Abstract
Most of the core proteins involved in the microRNA (miRNA) pathway in plants have been identified, and almost simultaneously hundreds of miRNA sequences processed in the Arabidopsis sporophyte have been discovered by exploiting next-generation sequencing technologies. However, there is very limited understanding about potentially distinct mechanisms of post-transcriptional regulation between different cell lineages. In this review the focus is on the Arabidopsis male gametophyte (pollen), where the germline differentiates after meiosis giving rise to the male gametes. Based on comparative analysis of miRNAs identified in sperm cells by in-depth sequencing, their possible functions during germ cell specification and beyond fertilization are discussed. In addition, 25 potentially novel miRNAs processed in sperm cells and pollen were identified, as well as enriched variations in the sequence length of known miRNAs, which might indicate subfunctionalization by association with a putative germline-specific Argonaute complex. ARGONAUTE 5 (AGO5), by close homology to AGO1 and localizing preferentially to the sperm cell cytoplasm in mature pollen, may be part of such a complex.
Keywords: Arabidopsis, male germ cells, microRNA, pollen, post-transcriptional gene silencing
Introduction
Post-transcriptional gene silencing was first observed over a decade ago, and since then numerous discoveries unveiled a fascinating and unexpectedly conserved cellular mechanism. In addition, several studies from plants to animals underlined distinct regulatory mechanisms that evolved throughout different cell lineages.
The microRNA (miRNA) pathway is an essential mechanism to regulate different biological processes. In plants it is involved in several fundamental processes such as development, response to biotic and abiotic stresses, and hormone responses. These small non-coding RNAs act through cleavage of highly complementary target mRNAs, but also through translational repression (Chen, 2004; Brodersen et al., 2008). More indirectly, miRNA-guided cleavage of TAS transcripts gives rise to trans-acting small interfering RNAs (ta-siRNAs) that regulate expression of other genes (Allen et al., 2005). miRNA biogenesis depends on processing of stem–loop precursors by DICER-Like 1 (DCL1), and subsequent cleavage of target mRNAs requires binding of mature miRNAs to ARGONAUTE 1 (AGO1) (Voinnet, 2009). Some miRNAs may also associate with AGO10, promoting translational repression of specific target transcripts (Brodersen et al., 2008). Based on data generated by numerous high-throughput small RNA sequencing studies, additional types of small RNAs resulting from known precursor stem–loops were identified (Vazquez et al., 2008; Chellappan et al., 2010; Zhang et al., 2010). Although processing of these miRNA-like RNAs is mostly dependent on the regular DCL1-based processing pathway (Zhang et al., 2010), it was shown that the production of some long miRNAs (23–27 nt) depends on a DCL3/RDR2/Pol IV pathway (Chellappan et al., 2010).
Although miRNA activity is well characterized in the Arabidopsis sporophyte, little is known about the role of miRNAs during male gametophyte development. The haploid microspores in higher plants undergo two mitotic divisions to form the gametes, which are nourished inside the pollen vegetative cell (VC) and passively transported to the embryo sac by the pollen tube. The male germline starts after pollen mitosis I (PMI), by asymmetric cell division in the microspore. The larger cell (VC) arrests cell cycle progression, while the smaller cell [germ cell (GC)] divides further in PMII to form two identical male gametes [sperm cells (SCs)] (Boavida et al., 2005). The genetic programmes driving male gametogenesis and pollen development have been studied and reported throughout the last decade mostly by means of microarray-based studies, but also with the characterization of loss-of-function mutations impairing gametophyte development (Becker and Feijó, 2007; Borg et al., 2009). These studies hold great promise for the identification of a core transcriptional programme driving plant germline differentiation and specification. However, understanding post-transcriptional regulation of gene expression in pollen and the gametes requires additional efforts.
This review aims to summarize recent discoveries on miRNA pathways and epigenetic reprogramming in the Arabidopsis germline, bridging novel findings with our current understanding of miRNA activity in plants. The comparative analysis of known miRNA families and potentially novel miRNAs identified in total pollen and isolated SCs by in-depth sequencing highlights their possible functions during gamete specification and fertilization.
Small RNA pathways during male gametogenesis
Perspective from transcriptomics
Arabidopsis pollen at anthesis is tricellular, having originated from a uninucleated microspore after two rounds of mitotic division. Honys and Twell (2004) have shown that throughout male gametogenesis there is a decrease in the number of genes expressed, accompanied by an enrichment of pollen-specific transcripts. Their data suggested activation of a pollen-specific transcriptional machinery, simultaneously with selective down-regulation of somatic genes as a possible result of miRNA activity. However, this early study used whole pollen and thus could not distinguish gene expression profiles between the two differentiated cell types in mature pollen grains. The understanding of general molecular functions acting in Arabidopsis SCs increased significantly over the last few years owing to SC purification methods developed for fluorescence-activated cell sorting (FACS) and microarray-based transcriptomics (Borges et al., 2008). Comparison of the expression profile of mature pollen grains with that of purified SCs clearly indicated that after PMI, the resulting VC and SCs activate distinct transcriptional programmes. The VC seems to be enriched in transcripts functionally skewed towards pollen germination and tube growth (Pina et al., 2005), and expression of repeats as a result of its chromatin decondensation (Slotkin et al., 2009). At this stage the twin SCs initiate a long DNA replication phase that will be sustained until the moment of fertilization. Concordantly, the SC transcriptome shows intricate control over DNA repair, cell cycle transitions, and ubiquitin-mediated protein degradation (Borges et al., 2008). These results showed that mRNAs accumulating in the male gametes are diverse and abundant, suggesting that the same might be true for other RNA species such as small non-coding RNAs.
