Abstract
Objective
Next-generation sequencing was performed to evaluate the effects of short-term application of dexamethasone on human gingiva-derived mesenchymal stem cells.
Methods
Human gingiva-derived stem cells were treated with a final concentration of 10−7 M dexamethasone and the same concentration of vehicle control. This was followed by mRNA sequencing and data analysis, gene ontology and pathway analysis, quantitative real-time polymerase chain reaction of mRNA, and western blot analysis of RUNX2 and β-catenin.
Results
In total, 26,364 mRNAs were differentially expressed. Comparison of the results of dexamethasone versus control at 2 hours revealed that 7 mRNAs were upregulated and 25 mRNAs were downregulated. The application of dexamethasone reduced the expression of RUNX2 and β-catenin in human gingiva-derived mesenchymal stem cells.
Conclusion
The effects of dexamethasone on stem cells were evaluated with mRNA sequencing, and validation of the expression was performed with qualitative real-time polymerase chain reaction and western blot analysis. The results of this study can provide new insights into the role of mRNA sequencing in maxillofacial areas.
Keywords: Dexamethasone, gingival, messenger RNA, stem cells
Introduction
Mesenchymal stem cells have the capacity for self-renewal and multilineage differentiation. These cells include three lineages that exhibit osteogenic, chondrogenic, and adipogenic differentiation, respectively1,2 Mesenchymal stem cells play a crucial role in tissue engineering and regenerative medicine.3 In recent years, stem cells have been derived from a variety of dental-related tissues such as the periodontal ligament, papilla, follicle, dental pulp of exfoliated deciduous and adult teeth, and maxillary sinus membrane, which represents a particularly rich source of mesenchymal stem cells.4,5 Stem cells from dental-related tissues display multifactorial advantages including a high proliferation rate, high viability, and easy induction to distinct cell lineages.4 Moreover, our group has reported that human gingiva-derived stem cells can be differentiated into osteoblasts under some conditions, including within osteogenic media, and that these cells may be applied to tissue engineering.6,7
Dexamethasone is a glucocorticoid drug that is broadly used to repress allergic reactions and treat autoimmune diseases.8 One study revealed that dexamethasone treatment induced osteonecrosis of the femoral head via inhibition of osteogenic differentiation of human bone marrow stem cells.9 Another study showed that osteoblastic MC3T3-E1 cell proliferation was directly inhibited by dexamethasone via deviant glucocorticoid receptor activation and subsequent P53 activation.10 Moreover, dexamethasone induces bone loss by inhibiting bone formation through suppression of osteoblast proliferation and collagen synthesis and by stimulating mature osteoclasts.11 As a result, high dosages of glucocorticoids can change bone remodeling and promote bone resorption, which can lead to osteoporosis.
In certain responses to a variety of physiological and pathological stimuli, mesenchymal stem cells can proliferate and differentiate into osteoblasts, chondrocytes, and adipocytes.12 Differentiation of multipotent mesenchymal stem cells to the osteoblast lineage and maturation of osteoprogenitors are regulated and activated by transcription factors.13 Among these transcription factors, runt-related transcription factor 2 (RUNX2) and the Wnt/β-catenin signaling pathway are crucial for osteoblast differentiation.14,15 RUNX2 is a bone-specific factor and the master regulator of bone formation.15 This transcription factor has been found to modulate a complex gene-regulatory network during osteoblastogenesis.16 Additionally, ablation of RUNX2 results in the absence of a mineralized skeleton in mice and the development of cleidocranial dysplasia in humans.17 The inactivation of β-catenin in mesenchymal progenitor cells exclusively blocks osteoblast differentiation.14,18 However, whole-level gene changes have not been well described.19 RNA sequencing can detect all coding and non-coding regions in the cell and determine their sequence and structure.20 Massive parallel sequencing can be performed using next-generation sequencing or second-generation sequencing.21 The present study was performed to evaluate the effects of short-term application of dexamethasone on human gingiva-derived mesenchymal stem cells with next-generation sequencing.
Materials and methods
Isolation and culture of human gingiva-derived stem cells
Gingival tissues were collected from healthy patients undergoing clinical crown-lengthening procedures. The design of the study was reviewed and approved by the Institutional Review Board of the Catholic University of Korea, College of Medicine (KC11SISI0348). Informed consent was obtained from all patients according to the Act on Legal Codes for Biomedical Ethics and Safety and the Declaration of Helsinki.
Human gingiva-derived stem cells were isolated and cultivated in accordance with the procedure described in a previous study by one of the authors.6 The gingival tissues were collected and contained in sterile phosphate-buffered saline (Welgene, Inc., Gyeongsan-si, Gyeongsangbuk-do, Korea) that included 100 U/mL penicillin and 100 µg/mL streptomycin (Sigma-Aldrich Co., St. Louis, MO, USA) at 4℃. The tissues were de-epithelialized, separated into 1- to 2-mm2 fragments, digested in 0.2-µm-pore filter, and modified in alpha minimum essential medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing dispase (1 mg/mL; Sigma-Aldrich Co.) and collagenase type IV (2 mg/mL; Sigma-Aldrich Co.) for 30 min at 37℃. The cell suspension was filtered with a 70-µm cell strainer (Falcon; BD Biosciences, Franklin Lakes, NJ, USA), and the cells were incubated at 37℃ in a humidified incubator with 5% CO2. After 24 hours, the nonadherent cells were washed with phosphate-buffered saline and replaced with fresh media every 3 to 4 days.
