Summary
High-risk neuroblastoma remains one of the deadliest childhood cancers. Identification of metabolic pathways that drive or maintain high-risk neuroblastoma may open new avenues of therapeutic interventions. Here we report the isolation and propagation of neuroblastoma sphere-forming cells with self-renewal and differentiation potential from tumors of TH-MYCN mice, an animal model of high-risk neuroblastoma with MYCN amplification. Transcriptional profiling reveals that mouse neuroblastoma sphere-forming cells acquire a metabolic program characterized by transcriptional activation of the cholesterol and serine-glycine synthesis pathways, primarily as a result of increased expression of sterol regulatory element-binding factors and Atf4, respectively. This metabolic reprogramming is recapitulated in high-risk human neuroblastomas and is prognostic for poor clinical outcome. Genetic and pharmacological inhibition of the metabolic program markedly decreases the growth and tumorigenicity of both mouse neuroblastoma sphere-forming cells and human neuroblastoma cell lines. These findings suggest a therapeutic strategy for targeting the metabolic program of high-risk neuroblastoma.
Introduction
Neuroblastoma is a common pediatric cancer of the sympathetic nervous system that arises in the adrenal medulla and paravertebral sympathetic ganglia (Brodeur, 2003; Cheung and Dyer, 2013; Maris et al., 2007). Neuroblastoma is classified into low-, intermediate-, and high-risk categories (Cohn et al., 2009). Patients with low- or intermediate-risk neuroblastoma have an overall survival rate of more than 90% following minimum or standard treatment. However, the overall survival rate for patients with high-risk neuroblastoma is less than 50% even after intensive, multimodal therapy in combination with bone marrow transplant (Park et al., 2013; Pinto et al., 2015). A better understanding of the molecular basis of high-risk neuroblastoma may suggest new therapeutic strategies.
The most common genetic alterations associated with high-risk neuroblastoma are MYCN amplification, 1p loss, 11q deletion, or 17q gain (Cohn et al., 2009). Neuroblastomas with amplification of the oncogene MYCN are generally classified as high-risk (Cohn et al., 2009), which are often associated with 1p loss and 17q gain (Bown, 2001; Caron, 1995; Cheng et al., 1995; Komuro et al., 1998). Approximately half of high-risk neuroblastomas carry no MYCN amplification, but are frequently harbor 11q deletion with or without 17q gain (Attiyeh et al., 2005; Caren et al., 2010; Guo et al., 1999; Luttikhuis et al., 2001). In general, high-risk neuroblastomas show an unfavorable histology, comprising predominantly Schwannian stroma-poor, undifferentiated or poorly differentiated tumors (Cohn et al., 2009; Shimada et al., 1999).
TH-MYCN transgenic mice have been widely used as an animal model for high-risk neuroblastomas with MYCN amplification (Dyer, 2004). These mice express human MYCN in migrating neural crest cells under control of the rat tyrosine hydroxylase (TH) gene promoter (Weiss et al., 1997), and develop tumors that, in most cases, are histologically undifferentiated or poorly differentiated (Moore et al., 2008). Gene expression profiling has revealed that TH-MYCN tumors are molecularly similar to International Neuroblastoma Staging System (INSS) stage 3-4 human neuroblastomas with MYCN amplification (Teitz et al., 2011). Neuroblastoma development in TH-MYCN mice begins with multifocal hyperplasia in early postnatal sympathetic ganglia characterized as clusters of small round blue cells in hematoxylin and eosin (H&E) staining. These hyperplastic lesions either regress spontaneously or develop into neuroblastomas (Hansford et al., 2004). Examination of stage- and lineage-specific markers has revealed that the hyperplasia is composed predominantly of highly proliferative Phox2B+ neuronal progenitors with undetectable expression of differentiation markers, whereas neuroblastoma tumors contain several distinct cell subpopulations, including Phox2B+TH-, Phox2B+TH+, and Phox2B-TH+ cells (Alam et al., 2009). Phox2B (paired like homeobox 2b) is a transcription factor that is expressed in sympathetic progenitors and is essential for embryonic development of the sympathetic nervous system (Pattyn et al., 1999), and TH is the first and rate-limiting enzyme in catecholamine biosynthesis that is highly expressed in differentiated sympathetic neurons (Goridis and Rohrer, 2002). Thus, TH-MYCN tumors are heterogeneous at the cellular level, consisting of tumor cells with varying degrees of differentiation.
It has been reported that histologically poorly differentiated tumors, regardless of their tissue origins, display a molecular similarity to embryonic stem (ES) cells, as evidenced by the coordinated up-or downregulation of gene sets associated with ES cell identity (Ben-Porath et al., 2008). This led us to hypothesize that gene expression profiling of undifferentiated TH-MYCN tumor cells with stem cell properties might help uncover the molecular mechanisms underlying high-risk neuroblastoma. Our investigation revealed that TH-MYCN neuroblastoma sphere-forming cells and high-risk human neuroblastomas share a common metabolic program for growth and tumorigenicity.
Results
TH-MYCN Neuroblastoma Sphere-Forming Cells Possess Self-Renewal Capacity
Sphere-forming assays have been widely used to isolate, propagate, and characterize normal and cancer stem cells (Pastrana et al., 2011). In this assay, stem cells grow as spheres in serum-free medium containing basic fibroblast growth factor and epidermal growth factor, with or without various tissue extracts and supplements (Reynolds and Weiss, 1992; Singh et al., 2003). In spite of extensive efforts, we were unable to obtain long-term (> 2 months) sphere culture from primary TH-MYCN tumors (n > 20) using various serum-free culture systems or ATCC mouse embryonic stem cell medium containing 10-20% fetal bovine serum (FBS). We next tried a culture medium for primary neural crest cells that contains 12% FBS plus other supplements (Etchevers, 2011), based on the reasoning that neuroblastoma arises in tissues originating from the neural crest (Le Douarin and Dupin, 1993) and that sympathetic neural crest cells or progenitors are likely to be the cell of origin for neuroblastoma (Cheung and Dyer, 2013; Marshall et al., 2014; Schulte et al., 2013). With this culture system, we were able to grow and propagate spheres from all 28 primary TH-MYCN tumors examined so far (Figure 1A). In all cases, only a minor population of primary tumor cells, presumably those with neural crest cell properties, were able to survive and proliferate in culture. In general, primary spheres appeared after 2 weeks of culture and then underwent rapid proliferation (Figure 1B). The sphere culture could be passaged continuously for at least 6 months. Immunofluorescence staining showed that a majority of sphere cells displayed nuclear expression of MYCN and Phox2B (Figures S1A and S1B, left panels), which are specific markers for progenitor cells or neuroblasts in TH-MYCN neuroblastoma tumors (Alam et al., 2009). In addition, we observed high levels of TH expression in a minor population of sphere cells (Figures S1A and S1B, left panels). Together, these data provide evidence for neuroblastoma origin of these sphere cells.
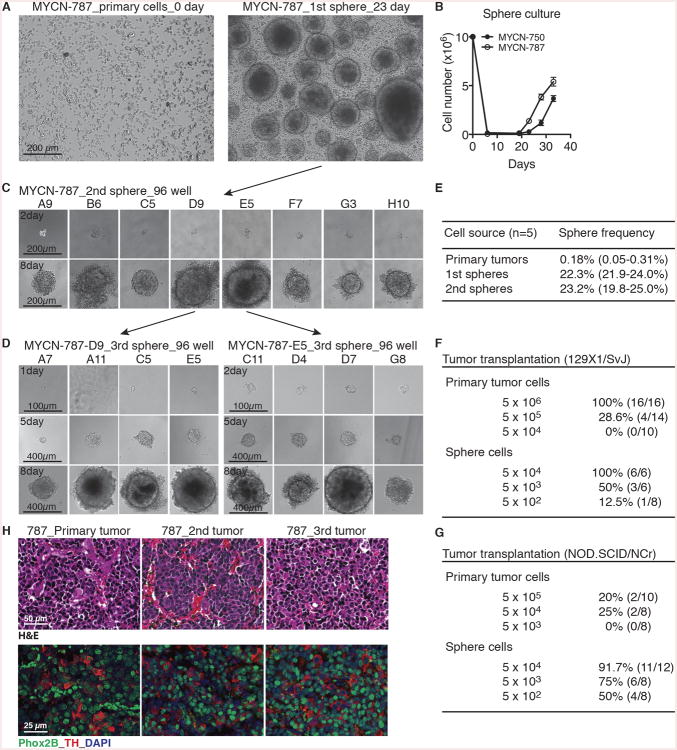
Figure 1. TH-MYCN neuroblastoma sphere-forming cells possess self-renewal and tumorigenic potential.