MicroRNA activity in pollen
Primary evidence indicating that miRNAs could be actively controlling male gametophytic development came from the study by Kidner and Martienssen (2005) in Arabidopsis. This work characterized a mutant allele of AGO1 (ago1-10) that is poorly transmitted through the male gametes. More recently, miRNAs were shown to be present and functional in mature pollen, along with expression of some of the most important genes involved in the miRNA silencing pathway such as DCL1, AGO1, and RDR6, which are consistently expressed during pollen development (Grant-Downton et al., 2009a). For details on small RNA pathways and their components, see the review by Le Trionnaire et al. (2011). Two independent studies reported the miRNA repertoire in Arabidopsis pollen using microarrays, quantitative reverse transcription-PCR (qRT-PCR), and pyrosequencing, resulting in the identification of ∼33 known miRNAs (Chambers and Shuai, 2009; Grant-Downton et al., 2009b). Grant-Downton et al. (2009b) also identified seven putative novel miRNAs, including one that targets an F-box protein specifically expressed in pollen and co-targeted by miR774. Their work showed that ta-siRNAs derived from miR173-directed cleavage of TAS transcripts also accumulate in pollen, but intriguingly precursor TAS transcripts could not be detected. These results clearly indicate that mature miRNAs and products of miRNA-guided cleavage are present in mature pollen, but the spatial temporal regulation of miRNA activity during pollen development and in plant GCs remained unexplored.
A potentially unique small RNA silencing pathway during germ cell differentiation and specification
While some of the key genes encoding proteins required for miRNA processing in Arabidopsis seem to be expressed in pollen and SCs, many other genes involved in small RNA pathways are either not expressed or depleted (Borges et al., 2008). In contrast, particular genes associated with small RNA activity and DNA methylation, such as AGO9, MET1, and DDM1, as well as AGO5, are highly enriched in SCs in comparison with total pollen and sporophytic tissues (Borges et al., 2008; Slotkin et al., 2009), suggesting that distinct genetic and epigenetic mechanisms might be established preferentially in the germline. AGO1 and, to some extent, AGO10 are the main regulators of miRNA-directed target cleavage in Arabidopsis (Vaucheret et al., 2004; Brodersen et al., 2008), but both seem to be down-regulated in the SCs. While AGO9 has been recently implicated in siRNA-mediated transposon inactivation (Olmedo-Monfil et al., 2010), AGO5 is a closely related homologue of AGO1 and AGO10 (Fig. 1) and could thus be involved in miRNA activity in the gametes. As predicted by the sperm transcriptome data, AGO5 is preferentially accumulated in the SC cytoplasm in mature pollen and growing pollen tubes (Fig. 2, Supplementary Videos S1 and S2 available at JXB online). The closest homologue in rice (MEL1) was shown to be required for correct progression through meiosis (Nonomura et al., 2007), but the biological function of AtAGO5 and OsMEL1 and their role in the small RNA pathways remain to be elucidated. In addition to the role of Argonaute proteins in small RNA-directed DNA methylation of heterochromatic regions, and miRNA-guided slicing and translational repression of target transcripts, it was recently shown that the catalytic activity of the mammalian Argonaute2 is responsible for a Dicer-independent pathway of miRNA processing (Cifuentes et al., 2010; Yang et al., 2010). As Argonaute proteins are widely conserved among eukaryotes, it might be worthwhile investigating whether Argonaute proteins in plants could also process miRNA stem–loop precursors.
Fig. 1.
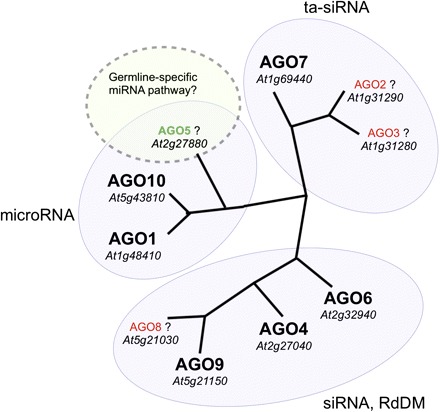
Argonaute protein family in Arabidopsis. Phylogenetic tree illustrating the 10 Argonaute proteins in Arabidopsis, subdivided into the three main functional classes based on sequence homology: miRNA-guided slicing and translational repression of target transcripts, trans-acting siRNA (ta-siRNA) activity, and chromatin remodelling by siRNA-directed DNA methylation (RdDM). AGO2, AGO3, AGO8 (red), and AGO5 (green) have unknown function. AGO5 is a close relative of AGO1 and AGO10, and could be involved in a novel miRNA pathway in the germline (see Fig. 2). (adapted from Yigit et al., 2006).
Fig. 2.
