gao Y, Kwon BK, Labhasetwar V. Journal of Controlled Release. 2015;219:141–154. doi: 10.1016/j.jconrel.2015.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorman AM. Journal of cellular and molecular medicine. 2008;12:2263–2280. doi: 10.1111/j.1582-4934.2008.00402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haston KM, Finkbeiner S. Annual review of pharmacology and toxicology. 2016;56:489–510. doi: 10.1146/annurev-pharmtox-010715-103548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trounson A, McDonald C. Cell Stem Cell. 2015;17:11–22. doi: 10.1016/j.stem.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Srikanth P, Young-Pearse TL. Journal of neurogenetics. 2014;28:5–29. doi: 10.3109/01677063.2014.881358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gazdic M, Volarevic V, Arsenijevic A, Erceg S, Moreno-Manzano V, Arsenijevic N, Stojkovic M. International Journal of Molecular Sciences. 2016;18:6. doi: 10.3390/ijms18010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue H, Yamanaka S. Clinical Pharmacology & Therapeutics. 2011;89:655–661. doi: 10.1038/clpt.2011.38. [DOI] [PubMed] [Google Scholar]
- 10.Nakaji-Hirabayashi T, Kato K, Iwata H. Bioconjugate chemistry. 2013;24:1798–1804. doi: 10.1021/bc400005m. [DOI] [PubMed] [Google Scholar]
- 11.Cooke M, Vulic K, Shoichet M. Soft Matter. 2010;6:4988–4998. [Google Scholar]
- 12.Hess DC, Fakhri N, West FD. Cell Therapy for Brain Injury. Springer; 2015. pp. 129–146. [Google Scholar]
- 13.Führmann T, Tam R, Ballarin B, Coles B, Donaghue IE, van der Kooy D, Nagy A, Tator C, Morshead C, Shoichet M. Biomaterials. 2016;83:23–36. doi: 10.1016/j.biomaterials.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 14.Jiang Y, Chen J, Deng C, Suuronen EJ, Zhong Z. Biomaterials. 2014;35:4969–4985. doi: 10.1016/j.biomaterials.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Bidarra SJ, Barrias CC, Granja PL. Acta biomaterialia. 2014;10:1646–1662. doi: 10.1016/j.actbio.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Collins MN, Birkinshaw C. Carbohydrate polymers. 2013;92:1262–1279. doi: 10.1016/j.carbpol.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 17.Tahrir FG, Ganji F, Ahooyi TM. Recent patents on drug delivery & formulation. 2015;9:107–120. doi: 10.2174/1872211308666141028145651. [DOI] [PubMed] [Google Scholar]
- 18.Lee SW, Lee HJ, Hwang HS, Ko K, Han DW, Ko K. Animal Cells and Systems. 2015;19:175–180. [Google Scholar]
- 19.Ramos-Hryb AB, Da-Costa MC, Trentin AG, Calloni GW. International Journal of Developmental Biology. 2014;57:885–890. doi: 10.1387/ijdb.130206gw. [DOI] [PubMed] [Google Scholar]
- 20.Jang JM, Tran SHT, Na SC, Jeon NL. ACS applied materials & interfaces. 2015;7:2183–2188. doi: 10.1021/am508292t. [DOI] [PubMed] [Google Scholar]
- 21.Terraf P, Babaloo H, Kouhsari SM. Molecular neurobiology. 2017;54:1119. doi: 10.1007/s12035-016-9726-4. [DOI] [PubMed] [Google Scholar]