Cells were plated onto a 100-mm dish (passage 4) at a density of 1.0 × 106 cells/well and cultured in a growth medium including alpha minimum essential medium (Gibco), ascorbic acid (Sigma-Aldrich Co.), fetal bovine serum (Gibco), L-glutamine (Sigma-Aldrich Co.), penicillin, and streptomycin (Sigma-Aldrich Co.). Dexamethasone (Sigma-Aldrich Co.) was dissolved in 95% ethanol to a concentration of 2 × 10−3 M (stock solution). The cells were then treated with a final concentration of 10−7 M dexamethasone and the same concentration of vehicle control (95% ethanol) for 2 and 24 hours. The doses of dexamethasone and the time points used in the present study were based on those used in previously published studies.22,23
Total RNA extraction
The human gingiva-derived stem cells were harvested at 2 and 24 hours. Total RNA was isolated using Trizol reagent (Molecular Research Center, Inc., Cincinnati, OH, USA). RNA quality was assessed with a bioanalyzer (Agilent 2100; Agilent Technologies, Amstelveen, The Netherlands) using the RNA 6000 Nano Chip (Agilent Technologies), and the quantity was determined by spectrophotometer (ND-2000; Thermo Fisher Scientific, Inc.) with a ratio of absorbance of >1.8 at 260 and 280 nm. To confirm high RNA quality for all samples, we checked whether the RNA integrity number was >7.0.
Sequencing of mRNA and analysis of data
A library of control and test RNAs was constructed using SENSE mRNA-Seq Library Prep Kit (Lexogen, Inc., Vienna, Austria) according to the manufacturer’s instructions. Briefly, each 2 µg of total RNA was prepared and incubated with magnetic beads decorated with oligo-dT, and other RNAs (excluding mRNA) were then removed with a washing solution. Library production was initiated by the random hybridization of starter/stopper heterodimers to the poly(A) RNA still bound to the magnetic beads. These starter/stopper heterodimers contained linker sequences compatible with Illumina (San Diego, CA, USA). A single-tube reverse transcription and ligation reaction extends the starter to the next hybridized heterodimer, where the newly synthesized cDNA insert is ligated to the stopper. Second-strand synthesis is performed to release the library from the beads, and the library is then amplified. Barcodes were introduced when the library was amplified. High-throughput sequencing was performed as paired-end 100-base-pair sequencing using HiSeq 2500 (Illumina).
RNA-Seq reads were mapped using the Top Hat software tool (Toronto, ON, Canada) to obtain the alignment file. The alignment file was used to assemble transcripts, estimate their abundances, and detect differential expressions of genes or isoforms using cufflinks. Gene classification was based on searches performed using the BioCarta (http://www.biocarta.com/), GenMAPP (http://www.genmapp.org/), DAVID (http://david.abcc.ncifcrf.gov/), and Medline databases (http://www.ncbi.nlm.nih.gov/).
Gene ontology and pathway analysis
Gene ontology analysis was performed and included osteoblast differentiation, bone remodeling, and Wnt signaling. P values were used to denote significance of gene ontology term enrichment (P < 0.05 was considered statistically significant). Pathway analysis was also performed for differentially expressed genes based on the PATHWAY database of the Kyoto Encyclopedia of Genes and Genomes. A fold change of 1.3 was applied in this study, and a log2 normalized read count of ≥4 was applied to minimize false counts. A P value of <0.05 was considered statistically significant.
Quantification by real-time polymerase chain reaction
Total RNA was isolated from cultured cells using GeneJET RNA Purification kit (Thermo Fisher Scientific, Inc.) and reverse-transcribed. The sense and antisense primers were designed based on GenBank. Primer sequences were as follows: RUNX2 Forward 5′-AAT GAT GGT GTT GAC GCT GA-3′, Reverse 5′-TTG ATA CGT GTG GGA TGT GG-3′; β-catenin Forward 5′-GAG GGG TGG GCT GGT ATC TC-3′, Reverse 5′-CTC GAC CAA AAA GGA CCA GA-3′; and β-actin Forward 5′-TGGCACCCAGCACAATGAA-3′, Reverse 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′. β-actin served as a housekeeping gene for normalization. The expression of mRNA was detected by real-time polymerase chain reaction (RT-PCR) using SYBR Green Real-Time PCR Master Mixes (Enzynomics, Daejeon, Korea) according to the manufacturer’s protocol. Quantitative RT-PCR experiments were repeated three times.
Western blot analysis
Cells were lysed in ice-cold RIPA lysis and extraction buffer (Thermo Fisher Scientific, Inc.) according to the manufacturer’s protocols. Whole cell lysates were quantified using the BCA assay (Thermo Fisher Scientific, Inc.). Protein samples were loaded and then transferred for immunoblotting. The membranes were incubated with the following primary antibodies overnight at 4℃: anti-RUNX2 antibody (Abcam, Cambridge, UK), anti-β-catenin (BD Biosciences, San Jose, CA, USA), or anti-GAPDH antibody (Abcam). After washing, membranes were incubated with secondary antibody (Abcam) for 1 hour at room temperature.