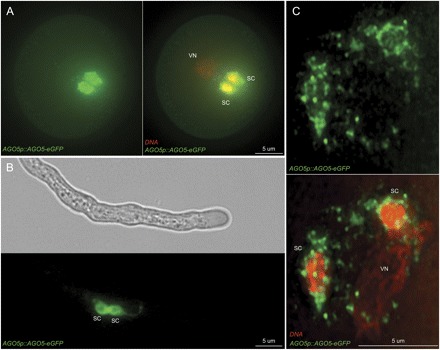
AGO5 expression in Arabidopsis pollen. (A) Transgene expression of AGO5 protein using the native promoter region (1000 bp upstream of the 5'-untranslated region) and the genomic coding sequence translationally fused to eGFP. AGO5p::AGO5–eGFP expression in mature pollen localizes preferentially in the sperm cell (SC) cytoplasm (B) remaining during pollen tube growth. (C) A magnification of the male germ unit shows that AGO5–GFP localizes in the SC cytoplasm and not in the nucleus, extending through the cytoplasmic connection that links the SCs with the vegetative nucleus (VN). DAPI-stained DNA shows SCs and VNs in A and C.
Small RNA activity in GCs has been studied in more detail in animals, leading to the discovery of divergent mechanisms of post-transcriptional regulation between soma and germline (Mishima et al., 2006; Kedde et al., 2007). General loss of miRNAs caused by Dicer mutations has been shown to impair germline maintenance in Caenorhabditis elegans and Drosophila (Knight and Bass, 2001; Hatfield et al., 2005), and differentiation defects in mouse GCs (Hayashi et al., 2008; Maatouk et al., 2008), but apparently miRNAs are not required for GC proliferation in zebrafish (Giraldez et al., 2005).
Evidence for small RNA silencing acting in Arabidopsis sperm cells
Epigenetic silencing of transposable elements
GCs developed specific mechanisms associated with chromatin remodelling and resetting of epigenetic marks that must be properly established before being transmitted to the next generation. For example, animals have a GC-specific class of small RNAs termed piRNAs (Piwi-interacting RNAs), associated with transposable element (TE) silencing and heterochromatin formation (Vagin et al., 2006; Carmell et al., 2007). Plants lack piRNAs and Piwi proteins, but, alternatively, particular TE silencing mechanisms are also specifically established in the germline (Slotkin et al., 2009; Olmedo-Monfil et al., 2010). A first insight into small RNA species in Arabidopsis SCs resulted from studying TE activity in pollen (Slotkin et al., 2009). It was shown that transposons are expressed and transpose in the vegetative nucleus (VN), concordantly with down-regulation of DECREASED DNA METHYLATION 1 (DDM1) and gain of certain classes of TE-derived siRNAs. However, the same 21 nt siRNAs are also abundantly accumulated in the SCs where the corresponding TE loci are highly methylated and transcriptionally silenced (Slotkin et al., 2009). These results strongly suggest an active communication between the VC and SCs, reviving earlier observations of a cytoplasmic tail that remains after pollen mitosis, connecting the gametes directly to the VN to engender the male germ unit (for a review see McCue et al., 2011). Along with siRNA movement between neighbouring cells, several lines of evidence indicate now that miRNAs are also on the move (Martienssen, 2010). It was recently discovered that 21 nt miRNAs produced in root cells can move into adjacent cells possibly as a miRNA duplex, and regulate expression of target genes post-transcriptionally (Carlsbecker et al., 2010). Therefore, it is possible that miRNAs in Arabidopsis pollen could also move between VC and SCs, as predicted for TE-derived siRNAs. Indeed, by expressing artificial miRNAs (amiRNAs) against green fluorescent protein (GFP) under the control of the VC-specific promoter LAT52, it was possible to target GFP expression in the SCs (Slotkin et al., 2009). Future studies should help in understanding the functional purpose and extent of an intercellular cross-talk during pollen development and tube growth.
Natural cis-antisense siRNAs regulating expression of endogenous genes
Natural cis-antisense siRNAs (cis-nat-siRNAs) were recently described in Arabidopsis SCs as playing a crucial role in double fertilization. These siRNAs are produced from the overlapping region of KPL and ARI14 transcripts, and were shown to be involved in ARI14 down-regulation (Ron et al., 2010). The authors proposed an interesting model in which ARI14 lost its E3 ubiquitin ligase activity due to amino acid mutations along with gene duplication of ARI 13/14/15, thus requiring that ARI14 is efficiently silenced post-transcriptionally to avoid competing with ARI13 protein that is also expressed in sperm. In addition, this study showed that KPL–ARI14 nat-siRNAs are processed by DCL1, and dependent on a RDR2/SGS3/Pol IV pathway, thus correlating with other previously described nat-siRNAs (Borsani et al., 2005; Katiyar-Agarwal et al., 2006). siRNAs matching the KPL transcript region were detected by co-expressing KPL and ARI14 ectopically, as well as other 21 nt siRNAs derived from the overlapping region. However, only one 21 nt siRNA that could target ARI14 was detected in the SC small RNA data set, originating from the KPL–ARI14 overlap region but outside the predicted area (Ron et al., 2010). The fact that other siRNAs were not detected may be explained by their low abundance. Alternatively, this could be a process that occurs earlier during pollen development, thus explaining why ARI14 transcripts are absent from SCs at the mature pollen stage (Borges et al., 2008).