- 22.Nery FC, da Hora CC, Yaqub U, Zhang X, McCarthy DM, Bhide PG, Irimia D, Breakefield XO. Journal of neuroscience methods. 2015;239:80–84. doi: 10.1016/j.jneumeth.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun G, Liu W, Fan Z, Zhang D, Han Y, Xu L, Qi J, Zhang S, Gao BT, Bai X. Neural plasticity. 2016;2016 doi: 10.1155/2016/4280407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam J, Lowry WE, Carmichael ST, Segura T. Advanced functional materials. 2014;24:7053–7062. doi: 10.1002/adfm.201401483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nih LR, Moshayedi P, Llorente IL, Berg AR, Cinkornpumin J, Lowry WE, Segura T, Carmichael ST. Data in Brief. 2017;10:202–209. doi: 10.1016/j.dib.2016.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Francis NL, Bennett NK, Halikere A, Pang ZP, Moghe PV. ACS Biomaterials Science & Engineering. 2016;2:1030–1038. doi: 10.1021/acsbiomaterials.6b00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei Y, Jeong D, Xiao J, Schaffer DV. Cellular and molecular bioengineering. 2014;7:172–183. doi: 10.1007/s12195-014-0333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karumbaiah L, Bellamkonda R. Neural Engineering. Springer; 2013. pp. 765–794. [Google Scholar]
- 29.Moshayedi P, Nih LR, Llorente IL, Berg AR, Cinkornpumin J, Lowry WE, Segura T, Carmichael ST. Biomaterials. 2016;105:145–155. doi: 10.1016/j.biomaterials.2016.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu X, Ding F, Williams DF. Biomaterials. 2014;35:6143–6156. doi: 10.1016/j.biomaterials.2014.04.064. [DOI] [PubMed] [Google Scholar]
- 31.Baiguera S, Del Gaudio C, Lucatelli E, Kuevda E, Boieri M, Mazzanti B, Bianco A, Macchiarini P. Biomaterials. 2014;35:1205–1214. doi: 10.1016/j.biomaterials.2013.10.060. [DOI] [PubMed] [Google Scholar]
- 32.Wobma H, Vunjak-Novakovic G. Tissue Engineering Part B: Reviews. 2016;22:101–113. doi: 10.1089/ten.teb.2015.0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gefen A, Gefen N, Zhu Q, Raghupathi R, Margulies SS. Journal of neurotrauma. 2003;20:1163–1177. doi: 10.1089/089771503770802853. [DOI] [PubMed] [Google Scholar]
- 34.Margulies SS, Prange M. J Biomech Eng. 2002;124:244–252. doi: 10.1115/1.1449907. [DOI] [PubMed] [Google Scholar]
- 35.Foster AA, Marquardt LM, Heilshorn SC. Current Opinion in Chemical Engineering. 2017;15:15–23. doi: 10.1016/j.coche.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sur S, Newcomb CJ, Webber MJ, Stupp SI. Biomaterials. 2013;34:4749–4757. doi: 10.1016/j.biomaterials.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.An B, Tang-Schomer MD, Huang W, He J, Jones JA, Lewis RV, Kaplan DL. Biomaterials. 2015;48:137–146. doi: 10.1016/j.biomaterials.2015.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teixeira AI, Ilkhanizadeh S, Wigenius JA, Duckworth JK, Inganäs O, Hermanson O. Biomaterials. 2009;30:4567–4572. doi: 10.1016/j.biomaterials.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 39.Sun Y, Yong KMA, Villa-Diaz LG, Zhang X, Chen W, Philson R, Weng S, Xu H, Krebsbach PH, Fu J. Nature materials. 2014;13:599–604. doi: 10.1038/nmat3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lantoine J, Grevesse T, Villers A, Delhaye G, Mestdagh C, Versaevel M, Mohammed D, Bruyère C, Alaimo L, Lacour SP. Biomaterials. 2016;89:14–24. doi: 10.1016/j.biomaterials.2016.02.041. [DOI] [PubMed] [Google Scholar]
- 41.Georges PC, Miller WJ, Meaney DF, Sawyer ES, Janmey PA. Biophysical journal. 2006;90:3012–3018. doi: 10.1529/biophysj.105.073114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, Healy KE. Biophysical journal. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Georges XJPC, Li B, Du Y, Kutzing MK, Previtera ML, Langrana NA, Firestein BL. The Open Neuroscience Journal. 2007;1:7–14. [Google Scholar]