(A) Phase-contrast images of MYCN-787 primary tumor cells at the day of plating (0 day, left panel) and primary (1st) spheres after culture for 23 days (right panel).
(B) Growth assays of MYCN-750 and -787 primary tumor cells. Spheres appeared after ∼2 weeks of culture. Viable cells were quantified by trypan blue staining. Error bars represent SD (n = 4).
(C-D) Phase-contrast images of serial sphere-forming assays of single cell-derived MYCN-787 secondary (C, 2nd) and tertiary (D, 3rd) spheres.
(E) Quantification of sphere-forming cell frequency in primary tumors, 1st and 2nd spheres.
(F-G) Tumor transplantation assays for primary tumor cells and primary sphere cells in syngeneic 129×1/SvJ (F) a nd NOD.SCID/NCr (G) mice.
(H) Serial tumor transplantation assays with H&E (top panel) and immunofluorescence (bottom panel) staining of MYCN-787 primary tumor, a representative 2nd tumor generated by MYCN-787 primary spheres, and a representative 3rd tumor by spheres from the 2nd tumor. Nuclei were stained with DAPI.
See also Figures S1.
To determine whether the sphere cells possess self-renewal potential, we performed serial sphere-forming assays with single sphere cells. In principle, a sphere is dissociated into single cells and replated at clonal density or a single cell per well. Formation of new spheres that maintain the ability to differentiate demonstrates self-renewal (Pastrana et al., 2011; Reynolds and Weiss, 1992; Singec et al., 2006). Primary spheres (Figure 1A, 1st sphere) were mechanically dissociated into single cells, which were then plated at the frequency of 1 cell per well in 96-well plates. Those wells with a single cell were confirmed by microscopy and marked. The resultant secondary spheres (Figure 1C, 2nd sphere) were again dissociated into single cells and replated at 1 cell per well for the generation of tertiary spheres (Figure 1D, 3rd sphere). Similar to primary spheres, single-cell derived secondary and tertiary spheres were composed predominantly of MYCN- and Phox2B-positive cells with or without TH expression (Figures S1A and S1B, middle and right panels). These findings demonstrate, at the single-cell level, that a subpopulation of mouse neuroblastoma sphere cells possesses self-renewal and differentiation potential as evidenced by their capacity to generate new spheres composed of distinct subpopulations of progeny cells found in primary spheres. With these assays, we estimated that the frequency of sphere-forming cells within primary mouse tumors was 0.18% (n = 5), which was increased to 22.3% and 23.2% for primary and secondary spheres, respectively (Figure 1E).
We next performed serial tumorigenic transplantation assays to demonstrate the ability of mouse neuroblastoma spheres to self-renew in vivo (Clarke et al., 2006; Lathia et al., 2015). Compared to their parental tumor cells, clonal primary sphere cells showed a marked increase (>100-fold) in the capacity to generate new tumors in syngeneic wild-type 129×1/SvJ mice and in immunodeficient NOD.SCID/NCr mice (Figures 1F and 1G). The secondary tumors derived from the sphere cells displayed histopathological features and molecular marker expression identical to their parental primary tumors (Figures 1H and S1C-D, primary vs. 2nd tumor). Likewise, spheres derived from the secondary tumors were able to generate tertiary tumors that phenocopied the primary and secondary tumors (Figures 1H and S1C-D, 3rd tumor). Collectively, these data demonstrate that mouse neuroblastoma spheres contain a subpopulation of sphere-forming cells that are able to self-renew and generate new tumors with the phenotypic heterogeneity found in the parental tumors.
TH-MYCN Neuroblastoma Sphere-Forming Cells Undergo Spontaneous and Retinoic Acid-Induced Differentiation
As shown above (Figure S1B), single sphere cells were able to generate new spheres composed of distinct subpopulations of cells expressing different levels of the progenitor marker Phox2B with or without expression of the differentiation marker TH. This observation indicates that TH-MYCN neuroblastoma sphere-forming cells underwent spontaneous differentiation in culture. We further investigated the responsiveness of these sphere-forming cells to retinoic acid (RA), a differentiation inducer of neuroblastoma cells (Sidell, 1982) and neural crest cells (Dupin and Le Douarin, 1995; Huang et al., 2016). Treatment of mouse neuroblastoma spheres with RA induced growth arrest as evidenced by the marked reduction in sphere size and numbers, as well as the total number of sphere cells (Figures 2A-2C). Consistent with the growth arrest phenotype in culture, RA treatment markedly inhibited the tumorigenicity of sphere cells (Figures 2D and 2E).
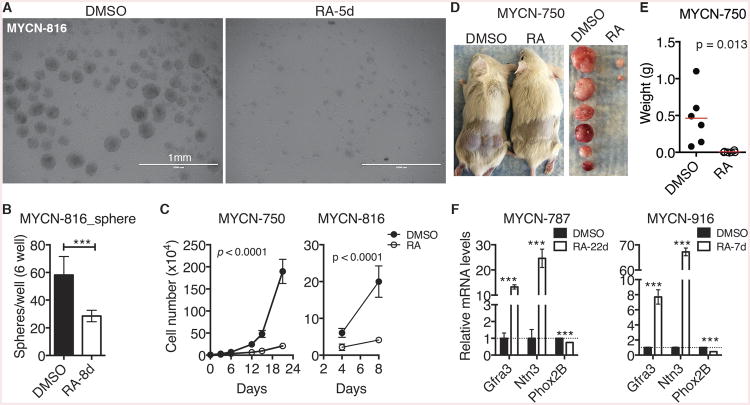
Figure 2. Retinoic acid induces growth arrest and differentiation of neuroblastoma sphere-forming cells.
(A-B) Phase-contrast images (A) and quantification (B) of spheres treated with DMSO or 10 μM RA. Error bars (B) represent SD (n = 3).
(C) Growth assays of spheres treated with DMSO or 10 μM RA. Viable cells were quantified by trypan blue staining. Error bars represent SD (n = 4). Data were analyzed by two-way ANOVA with p values indicated.
(D-E) Tumor transplantation assay of spheres treated with DMSO or 10 μM RA for 7 days. Tumor weight (E) was analyzed by scatter plot with horizontal lines indicating the mean.
(F) qRT-PCR analysis of mRNA expression in spheres treated with DMSO or 10 μM RA. Error bars represent SD (n = 3).
Unless indicated, data were analyzed with two-tailed Student's t-test. ***p < 0.001. See also Figure S2.
Most of the spheres that were treated with RA for an extended period of time became attached to the culture dish and displayed morphologic features of neuronal differentiation, such as extensive neurite outgrowth (Figure S2). Quantitative reverse-transcription PCR (qRT-PCR) revealed significant upregulation of neuronal differentiation genes, including Gfra3 and Ntn3 (Figure 2F), which are known to be upregulated in neuroblastoma cells undergoing neuronal differentiation (Mao et al., 2011; Wang et al., 2013; Wang et al., 2014). As expected, we also observed a significant decrease in Phox2B expression in spheres treated with RA (Figure 2F). It is known that Phox2B expression is downregulated during sympathetic neuron differentiation (Alam et al., 2009) and RA-induced neuroblastoma differentiation (Mao et al., 2011). Together, these findings demonstrate that mouse neuroblastoma sphere-forming cells possess the differentiation capacity of their cell of origin.
Cholesterol and Serine-Glycine Synthesis Pathways Are Transcriptionally Activated in Neuroblastoma Sphere-Forming Cells
To gain a molecular understanding of TH-MYCN neuroblastoma sphere-forming cells, we performed microarray gene expression profiling of three independent lines of sphere-forming cells in comparison with their parental primary tumors. A total of 2,376 genes showed significant changes in expression (≥ ±1.5 fold, P < 0.05), with 1,261 genes being downregulated and 1,115 genes upregulated in neuroblastoma spheres (Table S1). The downregulated genes are highly enriched for Gene Ontology (GO) terms associated with neuronal differentiation, including axonogenesis and neuron morphogenesis (Figure S3A and Table S2). We obtained similar results with PANTHER (Protein ANalysis THrough Evolutionary Relationships) analysis of the downregulated genes, which are highly enriched for PANTHER terms associated with neurogenesis and synaptic transmission (Figure S3B and Table S2). These findings provide further evidence, at the molecular level, that neuroblastoma sphere-forming cells are less differentiated than their parental primary tumor cells.