Comparative analyses of miRNAs accumulated in sperm cells
Several miRNA families were identified in the previously reported small RNA data sets of purified SCs and pollen by Illumina sequencing (Slotkin et al., 2009). From the 2 540 585 signatures sequenced in pollen and 1 925 202 in SCs, 283 561 and 256 787 sequences matched to known Arabidopsis miRNAs in pollen and sperm, respectively. In total, this corresponds to 75 known miRNA families expressed in pollen, and 83 in SCs (Fig. 3). The list of most known miRNAs in these two data sets is publicly available in the SBS database (Nakano et al., 2006). Comparing Illumina data with the recently available data set obtained by 454 sequencing of pollen small RNAs (Grant-Downton et al., 2009b) shows that out of the 31 different miRNA families in mature pollen detected by 454, only miR776 was not detected in the Illumina pollen sample (Fig. 3). A summary of normalized reads matching to the annotated form of known miRNAs in SC and pollen data sets in comparison with that of inflorescence is presented as Supplementary Table S1 at JXB online.
Fig. 3.
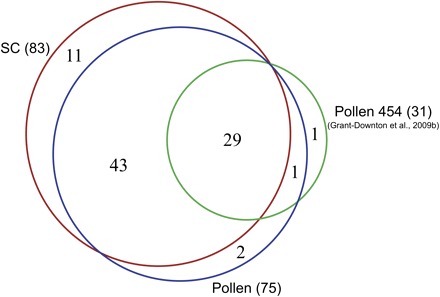
Venn diagram illustrating miRNA families detected in sperm cells and pollen. Overlap between known miRNA families detected in sperm cells (83) and pollen (75) by Illumina sequencing (Slotkin et al., 2009), and a 454 sequencing data set of pollen small RNAs (31) reported by Grant-Downton et al. (2009b). Numbers in parentheses represent total miRNA families identified in each data set.
Expression of most miRNA families seems to be distinct between total pollen and purified SCs (Fig. 4A), which was expected considering their different transcriptomes and cell fate (Borges et al., 2008). miR159a is particularly interesting as it is >5-fold enriched in SCs. Keeping in mind that miRNAs highly abundant in SCs should be identified in pollen samples despite a dilution effect, the significantly lower abundance of miR159a in pollen suggests that it could be sperm specific. miR159 was predicted to be involved in the regulation of several transcripts belonging to the MYB family of transcription factors, including DUO1 (Palatnik et al., 2007), a GC-specific transcription factor that is responsible for expression of several germline genes (Brownfield et al., 2009). miR159-guided cleavage of DUO1 transcripts was detected in mature pollen (Grant-Downton et al., 2009a), but recent work could show that depletion of miR159 in a miR159abc triple mutant does not seem to impair pollen development and fertilization (Allen et al., 2010), suggesting that post-transcriptional regulation of gene expression by miR159 either is not essential in pollen and the germline, or its closely related miR319 can compensate, to a certain extent, for the absence of miR159. This work has further shown that miR159 activity is essential during plant development, by uniquely controlling expression of the anther-specific MYB33 and MYB65 (Allen et al., 2010).
Fig. 4.
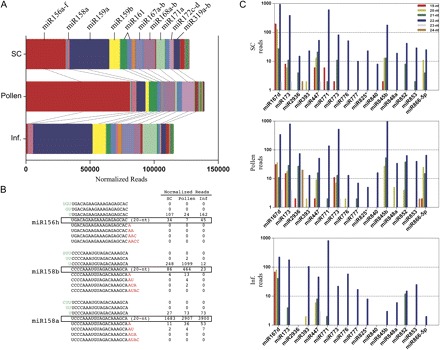
MiRNA families and variations in sequence length. (A) Relative abundance of known miRNAs detected in sperm cell (SC), pollen, and inflorescence data sets. (B) MiRNAs 156h and 158b have isoforms of 21 nt length with nucleotide extensions in the 5' terminus, for which the number of reads in SCs and pollen is higher than the annotated 20 nt isoforms. miR158a reads are presented to exemplify that these variations do not accumulate significantly for the majority of other miRNAs. (C) The 22 nt miRNAs that may function as siRNA triggers are differentially accumulated in SC, pollen, and inflorescence tissues.
MiRNA families such as miR156 and miR158 have isoforms in which the 5'-terminal nucleotide is a cytosine (miR156g and miR158b, respectively). Such isoforms were predicted to be associated preferentially with AGO5 (Mi et al., 2008; Takeda et al., 2008), which correlates with the fact that miR156g and miR158b levels are higher in pollen and sperm than in sporophytic tissues (Supplementary Table S1 at JXB online). However, it is intriguing that miR156a-f is so highly enriched in pollen and sperm (Fig. 4A), as the entire family is predicted to target the plant-specific SPL gene family of transcription factors (Rhoades et al., 2002). This possibly suggests that the same miRNA family may have distinct activity by association with specific AGO complexes. SPL genes are known to regulate several developmental transitions such as flowering and shoot development, but their importance in pollen and germline differentiation remains unexplored. Another example of a miRNA with a 5′ cytosine is miR845a, which is also highly enriched in pollen and sperm, but its targets are still unknown. Both miR845a and 845b are almost undetectable in inflorescence tissue (Supplementary Table S1), which suggests that they must be preferentially processed in SCs and pollen.