Data analysis
mRNA-Seq reads were mapped using Top Hat software to obtain the alignment file. Differentially expressed genes were determined based on counts from unique and multiple alignments using EdgeR within R version 3.2.2 (R development Core Team, 2011) with BIOCONDUCTOR version 3.0 (Gentleman et al., 2004). The alignment file was also used to assemble transcripts, estimate their abundances, and detect differential expression of genes or isoforms using cufflinks. We calculated the fragments per kilobase of exon per million fragments to determine the expression level of the gene regions. Global normalization was used for comparison between samples. Gene classification was based on searches done by DAVID (http://david.abcc.ncifcrf.gov/). The data are represented as mean ± standard deviation of the experiments. Either Student’s t-test or a two-way analysis of variance with a post-hoc test was performed to determine the differences between the groups using a commercially available program (SPSS 12 for Windows; SPSS Inc., Chicago, IL, USA). The level of significance was 0.05.
Results
Gene ontology
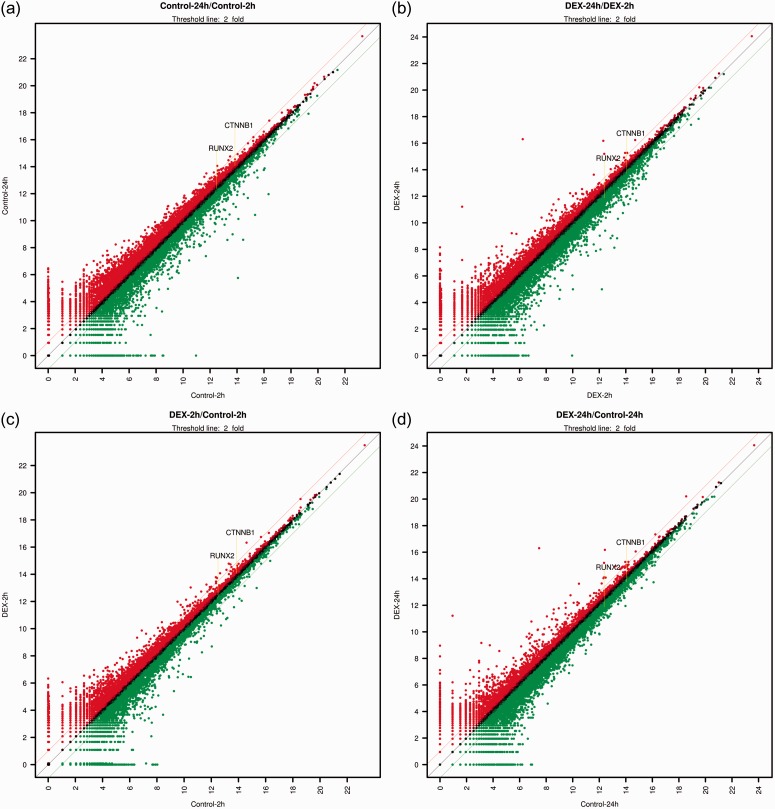
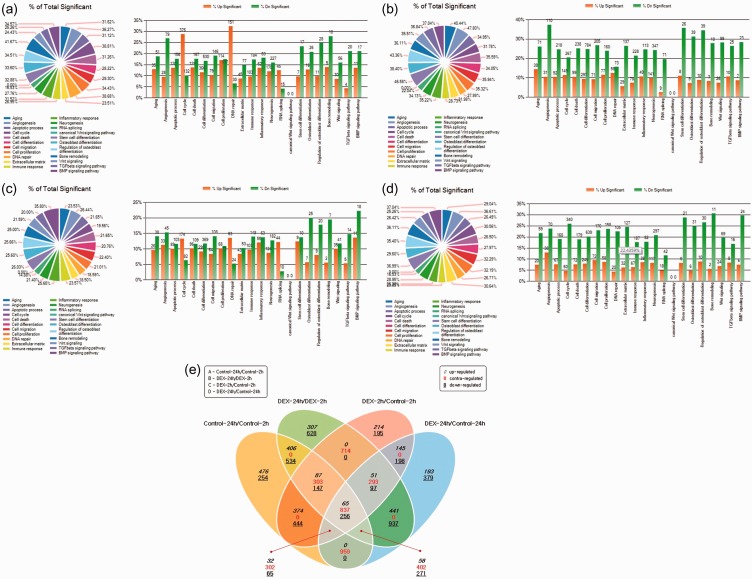
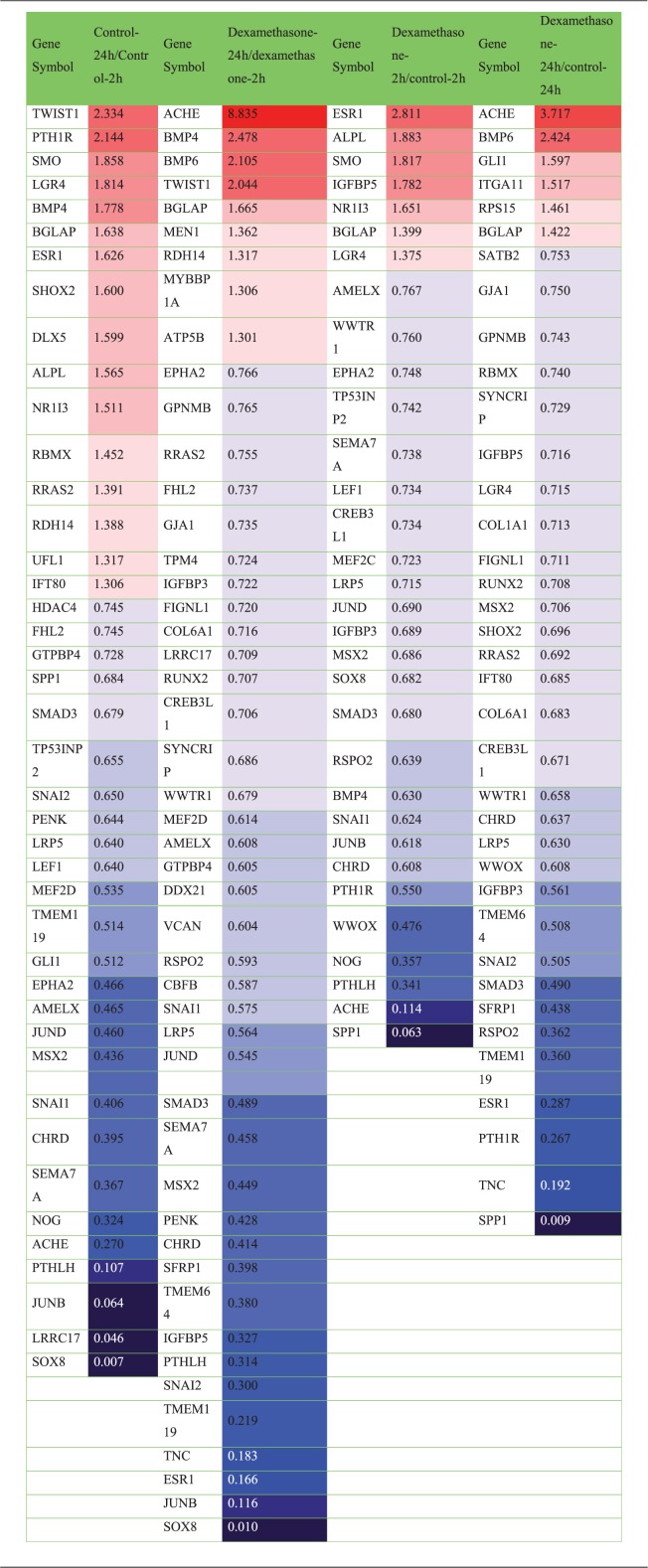
In total, 26,364 mRNAs were differentially expressed. Scatter plots, which differentially expressed the mRNAs, are shown in Figure 1. The gene ontology analysis results of the differentially expressed mRNAs are shown in Figure 2. We focused especially on osteoblast differentiation (fold change of 1.3, log2 normalized read count of ≥4 to minimize false counts, P < 0.05) (Table 1 and Figure 3). First, we assessed the effect of incubation time on the stem cells. Comparison