For genes upregulated in neuroblastoma sphere-forming cells, GO analysis revealed significant enrichment for GO terms associated with the biosynthesis of the serine family of amino acids and cholesterol (Figure 3A and Table S2). Expression levels of all genes within the two biosynthetic pathways were increased in neuroblastoma sphere-forming cells (Figures 3B and 3C). Gene set enrichment analysis (GSEA) of the microarray data also showed significantly higher expression of genes coding for enzymes involved in cholesterol biosynthesis (Figure 3D). We confirmed the microarray data by qRT-PCR, which showed significant downregulation of the neuronal genes Gfra3 and Ntn3, and upregulation of serine-glycine and cholesterol pathway genes, including Hmgcs1, Hmgcr, Mvk, Phgdh, Psat1, Psph, and Shmt2 (Figure 3E). Immunofluorescence staining of tumors generated by sphere-forming cells further confirmed the upregulation of serine-glycine pathway genes Phgdh and Psat1 in less differentiated TH- neuroblastoma cells in comparison with differentiated TH+ cells (Figure S3C).
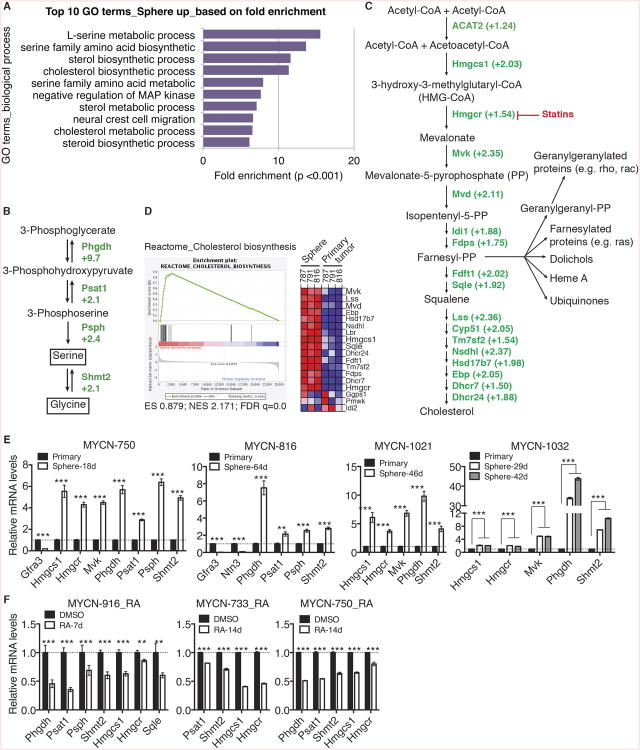
Figure 3. Transcriptional activation of the cholesterol and serine-glycine pathways in neuroblastoma sphere-forming cells.
(A) GO analysis of genes upregulated in neuroblastoma sphere-forming cells in comparison with their parental primary tumor cells. Shown are top 10 GO biological process terms based on fold enrichment.
(B-C) Fold changes in mRNA expression of genes in the serine-glycine (B) and cholesterol (C) pathways determined by microarray analysis.
(D) GSEA showing significant enrichment of the gene set involved in cholesterol synthesis, with most of the genes being upregulated in neuroblastoma sphere-forming cells.
(E-F) qRT-PCR analysis of mRNA expression of the indicated genes in neuroblastoma sphere-forming cells in comparison with their parental primary tumors (E) or treated with DMSO or 10 μM RA (F). Error bars represent SD (n = 3).
**p < 0.01, ***p < 0.001. See also Figure S3 and Tables S1 and S2.
In addition, our microarray data revealed transcriptional upregulation of genes involved in neural crest cell migration and anti-apoptosis including Ret and Bcl2 (Tables S1 and S2), which are known to be important in the pathogenesis of neuroblastoma (Cazes et al., 2014; Ham et al., 2016). The data provides further evidence for neural crest origin of TH-MYCN neuroblastoma sphere-forming cells and their relevance to neuroblastoma development.
We have shown previously that differentiation of human neuroblastoma cells is associated with transcriptional downregulation of metabolic pathways for cholesterol and serine-glycine synthesis (Wang et al., 2014) (see also Figure S3D). In agreement with the previous observation, qRT-PCR analysis of neuroblastoma sphere-forming cells treated with RA showed a marked reduction in mRNA expression of Hmgcs1, Hmgcr, Mvk, Phgdh, Psat1, Psph, and Shmt2 (Figure 3F).
Collectively, these data reveal a metabolic program in neuroblastoma sphere-forming cells that is characterized by increased transcriptional activation of serine-glycine and cholesterol synthesis pathway genes, in comparison with their parental primary tumors. This metabolic program is associated with the undifferentiated state of neuroblastoma sphere-forming cells.
Cholesterol and Serine-Glycine Synthesis Pathways Are Essential for Maintaining Neuroblastoma Sphere-Forming Cells
We next examined the biological significance of the metabolic program in neuroblastoma sphere-forming cells by blocking the serine-glycine and cholesterol synthesis pathways. We treated neuroblastoma sphere-forming cells with statins, a class of cholesterol-lowering drugs that inhibit Hmgcr, the rate-limiting enzyme in cholesterol synthesis (Figure 3C). Both Simvastatin and Fluvastatin markedly inhibited the ability of neuroblastoma sphere-forming cells to grow in culture (Figures 4A and S4A). Addition of mevalonate, the metabolite immediately downstream of Hmgcr (Figure 3C), fully rescued the anti-growth effect of Simvastatin (Figure 4B), demonstrating that the statins target Hmgcr to inhibit the growth of neuroblastoma sphere-forming cells. Moreover, Simvastatin treatment for 24 hours completely abolished the ability of neuroblastoma sphere-forming cells to generate tumors in immunodeficient mice (Figure 4C). Thus, the cholesterol synthesis pathway is essential for the growth and tumorigenicity of neuroblastoma sphere-forming cells.
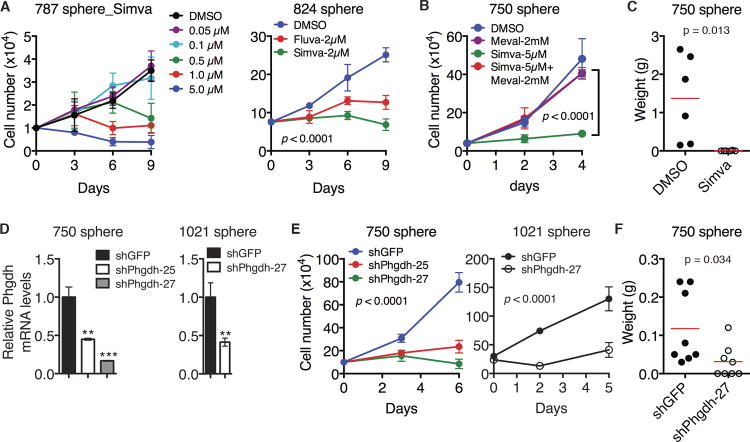
Figure 4. Cholesterol and serine-glycine pathways are essential for maintaining neuroblastoma sphere-forming cells.
(A) Growth assays of neuroblastoma sphere-forming cells treated with DMSO, increasing concentrations of Simvastatin (Simva) or 2 μM Fluvastatin (Fluva). Error bars represent SD (n = 4). Data were analyzed by two-way ANOVA with p values indicated.
(B) Growth assays of neuroblastoma sphere-forming cells treated with DMSO and Simva with or without mevalonate (Meval). Error bars represent SD (n = 4). Data were analyzed by two-way ANOVA with the p value indicated.
(C) Tumor transplantation assay of neuroblastoma sphere-forming cells treated with DMSO or 5 μM Simva for 24 hours. Tumor weight was analyzed by scatter plot with horizontal lines indicating the mean.