Novel miRNAs and natural variations in sequence length of known miRNA families
To screen for potentially new miRNAs in the SC and pollen small RNA data sets, the miRCat tool available online within the UEA plant sRNA toolkit (Moxon et al., 2008) was used. Based on correspondence with genomic loci that could encode stem–loop precursors, 25 small RNA sequences were identified as potential novel miRNAs. These results, including a number of predicted target transcripts for the novel miRNAs found, are presented in Table 1, and extended in Supplementary Table S2 at JXB online. Moreover, some of the miRNA sequences discovered seem to be isoforms of known miRNA families with variations in sequence length (Table 1). This is the case for miR158b that was detected with an additional uracil at the 5′ nucleotide terminus. Interestingly, this isoform is more abundant in pollen and SCs than the normal annotated version (Fig. 4B). The biological meaning of such variations in miRNA processing is not yet fully understood, but it was recently reported that single nucleotide extensions at the 5′ terminus of known miRNAs can lead to incorporation into different Argonaute complexes (Ebhardt et al., 2010). For this reason a sperm-enriched miR156h isoform with an extension at the 5′ end and higher abundance than the known 20 nt isoform deserves closer attention (Fig. 4B). This variation in natural sequence length of miR156h was analysed by Ebhardt et al. (2010), and curiously they observed that the known 20 nt version of miR156h is loaded into AGO1, while the miR156h plus 5′ U isoform seems to associate mainly with AGO5.
Table 1.
– Normalized reads for potentially novel miRNAs and isoforms with variations in the sequence length of known miRNA/miRNA* detected by miRCat tool, in sRNA datasets of Col pollen and sperm cells (SC) in comparison with inflorescence tissue (Inf)
| microRNA | Sequence | SC | Pollen | Inf | psRNA target(s) |
| Variations of known miRNA/miRNA*(a) | |||||
| miR156h | UUGACAGAAGAAAGAGAGCAC | 107 | 24 | 162 | SQUAMOSA PROMOTER BINDING PROTEIN-LIKE |
| miR158b | UCCCCAAAUGUAGACAAAGCA | 248 | 1099 | 12 | AT2G46590 |
| miR161a.1 | UUGAAAGUGACUACAUCGGGGu | 5605 | 7146 | 2066 | PPR (AT5G41170) |
| miR162 | AUCGAUAAACCUCUGCAUCCAGg | 4 | 58 | 1 | DICER-LIKE 1 (AT1G01040) |
| miR165* | gGAAUGUUGUCUGGAUCGAGGA | 46 | 102 | 9 | - |
| miR167c | uUAAGCUGCCAGCAUGAUCUUGU | 10 | 0 | 2 | - |
| miR408* | ACAGGGAACAAGCAGAGCATGG | 589 | 29 | 12 | AT2G47020, AT4G02940, AT1G04210, AT1G17180, AT4G03950 |
| miR773a | TTTGCTTCCAGCTTTTGTCTCC | 82 | 524 | 22 | MET2 (AT4G14140), AT4G05390, AT4G08990 |
| miR778* | ACAAACUCGGUGUACAUAGACcaaaccaag | 116 | 316 | 2 | AT5G22380, AT1G69610 |
| miR840* | UUGUUUAGGUCCCUUAGUUUCu | 21 | 27 | 9 | - |
| miR844a | AAUGGUAAGAUUGCUUAUAAGcu | 11 | 22 | 11 | AT2G13720, AT5G44120 |
| miR852 | AAGAUAAGCGCCUUAGUUCUGA | 42 | 63 | 15 | AT5G56650, AT5G56660 |
| miR852a | AAGAUAAGCGCCUUAGUUCUGA | 32 | 107 | 7 | - |
| miR860 | ucaAUAGAUUGGACUAUGUAUAUU | 17 | 9 | 10 | AT5G26030, AT3G12640 |
| miR863a | uugAGAGCAACAAGACAUAAUAAAGAG | 2 | 11 | 0 | - |
| miR870 | AAUCUAAUUUGGUGUUUCUUCGaug | 2 | 13 | 1 | AT3G55370 |
| Potentially novel miRNAs (b) | |||||
| miR2934(c) | CAUCCAAGGUGUUUGUAGAAA† | 101 | 392 | 6 | - |
| miR4240.2(d) | AUGGCUAGAGUGACUAGACCCG | 23 | 0 | 10 | - |
| miR447a.2 | UAUGGAAGAAAUUGUAGUAUU | 84 | 42 | 152 | AT1G42630, AT1G54710 |
| miR447c.2 | CCCCUUACAAUGUCGAGUAAA | 0 | 13 | 3 | - |
| miR868.2 | UCAUGUCGUAAUAGUAGUCAC | 32 | 92 | 5 | CMT1 (AT1G80740), CMT2 (AT4G19020) |
| miR5012 | UUUUACUGCUACUUGUGUUCC | 6 | 0 | 3 | AT1G53700, AT2G37678 |
| miR5013 | UUUGUGACAUCUAGGUGCUUU | 86 | 317 | 1 | AT3G60580 |
| miR5014 | UUGUACAAAUUUAAGUGUACG | 15 | 4 | 9 | - |
| miR5015.1 | UUGGUGUUAUGUGUAGUCUUC | 17 | 20 | 0 | - |
| miR5015.2 | UCUGUUGUUGUUGGUGUUAUG | 17 | 38 | 2 | AT2G38320, AT1G12860, AT2G01760, AT5G38740 |
| miR5016 | UUCUUGUGGAUUCCUUGGAAA | 2 | 5 | 0 | - |
| miR5017 | UUAUACCAAAUUAAUAGCAAA | 4 | 5 | 67 | - |
| miR5018 | UUAAAGCUCCACCAUGAGUCCAAU | 0 | 5 | 0 | - |
| miR5019 | UGUUGGGAAAGAAAAACUCUU | 6 | 40 | 2 | AT3G58810, AT1G14510, AT4G19550 |
| miR5020a | UGGAAGAAGGUGAGACUUGCA | 15 | 53 | 0 | - |
| miR5020b | AUGGCAUGAAAGAAGGUGAGA | 158 | 711 | 1 | - |
| miR5021 | UGAGAAGAAGAAGAAGAAAA | 0 | 7 | 0 | - |
| miR5022 | GUCAUGGGGUAUGAUCGAAUG† | 17 | 56 | 0 | - |
| miR5023 | AUUGGUAGUGGAUAAGGGGGC† | 0 | 18 | 0 | AT5G24950 |
| miR5024 | AUGACAAGGCCAAGAUAUAACA | 4 | 7 | 0 | - |
| miR5025 | ACUGUAUAUAUGUAAGUGACA | 4 | 9 | 0 | AT2G48010 |
| miR5026 | ACUCAUAAGAUCGUGACACGU | 10 | 16 | 2 | - |
| miR5027 | ACCGGUUGGAACUUGCCUUAA | 10 | 27 | 3 | - |
| miR5028 | AAUUGGGUUUAUGCUAGAGUU | 92 | 443 | 8 | - |
| miR5029 | AAUGAGAGAGAACACUGCAAA† | 84 | 288 | 1 | AT2G30070, AT4G02900 |
Underlined sequences indicate the mature form annotated in ASRP(a), including missing nucleotides (lower case). These variations were preferentially detected by miRCat since they are more abundant than the annotated isoforms of known miRNAs.