of the controls at 24 versus 2 hours revealed that 16 mRNAs were upregulated and 26 were downregulated. Comparison of the dexamethasone group at 24 versus 2 hours revealed that 9 mRNAs were upregulated and 37 were downregulated. Comparison of the data in both groups at 24 versus 2 hours showed that 4 genes were upregulated and 17 genes were downregulated. The four genes that were upregulated were TWIST1, BMP4, BGLAP, and RDH14, and the 17 genes that were downregulated were FHL2, GTPBP4, SMAD3, SNAI2, PENK, LRP5, MEF2D, EPHA2, AMELX, JUND, MSX2, CHRD, SEMA7A, PTHLH, JUNB, LRRC17, and SOX8. However, two genes (ESR1 and ACHE) showed opposite trends.
Figure 1.
Scatter plots showing the expression in untreated controls at 2 and 24 hours and dexamethasone-treated cells at 2 and 24 hours (x, y-axis: relative expression levels; Red: Expression level of y-value is higher than expression level of x-value; Green: Expression level of y-value is lower than expression level of x-value). (a) Control at 24 hours/control at 2 hours, (b) Dexamethasone at 24 hours/dexamethasone at 2 hours, (c) Dexamethasone at 2 hours/control at 2 hours, (d) Dexamethasone at 24 hours/control at 24 hours.
Figure 2.
Gene ontology analysis of mRNA expression. (a) Control at 24 hours/control at 2 hours. (b) Dexamethasone at 24 hours/dexamethasone at 2 hours. (c) Dexamethasone at 2 hours/control at 2 hours. (d) Dexamethasone at 24 hours/control at 24 hours. (e) Venn diagram analysis (fold change, 1.3; log2 normalized read counts of ≥4 were selected; P < 0.05).
Table 1.
Differentially expressed mRNA related to osteoblast differentiation (fold change, 1.3; log2 normalized read counts of ≥4 were selected; P < 0.05).
Figure 3.