(D) qRT-PCR analysis of Phgdh mRNA levels in neuroblastoma sphere-forming cells infected with lentiviruses expressing shRNA against GFP or Phgdh. Error bars represent SD (n = 3).
(E) Growth assays of neuroblastoma sphere-forming cells without or with Phgdh knockdown. Error bars represent SD (n = 4). Data were analyzed by two-way ANOVA with p values indicated.
(F) Tumor transplantation assay of neuroblastoma sphere-forming cells without or with Phgdh knockdown. Tumor weight was analyzed by scatter plot with horizontal lines indicating the mean.
**p < 0.01, ***p < 0.001. See also Figure S4.
We also found an essential role of the serine-glycine synthesis pathway in maintaining neuroblastoma sphere-forming cells. Small hairpin RNA (shRNA)-mediated knockdown of Phgdh expression (Figures 4D and S4B), the first enzyme in the pathway (Figure 3B), exerted a pronounced inhibitory effect on the growth (Figures 4E and S4C) and tumorigenicity (Figure 4F) of sphere-forming cells.
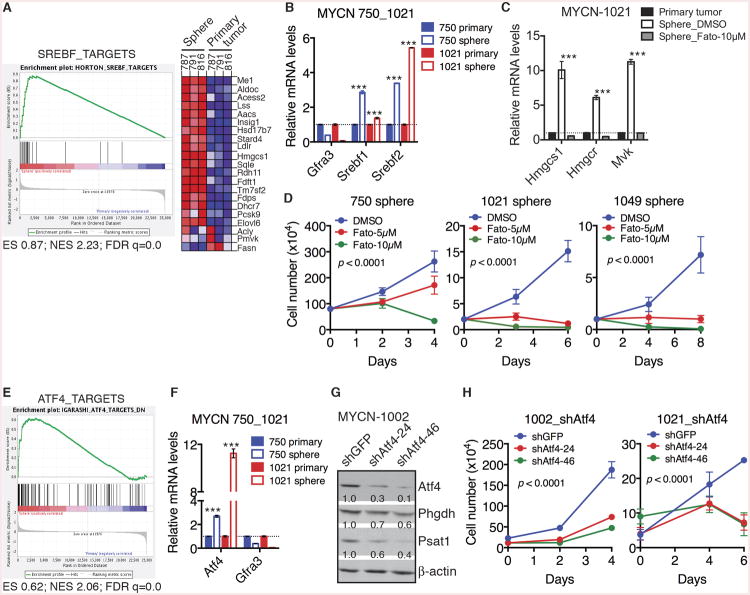
Srebf and Atf4 Activate the Metabolic Program in Neuroblastoma Sphere-Forming Cells
To gain a molecular understanding of the metabolic reprogramming in mouse neuroblastoma sphere-forming cells, we examined the microarray data by GSEA for identifying potential upstream transcription factors. The analysis revealed significant upregulation of the gene set consisting of direct targets of sterol regulatory element binding transcription factors (Srebf) in sphere-forming cells relative to their parental primary tumor cells (Figure 5A). Consistent with this finding, both microarray and qRT-PCR analyses showed increased expression of Srebf1 and Srebf2 in sphere-forming cells (Table S1 and Figure 5B). It is well established that Srebf2 has a major role in transcriptional activation of the cholesterol pathway (Brown and Goldstein, 1997; Horton et al., 2002; Horton et al., 2003). To determine whether the increased Srebf expression is responsible for transcriptional activation of the cholesterol pathway in sphere-forming cells, we treated these cells with Fatostatin, a small molecule inhibitor of Srebf activation (Kamisuki et al., 2009). Fatostatin treatment completely ablated the upregulation of key cholesterol pathway genes in sphere-forming cells (Figures 5C and S5A) and markedly inhibited their growth in culture (Figures 5D and S5B). We obtained essentially the same results with Betulin (Figures S5C and S5D), also a small-molecule inhibitor of Srebf activation (Tang et al., 2011). These data indicate that increased Srebf expression is essential for transcriptional activation of the cholesterol pathway in neuroblastoma sphere-forming cells.
Figure 5. Srebf and Atf4 mediate the metabolic reprogramming in neuroblastoma sphere-forming cells.
(A) GSEA showing significant enrichment of Srebf targets, with most genes being upregulated in neuroblastoma sphere-forming cells relative to parental primary tumor cells.
(B) qRT-PCR analysis of mRNA levels of Gfra3, Srebf1 and Srebf2 in neuroblastoma sphere-forming cells in comparison with their parental primary tumor cells. Error bars represent SD (n = 3).
(C) qRT-PCR analysis of mRNA levels of cholesterol pathway genes in parental primary tumor cells and neuroblastoma sphere-forming cells treated with DMSO or 10 μM Fatostatin (Fato) for 48 hours. Error bars represent SD (n = 3).
(D) Growth assays of neuroblastoma sphere-forming cells treated with DMSO or Fatostatin. Error bars represent SD (n = 4). Data were analyzed by two-way ANOVA with p values indicated.
(E) GSEA showing significant enrichment of Atf4 targets, with most genes being upregulated in neuroblastoma sphere-forming cells relative to parental primary tumor cells.
(F) qRT-PCR analysis of mRNA levels of Gfra3 and Atf4 in neuroblastoma sphere-forming cells relative to their parental primary tumor cells. Error bars represent SD (n = 3).
(G) Immunoblot analysis of Atf4, Phgdh and Psat1 levels relative to β-actin in neuroblastoma sphere-forming cells without or with Atf4 knockdown.
(H) Growth assays of neuroblastoma sphere-forming cells without or with Atf4 knockdown. Error bars represent SD (n = 4). Data were analyzed by two-way ANOVA with p values indicated.
**p < 0.01, ***p < 0.001. See also Figure S5 and Table S1.
GSEA also revealed significant upregulation of Activating Transcription Factor 4 (Atf4) and its target genes in neuroblastoma sphere-forming cells (Figures 5E and S5E). Atf4 can directly activate the transcription of serine-glycine pathway genes (Adams, 2007; Ding et al., 2013; Seo et al., 2009; Ye et al., 2012; Zhao et al., 2016). We confirmed the upregulation of Atf4 in sphere-forming cells by qRT-PCR (Figure 5F). Knockdown of Atf4 expression significantly decreased the expression of Phgdh and Psat1 (Figure 5G), and markedly inhibited the survival and proliferation of sphere-forming cells (Figures 5H and S5F). These findings suggest that increased Atf4 expression has a major role in transcriptional activation of the serine-glycine pathway in neuroblastoma sphere-forming cells.
Metabolic Program of Neuroblastoma Sphere-Forming Cells Is Essential for the Growth and Tumorigenicity of Human Neuroblastoma cells
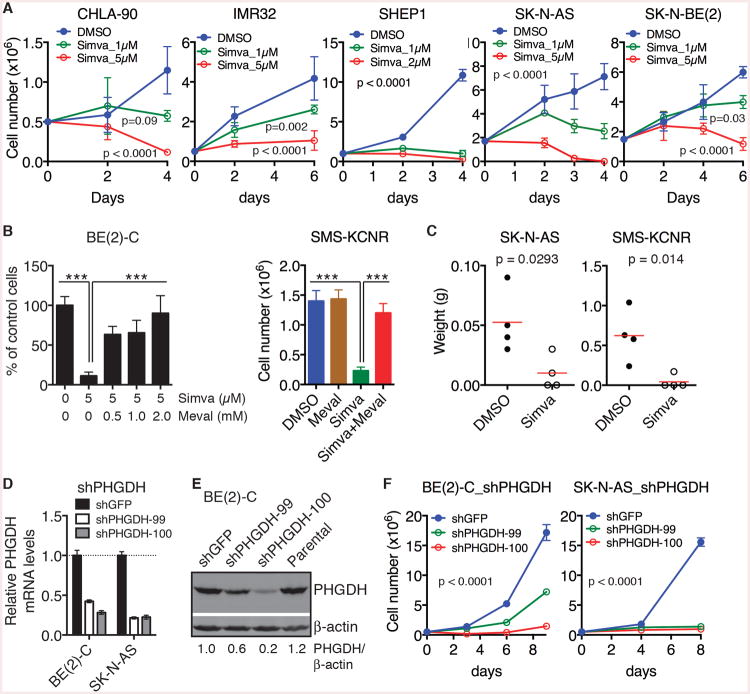
To test the relevance of above findings to human neuroblastoma, we first examined the functional significance of the metabolic program of mouse neuroblastoma sphere-forming cells in human neuroblastoma cell lines. Pharmacological inhibition of the cholesterol pathway by Simvastatin induced growth arrest or cell death in all human neuroblastoma cell lines examined (Figures 6A and S6A). Again, this inhibitory effect could be fully rescued by supplemental mevalonate (Figures 6B, S6B and S6C), demonstrating that Simvastatin targets the cholesterol pathway in human neuroblastoma cells. Treatment of SK-N-AS and SMS-KCNR cells with Simvastatin for 24 hours significantly attenuated their tumorigenicity in immunodeficient mice (Figure 6C). Of note, both MYCN-amplified (IMR32, SK-N-BE(2), and SMS-KCNR) and non-MYCN-amplified (CHLA-90, SHEP1, and SK-N-AS) neuroblastoma cell lines were sensitive to Simvastatin (Figures 6A and 6B).
