Abstract
The activity of the small GTPase, Rac1, plays a role in various cellular processes including cytoskeletal rearrangement, gene transcription, and malignant transformation. In this report constitutively active Rac1 (Rac V12) is shown to stimulate the activation of STAT3, a member of the family of signal transducers and activators of transcription (STATs). The activity of Rac1 leads to STAT3 translocation to the nucleus coincident with STAT3-dependent gene expression. The expression of Vav (Δ1–187), a constitutively active guanine nucleotide exchange factor for the Rho GTPases, or activated forms of Ras or Rho family members, leads to STAT3-specific activation. The activation of STAT3 requires tyrosine phosphorylation at residue 705, but is not dependent on phosphorylation of Ser-727. Our studies indicate that Rac1 induces STAT3 activation through an indirect mechanism that involves the autocrine production and action of IL-6, a known mediator of STAT3 response. Rac V12 expression results in the induction of the IL-6 and IL-6 receptor genes and neutralizing antibodies directed against the IL-6 receptor block Rac1-induced STAT3 activation. Furthermore, inhibition of the nuclear factor-κB activation or disruption of IL-6-mediated signaling through the expression of IκBα S32AS36A and suppressor of cytokine signaling 3 , respectively, blocks Rac1-induced STAT3 activation. These findings elucidate a mechanism dependent on the induction of an autocrine IL-6 activation loop through which Rac1 mediates STAT3 activation establishing a link between oncogenic GTPase activity and Janus kinase/STAT signaling.
Cells respond to a diverse array of extracellular stimuli that direct proliferation, growth arrest, differentiation, or apoptosis. Many cytokines elicit biological effects with the activation of a specific family of transcription factors known as signal transducers and activators of transcription (STATs; ref. 1). STATs reside latent in the cytoplasm and are activated after tyrosine phosphorylation by Janus kinases (JAKs) associated with cytokine receptors (2–5). Subsequent to tyrosine phosphorylation, STATs form dimers and translocate to the nucleus where they bind specific DNA targets and induce the transcription of responsive genes. Some members of the STAT family play a role in cellular proliferation. Moreover, accumulating evidence suggests that abnormal STAT regulation may be involved in oncogenic transformation. STAT3 has been identified as a potential oncogene, because a gain-of-function mutation renders cells transformed and tumorigenic (6). Additionally, constitutive activation of STAT3 has been reported in several human tumor cells and in cells transformed by various oncoproteins (7–9). Hence, identifying mediators of STAT3 activation may enhance our understanding of malignant transformation events.
Recent studies have suggested a link between STAT3 activity and small GTPases (10–13). Small GTPases comprise a family of more than 100 monomeric G proteins that function as molecular switches in cellular signaling (14). Small GTPases cycle between an active, GTP-bound, and an inactive, GDP-bound, state and are structurally classified into five subfamilies: Ras, Rho, Rab, Arf, and Ran. Members of the Rab, Arf, and Ran subfamilies function primarily in vesicular and nuclear/cytoplasmic trafficking and spindle microtubule assembly events, whereas Ras and Rho family members play key roles in gene induction and cytoskeletal rearrangement. Mutations of the Ras protooncogene that render a constitutively active oncogenic form are commonly identified in many types of tumors (15). Rac1 and Cdc42, Rho family members, have been shown to be involved in Ras-induced cellular transformation (16–19). Small GTPases are usually regulated by specific guanine nucleotide exchange factors (GEFs) that stimulate the exchange of GTP for GDP. Gain-of-function mutations in Vav (a Rho family GEF) also result in oncogenesis (20). Although the signaling pathways used by the small GTPases to elicit cellular transformation seem to involve the activation of a variety of kinases, reactive oxygen intermediates, and transcription factors, the mechanisms of malignant transformation are still not completely understood. In this report we present evidence that the JAK/STAT3 pathway is an indirect target of Ras and Rho GTPases. Persistent Rac1 activity leads to the autocrine production and signal transduction of IL-6. IL-6, a pleiotropic cytokine functioning in immune and inflammatory responses, is a known activator of STAT3 (21, 22). We show constitutively active Rac1 results in the induction of this cytokine and its receptor. Blocking the IL-6 signaling pathway inhibits Rac1-mediated STAT3-dependent gene expression. These findings identify an indirect link between two diverse signal transduction pathways as a result of oncogenic mutations.
Materials and Methods
Cells and Reagents.
Rat1, HeLa (ATCC, CL2), and HT1080 cells (ATCC) were grown in DMEM with 8% (vol/vol) FBS. Recombinant human and rat IL-6 and neutralizing anti-human IL-6 receptor antibodies were obtained from BioSource International (Camarillo, CA). Soluble human IL-6 receptor peptide was obtained from R & D Systems and was used in conjunction with exogenous IL-6 to improve the IL-6 responsiveness of HeLa cells.