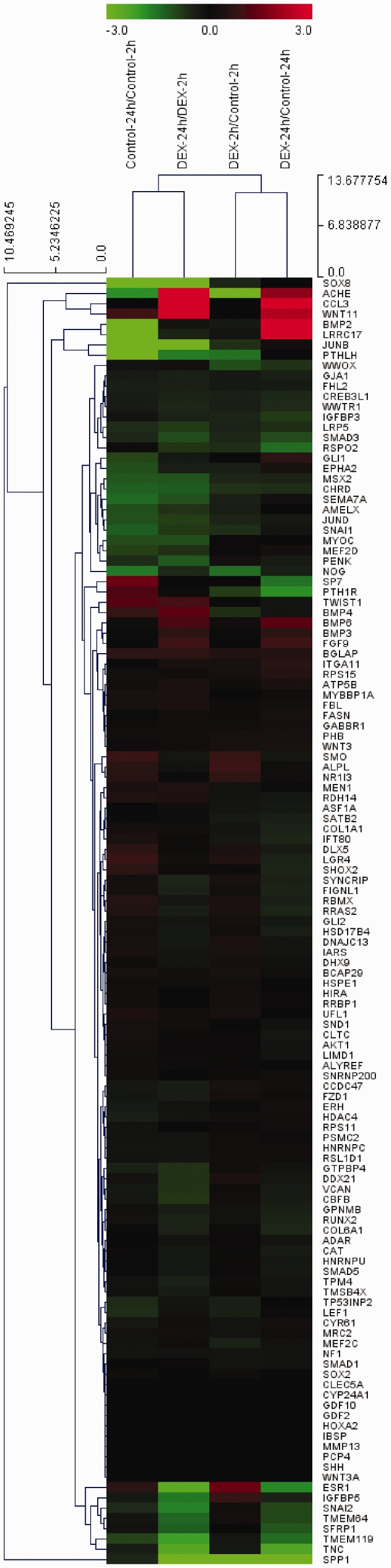
Clustering analysis of differentially expressed mRNA related to osteoblast differentiation (fold change, 1.3; log2 normalized read counts of ≥4 were selected; P < 0.05).
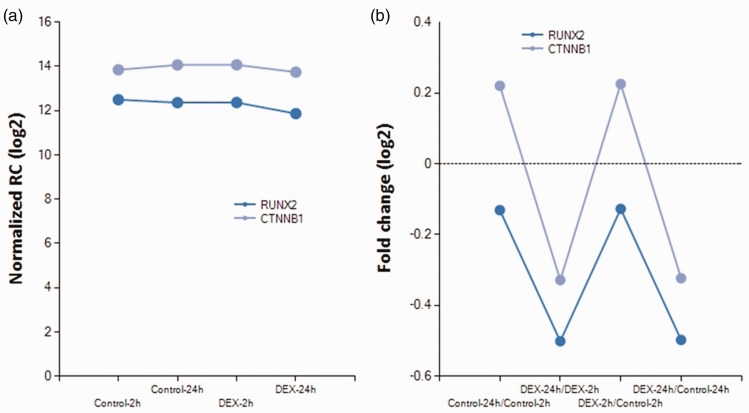
Next, we assessed the effect of dexamethasone. Comparison of the dexamethasone and control groups at 2 hours showed that 7 mRNAs were upregulated and 25 mRNAs were downregulated. Comparison of the dexamethasone and control groups at 24 hours revealed that 6 mRNAs were upregulated and 31 mRNAs were downregulated. In both groups, 1 gene (BGLAP) was upregulated and 11 genes (WWTR1, CREB3L1, LRP5, IGFBP3, MSX2, SMAD3, RSPO2, CHRD, PTH1R, WWOX, and SPP1) were downregulated. However, four genes (ESR1, IGFBP5, LGR4, and ACHE) showed opposite trends. Expression of RUNX2 was decreased in the dexamethasone group at 24 hours. Decreased expression of CTNNB1 (for β-catenin expression) was also noted in the dexamethasone group at 24 hours (Figure 4).
Figure 4.
Change in expression of RUNX2 and CTNNB1 (for β-catenin expression) (fold change, 1.3; log2 normalized read counts of ≥4 were selected; P < 0.05).
Validation of mRNA expression
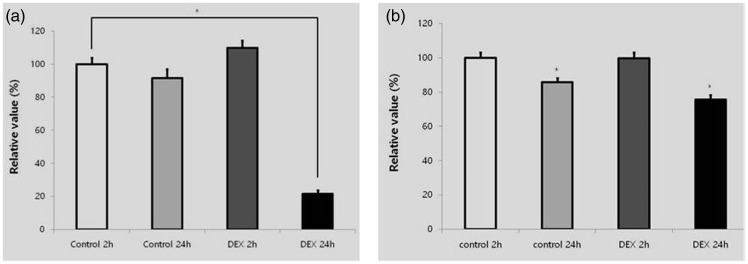
Quantitative RT-PCR revealed that the mRNA levels of RUNX2 and β-catenin were lower at 24 than at 2 hours (Figure 5). This change was more significant in the dexamethasone group at 24 hours.
Figure 5.
Validation of RUNX2 mRNA and β-catenin mRNA. (a) Expression of RUNX2. (b) Expression of β-catenin. *Statistically significant differences compared with the control at 2 hours.