Figure 6. Cholesterol and serine-glycine pathways are essential for the growth and tumorigenicity of human neuroblastoma cell lines.
(A) Growth assays of human neuroblastoma cell lines treated with DMSO or Simva. Error bars represent SD (n = 4). Data were analyzed by two-way ANOVA with p values indicated.
(B) Survival assays of human neuroblastoma cell lines treated with DMSO or 5 μM Simva in the absence or presence of mevalonate. Error bars represent SD (n = 4).
(C) Tumor transplantation assays of human neuroblastoma cell lines treated with 5 μM Simva for 24 hours. Tumor weight was analyzed by scatter plot with horizontal lines indicating the mean.
(D-E) qRT-PCR (D) and immunoblot (E) analyses of PHGDH expression in human neuroblastoma cell lines infected with lentiviruses expressing shGFP or shPHGDH. Error bars (D) represent SD (n = 3).
(F) Growth assay of human neuroblastoma cell lines without or with PHGDH knockdown. Error bars represent SD (n = 4). Data were analyzed by two-way ANOVA with p values indicated.
**p < 0.01, ***p < 0.001. See also Figure S6.
We also examined the effect of blocking the serine-glycine pathway. Knockdown of PHGDH expression by two independent shRNA constructs (Figures 6D and 6E) essentially arrested the growth of BE(2)-C and SK-N-AS cells in culture (Figure 6F). Collectively, these findings demonstrate that the metabolic program of mouse neuroblastoma sphere-forming cells is required for the growth and tumorigenicity of human neuroblastoma cell lines independent of their MYCN amplification status.
Metabolic Program of Neuroblastoma Sphere-Forming Cells Is Associated with High-Risk Neuroblastoma and Is a Predictor of Poor Clinical Outcome
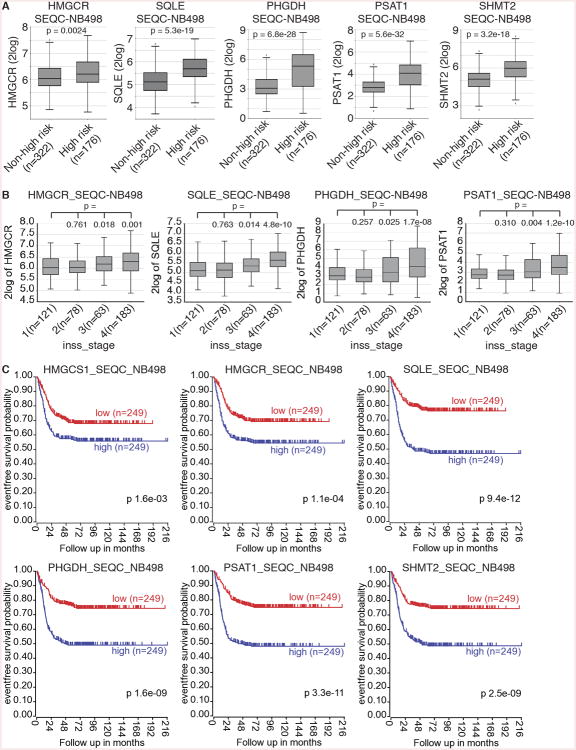
To further assess the clinical relevance of our findings with mouse neuroblastoma sphere-forming cells, we examined the correlation between the metabolic gene expression and clinical outcomes using gene expression data from two independent cohorts of neuroblastoma patients (n = 1,147). We focused our analysis on key genes in the cholesterol and serine-glycine pathways: HMGCS1 (catalyzing HMG-CoA synthesis), HMGCR (rate-limiting enzyme of the cholesterol pathway), SQLE (catalyzing the first cholesterol-specific step in the cholesterol pathway), PHGDH and PSAT1 (the first two enzymes in the serine-glycine pathway), and SHMT2 (linking glycine synthesis to one-carbon metabolism and nucleotide production). The RNA sequencing dataset from a clinical cohort of 498 neuroblastomas contains clinical data regarding to risk classification (high- or low-risk), tumor stages (INSS), and patient survival (Zhang et al., 2015). Analysis of the dataset revealed that high-risk neuroblastomas expressed these metabolic genes at significantly higher levels in comparison with low-risk neuroblastomas (Figure 7A), indicating that the metabolic program of mouse neuroblastoma sphere-forming cells is also activated in high-risk neuroblastomas. In line with this finding, we observed significantly higher expression of these metabolic genes in INSS stage 3-4 neuroblastomas (Figure 7B). Analysis of prognostic values of these metabolic genes revealed that higher expression was associated with a significant decrease in event-free survival (Figure 7C), as well as in overall survival (data not shown). We further validated differential expression of these metabolic genes in another cohort of 649 neuroblastomas (Kocak et al., 2013). Higher expression of these metabolic genes was significantly associated with advanced stages of neuroblastomas and lower event-free survival (Figures S7A and S7B).
Figure 7. The metabolic program of neuroblastoma sphere-forming cells is activated in high-risk neuroblastomas and is associated with poor clinical outcome.
(A-B) Box plots of individual metabolic gene expression in relation to neuroblastoma risk groups (A) and INSS stages (B) using the SEQC-NB498 dataset. Data were analyzed with two-tailed Student's t-test, with p values indicated.
(C) Event-free Kaplan-Meier survival curves for the SEQC cohort of 498 neuroblastoma patients based on differential expression of the metabolic genes. Log-rank test p values are shown.
See also Figure S7.
Given that the sphere-forming cells were isolated from tumors of TH-MYCN mice, a model of human high-risk neuroblastomas with MYCN amplification, we asked whether our findings are relevant to non-MYCN-amplified high-risk neuroblastomas. We further analyzed the NB498 dataset by excluding patients with MYCN-amplified neuroblastoma, which revealed significantly higher expression of HMGCR, SQLE, PSAT1, and SHMT2 in non-MYCN-amplified high-risk neuroblastomas in comparison with low-risk neuroblastomas (Figure S7C). In addition, higher expression of all 6 metabolic genes was significantly associated with worse prognosis for patients carrying non-MYCN-amplified neuroblastomas (Figure S7D).
Collectively, our analyses of human neuroblastoma gene expression data indicate that regardless of MYCN amplification status, high-risk and advanced neuroblastomas share the metabolic program of mouse neuroblastoma sphere-forming cells, characterized by transcriptional activation of the cholesterol and serine-glycine synthesis pathways. Our findings also suggest that the metabolic gene signature is of prognostic significance and predicts poor outcome.
Discussion
In this study, we report the functional and molecular characterization of TH-MYCN mouse neuroblastoma sphere-forming cells that possess self-renewal, differentiation, and tumorigenic potential. These cells acquire a metabolic program characterized by higher mRNA expression of genes coding for enzymes in the cholesterol and serine-glycine synthesis pathways, primarily as the result of increased expression of Srebf and Atf4. We present evidence that increased activation of these metabolic pathways is essential for maintaining neuroblastoma sphere-forming cells. Importantly, this metabolic program is also activated in high-risk human neuroblastoma and is required for the growth and tumorigenicity of human neuroblastoma cells. These findings suggest key metabolic pathways for high-risk neuroblastoma pathogenesis and maintenance, with therapeutic implications.