Plasmids.
Plasmids encoding constitutively active QL forms of Rac1, RhoG, Cdc42, and RhoA (23), and Rac V12, Rac V12H40, Rac V12L37, Rac N17 (17) were used. Plasmids encoding IκBα S32AS36A and SOCS3 have been described (24, 25). Site-directed mutagenesis with the Quick Change Site-Directed Mutagenesis kit (Stratagene) was performed to create the STAT3 mutants: STAT3 Y705F and STAT3 S727A. The STAT3 fusion protein with the green fluorescent protein tag (STAT3-GFP), contains the full-length cDNA of STAT3 with an inserted Kozak sequence and was generated by a PCR with Pfu polymerase (Stratagene) and subcloned into pEGFP-N1 (CLONTECH).
Transient Transfection Assays.
Cells were seeded (2 × 105 cells per well) in six-well plates and were transfected the next day by calcium phosphate-DNA precipitates (26). Experiments were performed with a total of 24 μg of DNA divided among three wells to serve as triplicate samples. Cells were cotransfected with luciferase reporter gene constructs (4 μg) regulated either by three copies of the STAT DNA-binding sequence from the IRF-1 promoter upstream of the minimal thymidine kinase promoter [(GAS)3-Luc] (27) or by five copies of the NF-κB DNA binding site [(NF-κB)5-Luc] (Stratagene). Cells were cotransfected with 5–7 μg of wild-type or mutant forms of STATs, Vav, Rho family GTPases, or pcDNA3 control vector. Data were normalized to the activity of a cotransfected simian virus 40 (SV40)-driven LacZ reporter gene (SV40-LacZ). Cell lysates were prepared for luciferase and β-galactosidase (β-gal) activity assays per the manufacturer's instructions 24–48 h after transfection (Promega).
Fluorescent Microscopy.
Cells seeded on glass coverslips were transfected with the STAT3-GFP fusion construct and evaluated 18–24 h after transfection. Cells were fixed with 4% (vol/vol) paraformaldehyde and visualized with a Zeiss Axioskop equipped for epifluorescence with a GFP filter set (Chroma Technology, Brattleboro, VT). Images were captured with a Diagnostic Instruments Spot 2 camera.
Reverse Transcription–PCR.
Total RNA was prepared by using Trizol (GIBCO/BRL). First-strand cDNA synthesis was performed with reverse transcriptase, 1 μg of total RNA, and oligo(dT) primers by using the Advantage RT-for PCR kit (CLONTECH). The cDNA was amplified by PCR with human IL-6, IL-6 receptor, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primer pairs (BioSource International, Camarillo, CA). The IL-6 primer pairs used were 5′-TTCAATGAGGAGACTTGCCTG-3′ and 5′-ACAACAACAATCTGAGGTGCC-3′. The IL-6 receptor primer pairs used were 5′-GTGAGGAAGTTTCAGAACAGTCCG-3′ and 5′-TGGGAGGCTTGTCGCATTTG-3′. The thermocycling parameters began with 94°C for 1 min 30 sec followed by 28–30 cycles of: 94°C 30 sec, 54°C 45 sec, 72°C 45 sec, followed by 7 min at 72°C. PCR products were separated on 2% agarose gels.
Results
Activation of STAT3-Dependent Gene Expression by Small GTPases.
To examine whether the activity of small GTPases could lead to STAT3 activation, STAT3-dependent gene expression was analyzed. Cells were cotransfected with a STAT-responsive luciferase reporter gene, a LacZ transfection control gene, STAT3, and activated forms of the small GTPases H-Ras V12 or K-Ras V12 (28). The constitutive activity of both Ras proteins resulted in STAT3-dependent gene induction (Fig. 1A). Because Ras regulates several downstream signaling molecules including the small GTPase Rac1 and the protein kinase Raf1, we tested the ability of these proteins to elicit STAT3-dependent promoter activity. The activated form of Raf1 (Raf CAAX) (29) had no effect on STAT-3 activation; however, the expression of the activated form of Rac1 (Rac V12) (17) resulted in a 10-fold increase in transcriptional activation of STAT3 (Fig. 1B). Because expression of the STAT-responsive reporter gene can be induced by any of the activated mammalian STAT proteins, we tested whether the effect of Rac V12 on transcriptional activation was specific to STAT3 in fibroblasts. When STAT family members STAT1, STAT2, STAT3, STAT5, or STAT6 were cotransfected with Rac V12, the STAT-driven gene induction in response to Rac V12 expression was specific for STAT3 (Fig. 2A).
Figure 1.