(a) – Annotation as in ASRP: The Arabidopsis Small RNA Project Database (http://asrp.cgrb.oregonstate.edu/).
(b) – No correspondence found in ASRP database. Predicted foldback structures and chromosome loci presented in Table S2.
(c) – Potentially mature miRNA sequence processed from ath-MIR2934, more abundant in our SC and pollen datasets than ath-miR2934 UCUUUCUGCAAACGCCUUGGA reported in Grant-Downton et al. (2009b), but classified in our study as ath-miR2934* (Table S1).
(d) – Processed from ath-MIR4240 (Ma et al., 2010), shifted 10nt from ath-miR4240 that was not detected in our SC and pollen datasets.
† Correspondent miRNA* sequence also detected in the sRNA dataset.
Isoforms of known miRNA families with nucleotide extensions at the 3′ terminus were also identified in the sperm and pollen data sets. For example, miR773 is annotated as a 21 nt miRNA and seems to be low abundant in both total pollen and SCs. However, a 22 nt miR773 with a C extension at the 3′ terminus is more abundant (Table 1), suggesting in this case activation of the recently discovered mechanism for triggering siRNA production from target transcripts (Chen et al., 2010; Cuperus et al., 2010). These studies showed that certain miRNAs are capable of triggering siRNA and ta-siRNA production only if presented to the target transcript in a 22 nt form, but the biological significance of this mechanism remains controversial. Analysing the abundance of all possible isoforms of known miRNA families from 19 nt to 24 nt in sequence length in the sperm and pollen data sets highlights other possible siRNA triggers (Supplementary Table S3 at JXB online). Among these are the 22 nt miRNA isoforms miR173, miR393, miR447, miR771, miR773, miR825*, miR167d, and miR828 (Fig. 4C) previously discussed (Chen et al., 2010; Cuperus et al., 2010). Interestingly miR773, targeting the DNA methyltransferase MET2, is more abundant in pollen, while miR771 that was confirmed to function as an siRNA trigger (Chen et al., 2010) is enriched in SCs, but the targets remain unknown. Other miRNA families that could potentially function as siRNA triggers (i.e. more abundant 22 nt isoform) are miR776, miR777, miR840, miR845b, miR848a, miR852, miR853, and miR2936 (Fig. 4C). MiR845 is highly enriched in pollen and SCs, and very low abundant in inflorescence tissue, suggesting preferential expression in the male gametophyte. This is a plant-specific and recently evolved miRNA family (Barakat et al., 2007), for which a biological function remains unclear.
Inheritance of parental miRNAs
A very exciting question is whether miRNAs accumulated in SCs could be delivered to the egg and central cell upon double fertilization, to play a role in the development of early zygote and endosperm, respectively. A similar question is being addressed in the animal field, originating from the discovery of miRNAs and siRNAs in human spermatozoa (Ostermeier et al., 2005). In mice, miRNAs delivered to the oocyte can be detected up to the 2-cell stage (Amanai et al., 2006), but miRNA function in general seems to play no role during oocyte maturation and pre-implantation development (Suh et al., 2010).
While the function of miRNAs delivered at fertilization is still a matter of debate (Dadoune, 2009), there is evidence that paternal mRNAs are transported to oocytes (Ostermeier et al., 2004) and that some have a function. For spermatozoon-derived PSG1 and HLA-E mRNA a possible functional role during early embryo development was demonstrated in humans (Avendano et al., 2009), while in mice the Kit transcript delivered by spermatozoa seems to act as an epigenetic modifier of early embryo development (Rassoulzadegan et al., 2006). In plants, the strongest indication for delivery and function of sperm-derived mRNAs upon fertilization to date came from the study on the YDA signalling pathway during embryonic patterning and the characterization of the SHORT SUSPENSOR (SSP) protein. It was found that SSP transcripts accumulate in Arabidopsis SCs, but are translationally repressed before fertilization, to be translated only during early zygotic development (Bayer et al., 2009). It is possible that in plants paternal miRNAs are also delivered at fertilization, as they could have important functions as signalling molecules, possibly triggering early zygotic patterning and endosperm development. MiRNAs would certainly provide an efficient mechanism of repro-gramming in the early zygote, before the gene expression programme that defines the maternal to zygotic transition is established. In the light of the parental conflict theory (Spielman et al., 2001) it is also possible that paternally inherited miRNAs could contribute as an immediate mechanism to regulate the maternally expressed inhibitors of embryo growth. However, small RNA sequencing or analysis of egg cells and early zygotes will be needed to determine if miRNAs accumulated in SCs are indeed inherited by the zygote. Moreover, the use of artificial miRNA target mimics (Todesco et al., 2010) to knock down specific miRNA families in SCs should clarify if their potential inheritance is of importance.