Neuroblastoma Sphere-Forming Cells
Our study provides evidence that within primary neuroblastoma tumors from TH-MYCN mice, there is a minor population of cells with the ability to grow as spheres in the culture medium for primary neural crest cells (Etchevers, 2011). As estimated by sphere-forming assays at clonal density, an average of 0.18% of primary tumor cells were sphere-forming cells. Single-cell based serial sphere-forming assays revealed that approximately 22-23% of sphere cells were able to generate new spheres. In addition to serial sphere-formation assays in vitro, we also performed serial tumor transplantation assays using clonal sphere cells, which demonstrated that neuroblastoma sphere-forming cells were able to generate new tumors in syngeneic and immunodeficient mice, exhibiting a marked increase (>100 fold) in tumorigenic potential in comparison with primary tumor cells. Thus, both in vitro and in vivo assays demonstrate the self-renewal capacity of mouse neuroblastoma sphere-forming cells.
For their differentiation potential, we show that the spheres were composed predominantly of MYCN+ and Phox2B+ cells, with a small number of sphere cells expressing high levels of the differentiation marker TH, with or without co-expression of MYCN and Phox2B. Given the single-cell origin of secondary and tertiary spheres, the finding of distinct cell subpopulations within the spheres indicates spontaneous differentiation of sphere-forming cells. Their differentiation potential was further demonstrated by their ability to undergo RA-induced neuronal differentiation. Finally, secondary and tertiary tumors generated by clonal sphere cells fully recapitulated the cellular heterogeneity found in the parental tumors, which is characterized by the presence of distinct subpopulations of Phox2B+TH-, Phox2B+TH+, and Phox2B-TH+ cells (Alam et al., 2009).
While our data indicate that only a minor population of primary tumor cells can give rise to spheres in culture, we would like to point out that there are alternative explanations for the low frequency, such as adaptation to culture conditions and damage to cells as a result of mechanic stress during tumor tissue dissociation. Nevertheless, our estimation suggests that the neural crest cell culture system allowed for a more than 100-fold enrichment of sphere-forming cells over primary tumors (∼23% in spheres vs. 0.18% in primary tumors). This is consistent with data from tumorigenic assays, showing that the spheres had an over 100-fold increase in the number of tumorigenic cells relative to primary tumors. Efforts are under way to refine the culture system for minimizing spontaneous differentiation. We are also analyzing the gene expression data for identifying surface markers that allow us to prospectively purify neuroblastoma sphere-forming cells, which may also lead to more accurate quantification of these cells in primary tumors.
Metabolic Program in Neuroblastoma Sphere-Forming Cells
It is well established that cancer cells reprogram their metabolism to meet the energetic and biosynthetic requirements of growth and proliferation. A major challenge is to identify the metabolic pathways that specifically support the survival and proliferation of cancer cells. Our isolation and transcriptional profiling of TH-MYCN neuroblastoma sphere-forming cells reveals a metabolic program activated in this population of tumorigenic cells, which is defined by increased expression of genes encoding enzymes within the cholesterol and serine-glycine synthesis pathways. Functional assays demonstrated that this metabolic program is essential for the maintenance of mouse neuroblastoma sphere-forming cells.
There are clearly advantages in transcriptional activation of these metabolic pathways for cell proliferation. The cholesterol pathway, also known as the mevalonate pathway, is crucial for the structure and function of cellular membranes and many membrane localized proteins (Goldstein and Brown, 1990; Silvente-Poirot and Poirot, 2012). Cholesterol is a structural component of the cell membrane. Other important metabolites generated from this pathway include ubiquinones, dolichols, and isoprenoids required for the prenylation and activation of Ras, Rho, and Rab families of proteins, which are known to promote cell proliferation (Berndt et al., 2011).
The serine-glycine pathway generates equimolar amounts of NADH, α-ketoglutarate (α-KG), serine, glycine, and 5,10-methylenetetrahydrofolate (5,10-MTHF). These metabolites are required for the production of proteins, nucleic acids, fatty acids, and membranes needed for cell proliferation (Amelio et al., 2014; DeBerardinis, 2011; Kalhan and Hanson, 2012; Locasale, 2013). Glycine donates carbon and nitrogen moieties for purine nucleotide synthesis. 5,10-MTHF is a coenzyme of thymidylate synthase, which is required for de novo thymidylate biosynthesis, and is a major source of one-carbon units for purine production. Increased activation of the serine-glycine pathway can also stimulate anaplerosis by supplying α-KG, which replenishes TCA cycle intermediates for biosynthesis (DeBerardinis et al., 2007). In addition, serine-driven one-carbon metabolism is a major pathway of NADPH production in proliferating cells (Fan et al., 2014). NADPH is required for the synthesis of nucleotides, amino acids, and lipids, and has a pivotal role in maintaining the cellular redox balance (Schulze and Harris, 2012).
Our findings further suggest a molecular mechanism for the increased activation of the metabolic program in neuroblastoma sphere-forming cells. Transcriptional profiling and qRT-PCR showed a significant increase in the expression of Srebf1, Srebf2, and Atf4 in neuroblastoma sphere-forming cells relative to their parental primary tumor cells. Srebf2 has a major role in transcriptional activation of the cholesterol pathway (Brown and Goldstein, 1997; Horton et al., 2002; Horton et al., 2003), and Atf4 is a key transcriptional activator of serine-glycine pathway genes (Adams, 2007; Ding et al., 2013; Seo et al., 2009; Ye et al., 2012; Zhao et al., 2016). In line with this model, pharmacological inhibition of Srebf activation and shRNA-mediated silencing of Atf4 were able to abrogate the transcriptional activation of the cholesterol and serine-glycine pathways in neuroblastoma sphere-forming cells, leading to a marked reduction in their growth and tumorigenicity. There is evidence suggesting that SREBFs and ATF4 may also play a role in activation of the metabolic program in human neuroblastoma. We have shown previously that ATF4 is required for the proliferation of human neuroblastoma cells (Ding et al., 2013; Zhao et al., 2016), and high expression of ATF4 and SREBF2 is significantly associated with poor prognosis in neuroblastoma patients (data not shown).
Targeting High-Risk Neuroblastoma Metabolism for Therapy
A key finding of our study is that the metabolic program identified in mouse neuroblastoma sphere-forming cells is also activated in high-risk neuroblastomas, regardless of MYCN amplification status. Our analysis of gene expression profiles of 1,147 neuroblastoma patients revealed that high-risk neuroblastomas have significantly higher expression of cholesterol and serine-glycine synthesis pathway genes in comparison with low-risk neuroblastomas. Higher expression of these metabolic genes is also significantly associated with advanced stages of neuroblastomas and is a predictor of poor clinical outcome. Moreover, this metabolic gene signature could potentially serve as a biomarker for the differentiation status of neuroblastomas, as evidenced by the observations that high-risk neuroblastomas are histologically undifferentiated or poorly differentiated and that expression of these metabolic genes are downregulated during differentiation. In further support of the clinical significance of the metabolic program, we found that pharmacological inhibition or genetic silencing of either metabolic pathway is sufficient to inhibit the growth and tumorigenicity of human neuroblastoma cell lines. These findings suggest that transcriptional activation of the cholesterol and serine-glycine synthesis pathways is crucial for the pathogenesis and progression of high-risk neuroblastoma.
Our findings have implications for the development of new therapeutic strategies that target this metabolic program in high-risk neuroblastoma. The cholesterol/mevalonate pathway has been implicated in cancer development (Freed-Pastor et al., 2012; Gruenbacher and Thurnher, 2015; Silvente-Poirot and Poirot, 2012), and statins have been shown to exhibit anti-tumor effects (Clendening and Penn, 2012; Thurnher et al., 2012). Our findings present a compelling case for statins as potential therapeutics for high-risk neuroblastoma. Similarly, blocking the serine-glycine pathway, either alone or in combination with other therapies, may represent a promising therapeutic strategy for high-risk neuroblastoma.