Western blot analysis
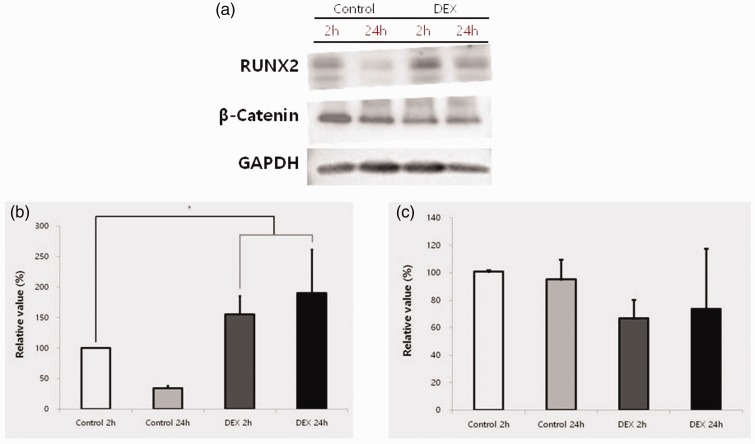
Western blot analysis was performed to detect protein expression following treatment with dexamethasone at 2 and 24 hours compared with the untreated group at 2 and 24 hours. Normalization of the protein expressions revealed 34.3% ± 4.3% expression of RUNX2 in the control group at 24 hours and 155.5% ± 29.8% and 190.3% ± 71.3% expression of RUNX2 in the dexamethasone group at 2 and 24 hours, respectively, when the control value at 2 hours was considered to be 100% (P < 0.05) (Figure 6). The expression of β-catenin in the control group was not significantly different between 2 and 24 hours. Normalization of the protein expression revealed 95.2% ± 14.5% expression of β-catenin in the control group at 24 hours and 66.8% ± 13.5% and 73.6% ± 43.9% expression of β-catenin in the dexamethasone group at 2 and 24 hours, respectively, when the control value at 2 hours was considered to be 100% (Figure 6).
Figure 6.
Western blot analysis for expression of RUNX2 and β-catenin. (a) Evaluation of protein expressions of RUNX2, β-catenin, and GAPDH. (b) Quantitative analysis of protein expressions of RUNX2 after normalization with GAPDH levels by densitometry. (c) Quantitative analysis of protein expressions of β-catenin after normalization with GAPDH levels by densitometry. *Statistically significant differences compared with the control at 2 hours.
Discussion
In this report, we examined the effects of predetermined concentrations of dexamethasone on stem cells at 2 and 24 hours. Sequencing of mRNA and validation of the expression was performed with qualitative RT-PCR and western blot analysis. The application of dexamethasone clearly resulted in reduced expression of RUNX2 and β-catenin in human gingiva-derived mesenchymal stem cells.
Stem cells derived from the gingiva were used in the present study; these cells have also been applied to tissue engineering.24 Human gingiva-derived mesenchymal stem cells from the maxillofacial region can be considered a favorable source of mesenchymal stem cells because harvesting these cells from the mandible or maxilla can easily be performed under local anesthesia.6,25 Moreover, one study revealed that human gingiva-derived mesenchymal stem cells are superior to bone marrow-derived mesenchymal stem cells for cell therapy in regenerative medicine.26
Two time points (2 and 24 hours) were selected to evaluate the effects of the dexamethasone incubation time. The immediate effects of dexamethasone were evaluated using the 2-hour results, and the more sustained effects were evaluated by using the 24-hour results. More pronounced effects were observed at 24 hours because of the cumulative effects of dexamethasone. Short-term, high-dose dexamethasone reportedly predisposes patients to complicated fracture union, especially patients with angle fractures.27 However, long-term physiologic concentrations of dexamethasone resulted in more intense staining for alkaline phosphatase in human bone-derived cells.28
Next-generation sequencing can be performed in a high-throughput manner and is more cost-effective than Sanger sequencing.21 This next-generation sequencing can be applied in several ways: gene expression profiling, chromatin immunoprecipitation sequencing, DNA methylation, de novo genome sequencing, metagenomics, noninvasive prenatal testing, identification of disease-related genes, assessment of human disease and health, and single-molecule and long-read sequencing.29 The wide range of applications facilitated by automatic sequencing analyzers, the simple workflow, and a 100-fold decrease in cost have brought us significantly closer to understanding the links between genotype and phenotype and establishing the molecular basis of many diseases.30 HiSeq (Illumina), Roche 454 (Roche, Branford, CT, USA), and SOLiD (Life Technologies, Inc., Gaithersburg, MD, USA) systems are representative sequencers used for next-generation sequencing. Among them, the HiSeq is reportedly has particularly high-speed data quality and complete end-to-end sequencing solutions.31
The sequencing data showed the expression of a total of 26,364 mRNAs. Gene ontology analysis of mRNAs related to osteoblast differentiation was then performed. Previously, mRNA expression was measured by microarray methods or RT-PCR techniques.32 These techniques are limited to the detection of known transcripts and have a limited capacity to differentiate between transcript variants.33 Although RNA sequencing is still a technology under active development, it offers several advantages over existing technologies, including determination of all coding and non-coding regions; additionally, it is a popular method for genome analysis.20,34 Notably, however, genes with low expression abundance might not be detected.35
Western blot analysis was performed to detect the protein expression of RUNX2 and β-catenin and provide information regarding possible mechanisms. Canonical Wnt/β-catenin signaling pathways are reportedly crucial for the osteoblastic lineage.36 Wnt signaling suppresses mesenchymal stem cell commitment to the chondrogenic and adipogenic lineages and enhances the osteoblastic lineage, and both RUNX2 and β-catenin are major regulators of this signaling.37 RUNX2 regulates a complex gene-regulatory network during osteoblastogenesis and upregulates various osteoblast lineage-specific genes.38 One study revealed that RUNX2-targeting miRNAs might be essential to ensure that the RUNX2 level is attenuated at key stages of osteoblast lineage progression to accommodate the biological functions of RUNX2 in mesenchymal cell-fate determination and maturation of osteoblasts.13 Inactivation of β-catenin in mesenchymal stem cells was shown to totally block osteoblast differentiation as well as mesenchymal cell differentiation into chondrocytes in the perichondrium and calvarium.39 Dexamethasone has been shown to induce osteoporosis mainly by suppressing osteoblast-mediated osteogenesis; however, the precise mechanisms involved in these signaling pathways remain unclear.40 For this reason, we examined the canonical Wnt/β-catenin signaling pathways in the present study and focused on changes in RUNX2 and β-catenin. The expression of RUNX2 was decreased by 70% after 24 hours of incubation with dexamethasone; the result for β-catenin was similar to this. We performed quantitative RT-PCR and western blot to validate this result by comparing mRNA and protein levels. Moderate consistency was found between the sequencing data and the RT-PCR results.