- 44.Chen C, Jiang P, Xue H, Peterson SE, Tran HT, McCann AE, Parast MM, Li S, Pleasure DE, Laurent LC. Nature communications. 2014;5:4430. doi: 10.1038/ncomms5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen C, Kim WY, Jiang P. JCI insight. 2016;1:e88632. doi: 10.1172/jci.insight.88632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duan B, Yin Z, Kang LH, Magin RL, Butcher JT. Acta biomaterialia. 2016;36:42–54. doi: 10.1016/j.actbio.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu S, Duan B, Liu P, Zhang C, Qin X, Butcher JT. ACS Applied Materials & Interfaces. 2016;8:16950–16960. doi: 10.1021/acsami.6b05199. [DOI] [PubMed] [Google Scholar]
- 48.Laterza C, Merlini A, De Feo D, Ruffini F, Menon R, Onorati M, Fredrickx E, Muzio L, Lombardo A, Comi G. Nature communications. 2013;4:2597. doi: 10.1038/ncomms3597. [DOI] [PubMed] [Google Scholar]
- 49.Tang X, Zhou L, Wagner AM, Marchetto MC, Muotri AR, Gage FH, Chen G. Stem cell research. 2013;11:743–757. doi: 10.1016/j.scr.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ader M, Tanaka EM. Current opinion in cell biology. 2014;31:23–28. doi: 10.1016/j.ceb.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 51.Esposito G, Imitola J, Lu J, De Filippis D, Scuderi C, Ganesh VS, Folkerth R, Hecht J, Shin S, Iuvone T. Human Molecular Genetics. 2008;17:440–457. doi: 10.1093/hmg/ddm322. [DOI] [PubMed] [Google Scholar]
- 52.Guidi S, Bonasoni P, Ceccarelli C, Santini D, Gualtieri F, Ciani E, Bartesaghi R. Brain Pathology. 2008;18:180–197. doi: 10.1111/j.1750-3639.2007.00113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu J, Esposito G, Scuderi C, Steardo L, Delli-Bovi LC, Hecht JL, Dickinson BC, Chang CJ, Mori T, Sheen V. PloS one. 2011;6:e22126. doi: 10.1371/journal.pone.0022126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker T, Huang J, Young K. Stem cells international. 2016;12:1–222. doi: 10.1155/2016/1890568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yousefifard M, Rahimi-Movaghar V, Nasirinezhad F, Baikpour M, Safari S, Saadat S, Jafari AM, Asady H, Tousi SR, Hosseini M. Neuroscience. 2016;322:377–397. doi: 10.1016/j.neuroscience.2016.02.034. [DOI] [PubMed] [Google Scholar]
- 56.Mertens J, Marchetto MC, Bardy C, Gage FH. Nature Reviews Neuroscience. 2016;17:424–437. doi: 10.1038/nrn.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ross CA, Akimov SS. Human molecular genetics. 2014;23:R17–R26. doi: 10.1093/hmg/ddu204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drawnel FM, Boccardo S, Prummer M, Delobel F, Graff A, Weber M, Gérard R, Badi L, Kam-Thong T, Bu L. Cell reports. 2014;9:810–820. doi: 10.1016/j.celrep.2014.09.055. [DOI] [PubMed] [Google Scholar]
- 59.Sterneckert JL, Reinhardt P, Schöler HR. Nature Reviews Genetics. 2014;15:625–639. doi: 10.1038/nrg3764. [DOI] [PubMed] [Google Scholar]
- 60.Yan Y, Martin LM, Bosco DB, Bundy JL, Nowakowski RS, Sang QXA, Li Y. Biomaterials. 2015;73:231–242. doi: 10.1016/j.biomaterials.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 61.Cairns DM, Chwalek K, Moore YE, Kelley MR, Abbott RD, Moss S, Kaplan DL. Stem Cell Reports. 2016;7:557–570. doi: 10.1016/j.stemcr.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seidlits SK, Khaing ZZ, Petersen RR, Nickels JD, Vanscoy JE, Shear JB, Schmidt CE. Biomaterials. 2010;31:3930–3940. doi: 10.1016/j.biomaterials.2010.01.125. [DOI] [PubMed] [Google Scholar]