Experimental Procedures
Neuroblastoma Sphere Culture
Neuroblastoma tumors were removed from TH-MYCN mice, minced, and mechanically dissociated into single cells using a cell strainer (70 μm, Fisher Scientific 22363548). Red blood cells were lysed in ACK buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA, pH 7.3), and dead cells were removed by passing through Ficoll-Paque Plus (GE Healthcare 17-1440-02). Viable tumor cells were cultured in complete neural crest cell culture medium (Etchevers, 2011). The medium was replaced every 3 days. After 3 weeks of culture, primary spheres were mechanically dissociated into single cells by pipetting and were then expanded through weekly dissociation of spheres.
To assess sphere-forming cell frequency in primary tumors (n = 5), ∼50,000 cells from each primary tumor were plated at clonal density of 1 cell/μl (Coles-Takabe et al., 2008; Pastrana et al., 2011) (2,000 cells/well, 6-well plates). Primary (1st) spheres were counted after 15-21 days in culture. For serial sphere-forming assays, primary spheres were mechanically dissociated into single cells by pipetting, and ∼500 cells were plated at 1 cell/well into 96-well plates. Secondary (2nd) spheres were counted after 8 days in culture. Approximately 100 cells from each 2nd sphere (n = 3 for each primary tumor) were plated at 1 cell/well into 96-well plates for assessing their capacity to generate tertiary (3rd) spheres.
Tumor Transplantation
Freshly isolated primary tumor cells, sphere cells, or human neuroblastoma cell lines in 200 μl serum-free DMEM were injected subcutaneously into flanks of recipient 129×1/SvJ and NOD.SCID/NCr mice. For serial tumor transplantation assays, single primary spheres from the clonal culture described above were individually expanded for the generation of secondary tumors, which were then dissociated into single cells and plated at clonal density. Single secondary spheres were again individually expanded for the generation of tertiary tumors. The experiment was terminated when the first tumor(s) reached ∼0.5 cm in diameter or 4 months post-injection.
Patient Data
Patient data used in this study were described previously (Kocak et al., 2013; Zhang et al., 2015). All analyses were conducted online using R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl), and the resulting figures and p value were downloaded.
Statistics
Quantitative data are presented as mean ± SD and were analyzed for statistical significance by unpaired, two-tailed Student's t-test or two-way ANOVA using GraphPad Prism 6.0h for Mac.
Supplementary Material
Acknowledgments
We thank Drs. LesleyAnn Hawthorn, Sam Chang, and Eiko Kitamura of the Georgia Cancer Center Genomics Core for assistance in microarray gene expression profiling. The work was supported by grants from NIH (R01 CA190429) and DoD (W81XWH-12-1-0613) to H.-F.D., and by grants from the National Natural Science Foundation of China (number 81201981 and 81550031) to Y.Z.
Footnotes
Accession Number: The NCBI Gene Expression Omnibus (GEO) accession number for the microarray data reported in this paper is GSE80252.
Supplemental Information: Supplemental Information includes 7 figures, 3 tables, and Supplemental Experimental Procedures.
Author Contributions: M.L., Y.X., and J.D. independently performed the experiments with assistance from B.Y., E.Z., and A.A. J-H.C. and H-F.D. analyzed the microarray data. C.Y., Z.D., S.H., L.Y., and H.C. provided reagents and helped design the study. M.L., Y.X., J.D., Y.Z. and H-F.D. designed the study, analyzed the data, and wrote the paper. Y.Z. and H-F.D. supervised and provided funding for the project.
References
- Adams CM. Role of the transcription factor ATF4 in the anabolic actions of insulin and the anti-anabolic actions of glucocorticoids. J Biol Chem. 2007;282:16744–16753. doi: 10.1074/jbc.M610510200. [DOI] [PubMed] [Google Scholar]
- Alam G, Cui H, Shi H, Yang L, Ding J, Mao L, Maltese WA, Ding HF. MYCN promotes the expansion of Phox2B-positive neuronal progenitors to drive neuroblastoma development. Am J Pathol. 2009;175:856–866. doi: 10.2353/ajpath.2009.090019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio I, Cutruzzola F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39:191–198. doi: 10.1016/j.tibs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attiyeh EF, London WB, Mosse YP, Wang Q, Winter C, Khazi D, McGrady PW, Seeger RC, Look AT, Shimada H, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown N. Neuroblastoma tumor genetics: clinical and biological aspects. J Clin Pathol. 2001;54:897–910. doi: 10.1136/jcp.54.12.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Caren H, Kryh H, Nethander M, Sjoberg RM, Trager C, Nilsson S, Abrahamsson J, Kogner P, Martinsson T. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc Natl Acad Sci U S A. 2010;107:4323–4328. doi: 10.1073/pnas.0910684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron H. Allelic loss of chromosome 1 and additional chromosome 17 material are both unfavourable prognostic markers in neuroblastoma. Med Pediatr Oncol. 1995;24:215–221. doi: 10.1002/mpo.2950240402. [DOI] [PubMed] [Google Scholar]
- Cazes A, Lopez-Delisle L, Tsarovina K, Pierre-Eugene C, De Preter K, Peuchmaur M, Nicolas A, Provost C, Louis-Brennetot C, Daveau R, et al. Activated Alk triggers prolonged neurogenesis and Ret upregulation providing a therapeutic target in ALK-mutated neuroblastoma. Oncotarget. 2014;5:2688–2702. doi: 10.18632/oncotarget.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NC, Van Roy N, Chan A, Beitsma M, Westerveld A, Speleman F, Versteeg R. Deletion mapping in neuroblastoma cell lines suggests two distinct tumor suppressor genes in the 1p35-36 region, only one of which is associated with N-myc amplification. Oncogene. 1995;10:291–297. [PubMed] [Google Scholar]
- Cheung NK, Dyer MA. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer. 2013;13:397–411. doi: 10.1038/nrc3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer Stem Cells--Perspectives on Current Status and Future Directions: AACR Workshop on Cancer Stem Cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- Clendening JW, Penn LZ. Targeting tumor cell metabolism with statins. Oncogene. 2012;31:4967–4978. doi: 10.1038/onc.2012.6. [DOI] [PubMed] [Google Scholar]
- Cohn SL, Pearson ADJ, London WB, Monclair T, Ambros PF, Brodeur GM, Faldum A, Hero B, Iehara T, Machin D, et al. The International Neuroblastoma Risk Group (INRG) Classification System: An INRG Task Force Report. J Clin Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles-Takabe BL, Brain I, Purpura KA, Karpowicz P, Zandstra PW, Morshead CM, van der Kooy D. Don't look: growing clonal versus nonclonal neural stem cell colonies. Stem Cells. 2008;26:2938–2944. doi: 10.1634/stemcells.2008-0558. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ. Serine metabolism: some tumors take the road less traveled. Cell Metab. 2011;14:285–286. doi: 10.1016/j.cmet.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Li T, Wang X, Zhao E, Choi JH, Yang L, Zha Y, Dong Z, Huang S, Asara JM, et al. The Histone H3 Methyltransferase G9A Epigenetically Activates the Serine-Glycine Synthesis Pathway to Sustain Cancer Cell Survival and Proliferation. Cell Metab. 2013;18:896–907. doi: 10.1016/j.cmet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin E, Le Douarin NM. Retinoic acid promotes the differentiation of adrenergic cells and melanocytes in quail neural crest cultures. Dev Biol. 1995;168:529–548. doi: 10.1006/dbio.1995.1100. [DOI] [PubMed] [Google Scholar]
- Dyer MA. Mouse models of childhood cancer of the nervous system. J Clin Pathol. 2004;57:561–576. doi: 10.1136/jcp.2003.009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchevers H. Primary culture of chick, mouse or human neural crest cells. Nat Protoc. 2011;6:1568–1577. doi: 10.1038/nprot.2011.398. [DOI] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, Barsotti A, Chicas A, Li W, Polotskaia A, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Goridis C, Rohrer H. Specification of catecholaminergic and serotonergic neurons. Nat Rev Neurosci. 2002;3:531–541. doi: 10.1038/nrn871. [DOI] [PubMed] [Google Scholar]