Conclusions
Post-transcriptional gene silencing involves miRNA activity to eliminate transcripts of previous developmental processes, while modulating expression of active genes. In recent years, small RNA sequencing technologies allowed a robust temporal and spatial profiling of miRNAs processed in several species, tissues, and cell types, leading to the discovery of an ancient and widespread mechanism to control gene expression. Consequently, it can be assumed that the basis of miRNA processing and activity in eukaryotes is now known; however, we are just taking the first steps towards understanding its origins and evolution.
The male gametophyte of flowering plants constitutes a unique system to analyse specialized small RNA pathways and small RNA trafficking with cellular resolution. Post-meiotic pollen development in particular is an exciting biological process in plants for its structural simplicity and fast transitions over two mitotic divisions, which are coupled with germline differentiation and specification. Small RNA sequencing data have provided evidence that miRNAs are abundant and diverse in Arabidopsis SCs, and that after GC differentiation during PMI, the SCs and neighbouring VC activate distinct miRNA pathways that correlate with different cell fate and gene expression profiles. However, there are still several open questions. It is intriguing that certain miRNAs seem to be processed in an unusual manner in pollen and SCs in comparison with what is generally observed in sporophytic tissues. The observation that AGO5 is likely to be part of the unique miRNA-induced silencing complex established in the male germline supports the idea that distinct and still unknown cellular pathways might exist. These results should provide useful information for future studies, as a baseline comparison to dissect the many hypotheses discussed.
Supplementary Material
Acknowledgments
We thank Paulo Almeida for bioinformatics support and Elena Baena-González for critical reading of the manuscript. This work was supported by grants PTDC/AGR-GPL/103778/2008, PTDC/BIA-BCM/103787/2008, and PTDC/AGR-GPL/70592/2006 from Fundacão para a Ciência e a Tecnologia (FCT), Portugal. FB and PAP were supported by FCT PhD fellowships SFRH/BD/48761/2008 and SFRH/BD/63477/2009, respectively.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Allen RS, Li J, Alonso-Peral MM, White RG, Gubler F, Millar AA. MicroR159 regulation of most conserved targets in Arabidopsis has negligible phenotypic effects. Silence. 2010;1:18. doi: 10.1186/1758-907X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanai M, Brahmajosyula M, Perry AC. A restricted role for sperm-borne microRNAs in mammalian fertilization. Biology of Reproduction. 2006;75:877–884. doi: 10.1095/biolreprod.106.056499. [DOI] [PubMed] [Google Scholar]
- Avendano C, Franchi A, Jones E, Oehninger S. Pregnancy-specific β-1-glycoprotein 1 and human leukocyte antigen-E mRNA in human sperm: differential expression in fertile and infertile men and evidence of a possible functional role during early development. Human Reproduction. 2009;24:270–277. doi: 10.1093/humrep/den381. [DOI] [PubMed] [Google Scholar]
- Barakat A, Wall K, Leebens-Mack J, Wang YJ, Carlson JE, Depamphilis CW. Large-scale identification of microRNAs from a basal eudicot (Eschscholzia californica) and conservation in flowering plants. The Plant Journal. 2007;51:991–1003. doi: 10.1111/j.1365-313X.2007.03197.x. [DOI] [PubMed] [Google Scholar]
- Bayer M, Nawy T, Giglione C, Galli M, Meinnel T, Lukowitz W. Paternal control of embryonic patterning in Arabidopsis thaliana. Science. 2009;323:1485–1488. doi: 10.1126/science.1167784. [DOI] [PubMed] [Google Scholar]
- Becker JD, Feijó JA. How many genes are needed to make a pollen tube? Lessons from transcriptomics. Annals of Botany. 2007;100:1117–1123. doi: 10.1093/aob/mcm208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boavida LC, Becker JD, Feijó JA. The making of gametes in higher plants. International Journal of Developmental Biology. 2005;49:595–614. doi: 10.1387/ijdb.052019lb. [DOI] [PubMed] [Google Scholar]
- Borg M, Brownfield L, Twell D. Male gametophyte development: a molecular perspective. Journal of Experimental Botany. 2009;60:1465–1478. doi: 10.1093/jxb/ern355. [DOI] [PubMed] [Google Scholar]
- Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijó JA, Becker JD. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiology. 2008;148:1168–1181. doi: 10.1104/pp.108.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- Brownfield L, Hafidh S, Borg M, Sidorova A, Mori T, Twell D. A plant germline-specific integrator of sperm specification and cell cycle progression. PloS Genetics. 2009 doi: 10.1371/journal.pgen.1000430. 5, e1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Developmental Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Chambers C, Shuai B. Profiling microRNA expression in Arabidopsis pollen using microRNA array and real-time PCR. BMC Plant Biology. 2009;9:87. doi: 10.1186/1471-2229-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan P, Xia J, Zhou X, Gao S, Zhang X, Coutino G, Vazquez F, Zhang W, Jin H. siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Research. 2010;38:6883–6894. doi: 10.1093/nar/gkq590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proceedings of the National Academy of Sciences, USA. 2010;107:15269–15274. doi: 10.1073/pnas.1001738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nature Structural and Molecular Biology. 2010;17:997–1003. doi: 10.1038/nsmb.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadoune JP. Spermatozoal RNAs: what about their functions? Microscopy Research and Technique. 2009;72:536–551. doi: 10.1002/jemt.20697. [DOI] [PubMed] [Google Scholar]