- 63.Wang Y, Lapitsky Y, Kang CE, Shoichet MS. Journal of Controlled Release. 2009;140:218–223. doi: 10.1016/j.jconrel.2009.05.025. [DOI] [PubMed] [Google Scholar]
- 64.Mészár Z, Felszeghy S, Veress G, Matesz K, Székely G, Módis L. Brain Research Bulletin. 2008;75:414–418. doi: 10.1016/j.brainresbull.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 65.Kleinman HK, Martin GR. Seminars in Cancer Biology. Vol. 15. Elsevier; 2005. pp. 378–386. [DOI] [PubMed] [Google Scholar]
- 66.Bakshi A, Keck CA, Koshkin VS, LeBold DG, Siman R, Snyder EY, McIntosh TK. Brain Research. 2005;1065:8–19. doi: 10.1016/j.brainres.2005.09.059. [DOI] [PubMed] [Google Scholar]
- 67.Einstein O, Ben-Menachem-Tzidon O, Mizrachi-Kol R, Reinhartz E, Grigoriadis N, Ben-Hur T. Glia. 2006;53:449–455. doi: 10.1002/glia.20305. [DOI] [PubMed] [Google Scholar]
- 68.Iwasaki M, Wilcox JT, Nishimura Y, Zweckberger K, Suzuki H, Wang J, Liu Y, Karadimas SK, Fehlings MG. Biomaterials. 2014;35:2617–2629. doi: 10.1016/j.biomaterials.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 69.Lei Y, Schaffer DV. Proceedings of the National Academy of Sciences. 2013;110:E5039–E5048. doi: 10.1073/pnas.1309408110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nie Y, Bergendahl V, Hei DJ, Jones JM, Palecek SP. Biotechnology progress. 2009;25:20–31. doi: 10.1002/btpr.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kushchayev SV, Giers MB, Hom Eng D, Martirosyan NL, Eschbacher JM, Mortazavi MM, Theodore N, Panitch A, Preul MC. Journal of Neurosurgery: Spine. 2016;25:114–124. doi: 10.3171/2015.9.SPINE15628. [DOI] [PubMed] [Google Scholar]
- 72.Schizas N, Rojas R, Kootala S, Andersson B, Pettersson J, Hilborn J, Hailer NP. Journal of biomaterials applications. 2014;28:825–836. doi: 10.1177/0885328213483636. [DOI] [PubMed] [Google Scholar]
- 73.Lam J, Carmichael ST, Lowry WE, Segura T. Advanced healthcare materials. 2015;4:534–539. doi: 10.1002/adhm.201400410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chatterjee S, Fisher AB. Antioxidants & redox signaling. 2014;20:868–871. doi: 10.1089/ars.2013.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu W, An C, Chen WJ. BioMed research international. 2015;2015:486827. doi: 10.1155/2015/486827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fattahi P, Yang G, Kim G, Abidian MR. Advanced Materials. 2014;26:1846–1885. doi: 10.1002/adma.201304496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 78.Gefen A, Margulies SS. Journal of biomechanics. 2004;37:1339–1352. doi: 10.1016/j.jbiomech.2003.12.032. [DOI] [PubMed] [Google Scholar]
- 79.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PDP, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, Rehfeldt F. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dierssen M. Nature Reviews Neuroscience. 2012;13:844–858. doi: 10.1038/nrn3314. [DOI] [PubMed] [Google Scholar]
- 81.Costa AC, Scott-McKean JJ. CNS drugs. 2013;27:679–702. doi: 10.1007/s40263-013-0089-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