- Gruenbacher G, Thurnher M. Mevalonate metabolism in cancer. Cancer Lett. 2015;356:192–196. doi: 10.1016/j.canlet.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Guo C, White PS, Weiss MJ, Hogarty MD, Thompson PM, Stram DO, Gerbing R, Matthay KK, Seeger RC, Brodeur GM, et al. Allelic deletion at 11q23 is common in MYCN single copy neuroblastomas. Oncogene. 1999;18:4948–4957. doi: 10.1038/sj.onc.1202887. [DOI] [PubMed] [Google Scholar]
- Ham J, Costa C, Sano R, Lochmann TL, Sennott EM, Patel NU, Dastur A, Gomez-Caraballo M, Krytska K, Hata AN, et al. Exploitation of the Apoptosis-Primed State of MYCN-Amplified Neuroblastoma to Develop a Potent and Specific Targeted Therapy Combination. Cancer Cell. 2016;29:159–172. doi: 10.1016/j.ccell.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansford LM, Thomas WD, Keating JM, Burkhart CA, Peaston AE, Norris MD, Haber M, Armati PJ, Weiss WA, Marshall GM. Mechanisms of embryonal tumor initiation: distinct roles for MycN expression and MYCN amplification. Proc Natl Acad Sci U S A. 2004;101:12664–12669. doi: 10.1073/pnas.0401083101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Miller ML, McHenry LK, Zheng T, Zhen Q, Ilkhanizadeh S, Conklin BR, Bronner ME, Weiss WA. Generating trunk neural crest from human pluripotent stem cells. Sci Rep. 2016;6:19727. doi: 10.1038/srep19727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhan SC, Hanson RW. Resurgence of serine: an often neglected but indispensable amino Acid. J Biol Chem. 2012;287:19786–19791. doi: 10.1074/jbc.R112.357194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisuki S, Mao Q, Abu-Elheiga L, Gu Z, Kugimiya A, Kwon Y, Shinohara T, Kawazoe Y, Sato S, Asakura K, et al. A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem Biol. 2009;16:882–892. doi: 10.1016/j.chembiol.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Kocak H, Ackermann S, Hero B, Kahlert Y, Oberthuer A, Juraeva D, Roels F, Theissen J, Westermann F, Deubzer H, et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis. 2013;4:e586. doi: 10.1038/cddis.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro H, Valentine MB, Rowe ST, Kidd VJ, Makino S, Brodeur GM, Cohn SL, Look AT. Fluorescence in situ hybridization analysis of chromosome 1p36 deletions in human MYCN amplified neuroblastoma. J Pediatr Surg. 1998;33:1695–1698. doi: 10.1016/s0022-3468(98)90612-1. [DOI] [PubMed] [Google Scholar]
- Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin NM, Dupin E. Cell lineage analysis in neural crest ontogeny. J Neurobiol. 1993;24:146–161. doi: 10.1002/neu.480240203. [DOI] [PubMed] [Google Scholar]
- Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttikhuis ME, Powell JE, Rees SA, Genus T, Chughtai S, Ramani P, Mann JR, McConville CM. Neuroblastomas with chromosome 11q loss and single copy MYCN comprise a biologically distinct group of tumours with adverse prognosis. Br J Cancer. 2001;85:531–537. doi: 10.1054/bjoc.2001.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Ding J, Zha Y, Yang L, McCarthy BA, King W, Cui H, Ding HF. HOXC9 Links Cell-Cycle Exit and Neuronal Differentiation and Is a Prognostic Marker in Neuroblastoma. Cancer Res. 2011;71:4314–4324. doi: 10.1158/0008-5472.CAN-11-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- Marshall GM, Carter DR, Cheung BB, Liu T, Mateos MK, Meyerowitz JG, Weiss WA. The prenatal origins of cancer. Nat Rev Cancer. 2014;14:277–289. doi: 10.1038/nrc3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore HC, Wood KM, Jackson MS, Lastowska MA, Hall D, Imrie H, Redfern CP, Lovat PE, Ponthan F, O'Toole K, et al. Histological profile of tumours from MYCN transgenic mice. J Clin Pathol. 2008;61:1098–1103. doi: 10.1136/jcp.2007.054627. [DOI] [PubMed] [Google Scholar]
- Park JR, Bagatell R, London WB, Maris JM, Cohn SL, Mattay KK, Hogarty M Committee, C.O.G.N. Children's Oncology Group's 2013 blueprint for research: neuroblastoma. Pediatr Blood Cancer. 2013;60:985–993. doi: 10.1002/pbc.24433. [DOI] [PubMed] [Google Scholar]
- Pastrana E, Silva-Vargas V, Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8:486–498. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399:366–370. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, Nakagawara A, Berthold F, Schleiermacher G, Park JR, et al. Advances in Risk Classification and Treatment Strategies for Neuroblastoma. J Clin Oncol. 2015;33:3008–3017. doi: 10.1200/JCO.2014.59.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Schulte JH, Lindner S, Bohrer A, Maurer J, De Preter K, Lefever S, Heukamp L, Schulte S, Molenaar J, Versteeg R, et al. MYCN and ALKF1174L are sufficient to drive neuroblastoma development from neural crest progenitor cells. Oncogene. 2013;32:1059–1065. doi: 10.1038/onc.2012.106. [DOI] [PubMed] [Google Scholar]
- Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- Seo J, Fortuno ES, 3rd, Suh JM, Stenesen D, Tang W, Parks EJ, Adams CM, Townes T, Graff JM. Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes. 2009;58:2565–2573. doi: 10.2337/db09-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, Ambros IM, Dehner LP, Hata J, Joshi VV, Roald B, Stram DO, Gerbing RB, Lukens JN, Matthay KK, et al. The International Neuroblastoma Pathology Classification (the Shimada system) Cancer. 1999;86:364–372. [PubMed] [Google Scholar]
- Sidell N. Retinoic acid-induced growth inhibition and morphologic differentiation of human neuroblastoma cells in vitro. J Natl Cancer Inst. 1982;68:589–596. [PubMed] [Google Scholar]
- Silvente-Poirot S, Poirot M. Cholesterol metabolism and cancer: the good, the bad and the ugly. Curr Opin Pharmacol. 2012;12:673–676. doi: 10.1016/j.coph.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Singec I, Knoth R, Meyer RP, Maciaczyk J, Volk B, Nikkhah G, Frotscher M, Snyder EY. Defining the actual sensitivity and specificity of the neurosphere assay in stem cell biology. Nat Methods. 2006;3:801–806. doi: 10.1038/nmeth926. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Tang JJ, Li JG, Qi W, Qiu WW, Li PS, Li BL, Song BL. Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metab. 2011;13:44–56. doi: 10.1016/j.cmet.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Teitz T, Stanke JJ, Federico S, Bradley CL, Brennan R, Zhang J, Johnson MD, Sedlacik J, Inoue M, Zhang ZM, et al. Preclinical models for neuroblastoma: establishing a baseline for treatment. PLoS One. 2011;6:e19133. doi: 10.1371/journal.pone.0019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurnher M, Nussbaumer O, Gruenbacher G. Novel aspects of mevalonate pathway inhibitors as antitumor agents. Clin Cancer Res. 2012;18:3524–3531. doi: 10.1158/1078-0432.CCR-12-0489. [DOI] [PubMed] [Google Scholar]
- Wang X, Choi JH, Ding J, Yang L, Ngoka LC, Lee EJ, Zha Y, Mao L, Jin B, Ren M, et al. HOXC9 directly regulates distinct sets of genes to coordinate diverse cellular processes during neuronal differentiation. BMC Genomics. 2013;14:830. doi: 10.1186/1471-2164-14-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yang L, Choi JH, Kitamura E, Chang CS, Ding J, Lee EJ, Cui H, Ding HF. Genome-wide analysis of HOXC9-induced neuronal differentiation of neuroblastoma cells. Genom Data. 2014;2:50–52. doi: 10.1016/j.gdata.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. Embo J. 1997;16:2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, Thompson CB. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci U S A. 2012;109:6904–6909. doi: 10.1073/pnas.1204176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Yu Y, Hertwig F, Thierry-Mieg J, Zhang W, Thierry-Mieg D, Wang J, Furlanello C, Devanarayan V, Cheng J, et al. Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol. 2015;16:133. doi: 10.1186/s13059-015-0694-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao E, Ding J, Xia Y, Liu M, Ye B, Choi JH, Yan C, Dong Z, Huang S, Zha Y, et al. KDM4C and ATF4 Cooperate in Transcriptional Control of Amino Acid Metabolism. Cell Rep. 2016;14:506–519. doi: 10.1016/j.celrep.2015.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.