- Ebhardt HA, Fedynak A, Fahlman RP. Naturally occurring variations in sequence length creates microRNA isoforms that differ in argonaute effector complex specificity. Silence. 2010;1:12. doi: 10.1186/1758-907X-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Grant-Downton R, Hafidh S, Twell D, Dickinson HG. Small RNA pathways are present and functional in the angiosperm male gametophyte. Molecular Plant. 2009;2:500–512. doi: 10.1093/mp/ssp003. [DOI] [PubMed] [Google Scholar]
- Grant-Downton R, Le Trionnaire G, Schmid R, Rodriguez-Enriquez J, Hafidh S, Mehdi S, Twell D, Dickinson H. MicroRNA and tasiRNA diversity in mature pollen of Arabidopsis thaliana. BMC Genomics. 2009;10:643. doi: 10.1186/1471-2164-10-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One. 2008 doi: 10.1371/journal.pone.0001738. 3, e1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biology. 2004;5:R85. doi: 10.1186/gb-2004-5-11-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A., Jr.,, Zhu JK, Staskawicz BJ, Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proceedings of the National Academy of Sciences, USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. The role of ARGONAUTE1 (AGO1) in meristem formation and identity. Developmental Biology. 2005;280:504–517. doi: 10.1016/j.ydbio.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Trionnaire G, Grant-Downton R, Kourmpetli S, Dickinson H, Twell D. Small RNA activity and function in angiosperm gametophytes. Journal of Experimental Botany. 2011 doi: 10.1093/jxb/erq399. 62. [DOI] [PubMed] [Google Scholar]
- Ma Z, Coruh C, Axtell MJ. Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. The Plant Cell. 2010;22:1090–1103. doi: 10.1105/tpc.110.073882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatouk DM, Loveland KL, McManus MT, Moore K, Harfe BD. Dicer1 is required for differentiation of the mouse male germline. Biology of Reproduction. 2008;79:696–703. doi: 10.1095/biolreprod.108.067827. [DOI] [PubMed] [Google Scholar]
- Martienssen R. Molecular biology. Small RNA makes its move. Science. 2010;328:834–835. doi: 10.1126/science.1190510. [DOI] [PubMed] [Google Scholar]
- McCue AD, Cresti M, Feijo J, Slotkin RK. Cytoplasmic connection of sperm cells to the pollen vegetative cell nucleus: potential roles of the male germ unit revisited. Journal of Experimental Botany. 2011 doi: 10.1093/jxb/err032. 62. [DOI] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, Chen S, Hannon GJ, Qi Y. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5' terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Current Biology. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon S, Schwach F, Dalmay T, Maclean D, Studholme DJ, Moulton V. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics. 2008;24:2252–2253. doi: 10.1093/bioinformatics/btn428. [DOI] [PubMed] [Google Scholar]
- Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Research. 2006;34:D731–D735. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. The Plant Cell. 2007;19:2583–2594. doi: 10.1105/tpc.107.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil V, Duran-Figueroa N, Arteaga-Vazquez M, Demesa-Arevalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier GC, Goodrich RJ, Moldenhauer JS, Diamond MP, Krawetz SA. A suite of novel human spermatozoal RNAs. Journal of Andrology. 2005;26:70–74. [PubMed] [Google Scholar]
- Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature. 2004;429:154. doi: 10.1038/429154a. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Wollmann H, Schommer C, et al. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Developmental Cell. 2007;13:115–125. doi: 10.1016/j.devcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijó JA, Becker JD. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiology. 2005;138:744–756. doi: 10.1104/pp.104.057935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Ron M, Saez MA, Williams LE, Fletcher JC, McCormick S. Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes and Development. 2010;24:1010–1021. doi: 10.1101/gad.1882810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijó JA, Martienssen RA. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman M, Vinkenoog R, Dickinson HG, Scott RJ. The epigenetic basis of gender in flowering plants and mammals. Trends in Genetics. 2001;17:705–711. doi: 10.1016/s0168-9525(01)02519-7. [DOI] [PubMed] [Google Scholar]
- Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Current Biology. 2010;20:271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant and Cell Physiology. 2008;49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genetics. 2010 doi: 10.1371/journal.pgen.1001031. 6, e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes and Development. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Blevins T, Ailhas J, Boller T, Meins F., Jr. Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Research. 2008;36:6429–6438. doi: 10.1093/nar/gkn670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O'Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proceedings of the National Academy of Sciences, USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Zhang W, Gao S, Zhou X, Xia J, Chellappan P, Zhang X, Jin H. Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biology. 2010;11:R81. doi: 10.1186/gb-2010-11-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