Effect of H-Ras, K-Ras, Rac1, and Raf expression on the transcriptional activation of STAT3. Rat1 cells were cotransfected with (GAS)3-Luc, SV40-LacZ, and pCDNA3 (control) or STAT3 either alone (open bars) or with (A) H-Ras V12 and K-Ras V12 or (B) Rac V12 and Raf CAAX (solid bars). Cell lysates were prepared 48 h after transfection and used to measure luciferase and β-gal activity. The data presented are means ± SEM (n = 3) and represent one of three similar experiments.
Figure 2.
Effect of Rho family members and oncogenic Vav (Δ1–187) expression on STAT activation. (A) Rat1 cells were cotransfected with (GAS)3-Luc, SV40-LacZ, pCDNA3 (control), and STATs 1, 2, 3, 5a, and 6 either alone (open bars) or with Rac V12 or oncogenic Vav (Δ1–187) (solid bars). (B) Rat1 cells were similarly cotransfected with either constitutively active forms of Rac1, RhoA, Cdc42, and RhoG alone (open bars) or with STAT3 (solid bars). Cell lysates were prepared 48 h after transfection and used to measure both luciferase activity and β-gal activity. The data presented are means ± SEM (n = 3) and represent one of four similar experiments.
G proteins are usually regulated by GEFs which stimulate the exchange of GDP for GTP and thereby lead to the activation of the GTPase. Vav is an exchange factor that acts catalytically to stimulate several Rho family GTPases, including Rac1 (30). Because Rac V12 activates STAT3-dependent gene expression, a constitutively active form of Vav (Δ1–187) was expected to have a similar effect on transcriptional activation. This expectation was in fact the case, because coexpression of the oncogenic form of Vav (Δ1–187) with STAT3 led to more than a 10-fold induction of the responsive gene (Fig. 2A). Similarly, the constitutive activity of Vav-2 (31), a ubiquitously expressed Rho family GEF, resulted in STAT3-dependent gene expression (data not shown).
Because Vav can act as an exchange factor for several Rho family GTPases, we tested the ability of other Rho proteins to stimulate STAT3 activation. Constitutively active forms of Rac1, RhoG, Cdc42, and RhoA (23) were found to induce transcriptional activation of STAT3 (Fig. 2B). These results suggest that STAT3-specific gene induction may result from common signaling pathways used by various Rho family members.
Mutations That Affect Activity.
STAT transcription factors reside in a latent state in the cytoplasm of the cell and are activated by tyrosine phosphorylation. After activation the STATs dimerize and translocate to the nucleus to bind specific DNA response elements. The critical tyrosine residue for STAT3 phosphorylation is Y705 (32). It has recently been suggested that phosphorylation of Ser-727 may contribute to the maximal transcriptional potential of STAT3 (33, 34). By expressing the STAT3 mutants STAT3 Y705F or STAT3 S727A with Rac V12, we found tyrosine 705 to be critical for STAT3 activation in response to Rac V12 expression but not Ser-727 (Fig. 3A). Consistent with these findings, STAT3β, an alternate spliced form of STAT3 that lacks Ser-727 (35), was as transcriptionally active as STAT3 in the presence of Rac V12 expression (data not shown). The STAT3 S727A mutant was also equivalent in its response to IL-6 treatment as wild-type STAT3 in our system (data not shown).
Figure 3.
Functional analysis of mutants of STAT3 and Rac1 on transcriptional activation. Rat1 cells were cotransfected with (GAS)3-Luc, SV40-LacZ, and pCDNA3 (control) alone (open bars); (A) with wild-type STAT3, STAT3 Y705F, or STAT3 S727A in the presence of Rac V12 (solid bars); or (B) with Rac V12, Rac V12L37, or Rac V12H40 in the presence of STAT3 (solid bars). Cell lysates were prepared 48 h after transfection and used to measure luciferase and β-gal activity. The data shown are means ± SEM and are representative of similar experiments.
Several downstream effector molecules have been identified for Rac1, and specific targeted mutations have been characterized that inhibit interaction with these effectors. Rac1 mutants deficient in effector binding were evaluated to gain insight into downstream pathways necessary for Rac1-mediated STAT3 activation (17). The point mutant Rac V12H40 is unable to mediate the activation of c-Jun N-terminal kinase (JNK), whereas the Rac V12L37 mutant maintains the ability to activate JNK but is deficient in induction of actin polymerization (17). Assays in which both of these mutations of Rac1 were tested indicated that Rac V12H40 expression mediated STAT3 activation, whereas Rac V12L37 did not (Fig. 3B). The expression of these Rac1 mutants was confirmed by Western blot analysis (data not shown).
Evidence That Rac1 Induces STAT3 Activation by Autocrine IL-6 Signaling.