The effects of dexamethasone on stem cells were evaluated by mRNA sequencing, and the expression was validated by quantitative RT-PCR and western blot analysis. The results clearly showed that the application of dexamethasone reduced the expression of RUNX2 and β-catenin in human gingiva-derived mesenchymal stem cells. This study may provide new insights into the role of mRNA sequencing in maxillofacial areas.
Ethical approval statement
The design of the study was reviewed and approved by the Institutional Review Board of the Catholic University of Korea, College of Medicine (KC11SISI0348). Informed consent was obtained from all patients according to the Act on Legal Codes for Biomedical Ethics and Safety and the Declaration of Helsinki.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Declaration of conflicting interest
The Authors declare that there is no conflict of interest.
Funding
This research was partly supported by the Research Fund of Seoul St. Mary’s Hospital, The Catholic University of Korea and partly supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, Information and Communication Technology & Future Planning (NRF-2014R1A1A1003106).
References
- 1.Huang GJ, Gronthos S, Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dent Res 2009; 88: 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baksh D, Song L, Tuan R. Adult mesenchymal stem cells: characterization, differentiation, and application in cell and gene therapy. J Cell Mol Med 2004; 8: 301–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seo BM, Miura M, Gronthos S, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet 2004; 364: 149–155. [DOI] [PubMed] [Google Scholar]
- 4.Lei M, Li K, Li B, et al. Mesenchymal stem cell characteristics of dental pulp and periodontal ligament stem cells after in vivo transplantation. Biomaterials 2014; 35: 6332–6343. [DOI] [PubMed] [Google Scholar]
- 5.Hakki SS, Kayis SA, Hakki EE, et al. Comparison of mesenchymal stem cells isolated from pulp and periodontal ligament. J Periodontol 2015; 86: 283–291. [DOI] [PubMed] [Google Scholar]
- 6.Jin SH, Lee JE, Yun JH, et al. Isolation and characterization of human mesenchymal stem cells from gingival connective tissue. J Periodontal Res 2015; 50: 461–467. [DOI] [PubMed] [Google Scholar]
- 7.Kim BB, Ko Y, Park JB. Effects of risedronate on the morphology and viability of gingiva-derived mesenchymal stem cells. Biomed Rep 2015; 3: 845–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li T, Li H, Li T, et al. MicroRNA expression profile of dexamethasone-induced human bone marrow-derived mesenchymal stem cells during osteogenic differentiation. J Cell Biochem 2014; 115: 1683–1691. [DOI] [PubMed] [Google Scholar]
- 9.Cárcamo-Orive I, Gaztelumendi A, Delgado J, et al. Regulation of human bone marrow stromal cell proliferation and differentiation capacity by glucocorticoid receptor and AP-1 crosstalk. J Bone Miner Res 2010; 25: 2115–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Qian W, Weng X, et al. Glucocorticoid receptor and sequential P53 activation by dexamethasone mediates apoptosis and cell cycle arrest of osteoblastic MC3T3-E1 cells. PLoS One 2012; 7: e37030–e37030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laxman N, Rubin CJ, Mallmin H, et al. Second generation sequencing of microRNA in human bone cells treated with parathyroid hormone or dexamethasone. Bone 2016; 84: 181–188. [DOI] [PubMed] [Google Scholar]
- 12.Zhou S. TGF-β regulates β-catenin signaling and osteoblast differentiation in human mesenchymal stem cells. J Cell Biochem 2011; 112: 1651–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Xie RL, Croce CM, et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc. Natl. Acad. Sci. U.S.A 2011; 108: 9863–9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haxaire C, Haÿ E, Geoffroy V. Runx2 controls bone resorption through the down-regulation of the Wnt pathway in osteoblasts. Am J Pathol 2016; 186: 1598–1609. [DOI] [PubMed] [Google Scholar]
- 15.Kook SH, Heo JS, Lee JC. Crucial roles of canonical Runx2-dependent pathway on Wnt1-induced osteoblastic differentiation of human periodontal ligament fibroblasts. Mol Cell Biochem 2015; 402: 213–223. [DOI] [PubMed] [Google Scholar]
- 16.Hecht J, Seitz V, Urban M, et al. Detection of novel skeletogenesis target genes by comprehensive analysis of a Runx2(−/−) mouse model. Gene Expr Patterns 2007; 7: 102–112. [DOI] [PubMed] [Google Scholar]
- 17.Otto F, Kanegane H, Mundlos S. Mutations in the RUNX2 gene in patients with cleidocranial dysplasia. Human mutat 2002; 19: 209–216. [DOI] [PubMed] [Google Scholar]
- 18.Day TF, Guo X, Garrett-Beal L, et al. Wnt/β-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev cell 2005; 8: 739–750. [DOI] [PubMed] [Google Scholar]
- 19.Laxman N, Rubin CJ, Mallmin H, et al. Second generation sequencing of microRNA in human bone cells treated with parathyroid hormone or dexamethasone. Bone 2016; 84: 181–188. [DOI] [PubMed] [Google Scholar]
- 20.t' Hoen PA, Friedländer MR, Almlöf J, et al. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat Biotechnol 2013; 31: 1015–1022. [DOI] [PubMed] [Google Scholar]
- 21.Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods 2008; 5: 16–18. [DOI] [PubMed] [Google Scholar]
- 22.Park JB. The effects of dexamethasone, ascorbic acid, and beta-glycerophosphate on osteoblastic differentiation by regulating estrogen receptor and osteopontin expression. J Surg Res 2012; 173: 99–104. [DOI] [PubMed] [Google Scholar]
- 23.Park JB. Effects of the combination of dexamethasone and fibroblast growth factor2 on differentiation of osteoprecursor cells. Mol Med Rep 2014; 9: 659–662. [DOI] [PubMed] [Google Scholar]
- 24.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5: R80. [DOI] [PMC free article] [PubMed]
- 25.Jin SH, Kweon H, Park JB, et al. The effects of tetracycline-loaded silk fibroin membrane on proliferation and osteogenic potential of mesenchymal stem cells. J Surg Res 2014; 192: e1–e9. [DOI] [PubMed] [Google Scholar]
- 26.Park JB, Kim YS, Lee G, et al. The effect of surface treatment of titanium with sand-blasting/acid-etching or hydroxyapatite-coating and application of bone morphogenetic protein-2 on attachment, proliferation, and differentiation of stem cells derived from buccal fat pad. Tissue Eng Regen Med 2013; 10: 115–121. [Google Scholar]
- 27.Tomar GB, Srivastava RK, Gupta N, et al. Human gingiva-derived mesenchymal stem cells are superior to bone marrow-derived mesenchymal stem cells for cell therapy in regenerative medicine. Biochem Biophys Res Commun 2010; 393: 377–383. [DOI] [PubMed] [Google Scholar]
- 28.Snall J, Apajalahti S, Suominen AL, et al. Influence of perioperative dexamethasone on delayed union in mandibular fractures: a clinical and radiological study. Med Oral Patol Oral Cir Bucal 2015; 20: e621–e626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong MM, Rao LG, Ly H, et al. Long-term effects of physiologic concentrations of dexamethasone on human bone-derived cells. J Bone Miner Res 1990; 5: 803–813. [DOI] [PubMed] [Google Scholar]
- 30.Buermans HPJ, den Dunnen JT. Next generation sequencing technology: advances and applications. BBA Mol Basis Dis 2014; 1842: 1932–1941. [DOI] [PubMed] [Google Scholar]
- 31.Janitz M. Next-generation genome sequencing: towards personalized medicine, Weinheim, Germany: John Wiley & Sons, 2011. [Google Scholar]
- 32.Levy SE, Myers RM. Advancements in next-generation sequencing. Annu Rev Genomics Hum Genet 2016; 17: 95–115. [DOI] [PubMed] [Google Scholar]
- 33.Mutz KO, Heilkenbrinker A, Lönne M, et al. Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol 2013; 24: 22–30. [DOI] [PubMed] [Google Scholar]
- 34.Zhou W, Calciano MA, Jordan H, et al. High resolution analysis of the human transcriptome: detection of extensive alternative splicing independent of transcriptional activity. BMC Genet 2009; 10: 63–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarazona S, Garcia-Alcalde F, Dopazo J, et al. Differential expression in RNA-seq: a matter of depth. Genome Res 2011; 21: 2213–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regard JB, Zhong Z, Williams BO, et al. Wnt signaling in bone development and disease: making stronger bone with Wnts. Cold Spring Harb Perspect Biol 2012; 4: a007997–a007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med 2013; 19: 179–192. [DOI] [PubMed] [Google Scholar]
- 39.Wu H, Whitfield TW, Gordon JA, et al. Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biol 2014; 15: 1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hill TP, Später D, Taketo MM, et al. Canonical Wnt/β-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell 2005; 8: 727–738. [DOI] [PubMed] [Google Scholar]
- 41.Hayashi K, Yamaguchi T, Yano S, et al. BMP/Wnt antagonists are upregulated by dexamethasone in osteoblasts and reversed by alendronate and PTH: potential therapeutic targets for glucocorticoid-induced osteoporosis. Biochem Biophys Res Commun 2009; 379: 261–266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article.