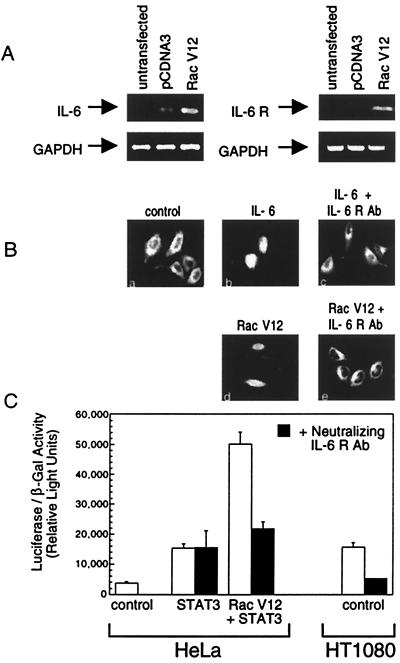
IL-6 is known to stimulate the JAK/STAT3 pathway, and a recent study of mast cells described the induction of various cytokines in response to oncogenic Vav expression (36). To test the possibility that Vav-induced activation of Rac1 in fibroblasts could lead to the production and subsequent autocrine action of IL-6, we investigated whether Rac V12 expression would induce the IL-6 gene. Total RNA isolated from untransfected cells, control-transfected cells (pCDNA3), and Rac V12-transfected cells was used to generate cDNA. We evaluated IL-6 mRNA levels in response to Rac1 activity by PCR analysis by using specific primers corresponding to the IL-6 gene or to a control gene, GAPDH. Although low levels of IL-6 mRNA were detected in untransfected and control-transfected cells, Rac V12 expression led to a notable increase in IL-6 mRNA (Fig. 4A Left). Moreover, we investigated whether the IL-6 receptor gene was regulated by Rac V12 expression and found an apparent induction of IL-6 receptor mRNA (Fig. 4A Right). These results suggested that the activity of Rac1 can induce the IL-6 gene along with its cognate receptor and consequently elicit an autocrine IL-6 activation loop for STAT3. Because STAT proteins reside in the cytoplasm of the cell in a latent state and accumulate in the nucleus after tyrosine phosphorylation and activation, we visually inspected the cellular localization of a fusion protein of STAT3 with GFP (STAT3-GFP). IL-6 treatment stimulated the nuclear accumulation of STAT3-GFP, and this localization was inhibited by addition of neutralizing antibodies to the IL-6 receptor (Fig. 4B). When cells were cotransfected with STAT3-GFP and Rac V12, STAT3-GFP accumulated in the nucleus similar to the response to IL-6. To test whether STAT3-GFP responded to autocrine IL-6 induced by Rac V12, neutralizing IL-6 receptor antibodies were added to the media. The neutralizing IL-6 receptor antibodies effectively blocked the nuclear accumulation of STAT3 in response to Rac V12 expression (Fig. 4B), whereas control antibodies had no effect (data not shown). Together these results provide strong evidence that autocrine-acting IL-6 is produced and required for Rac1-mediated stimulation of the JAK/STAT3 pathway. We have also investigated whether the other Rho family members mediate STAT3 activation in an IL-6-dependent pathway. The expression of constitutively active RhoG, Cdc42, and RhoA caused the translocation from the cytoplasm to the nucleus of cotransfected STAT3-GFP. This GTPase-induced STAT3 translocation was blocked to varying degrees by neutralizing IL-6 receptor antibodies, supporting a role for autocrine IL-6 in Rho family-induced STAT3 activation (data not shown).
Figure 4.
Effect of Rac V12 expression on STAT3 activation and specific gene induction. (A) HeLa cells were either untransfected or transfected with either pCDNA3 (as a transfection control) or Rac V12. Total RNA was collected and examined by reverse transcription–PCR for IL-6, IL-6 receptor, and GAPDH mRNA expression. The ethidium bromide-stained 2% agarose gel shows the IL-6 and GAPDH cDNA fragments (Left) and IL-6 receptor and GAPDH cDNA fragments (Right) amplified from the same amounts of cDNA. (B) HeLa cells were transfected with STAT3-GFP alone or with Rac V12 and were incubated for 18 h in serum-free DMEM in the presence of neutralizing IL-6 receptor antibody (1 μg/ml). Cells either were left untreated (a, d, and e) or were treated with soluble IL-6 receptor protein (100 ng/ml) (to enhance IL-6 responsiveness) and IL-6 (20 ng/ml) for 20 min (b and c). Cells were fixed and examined by fluorescent microscopy. (C) HeLa cells and HT1080 cells were cotransfected with (Gas)3-Luc, SV40-LacZ, and either pCDNA3 (control) or STAT3 alone or with Rac V12 and placed in serum-free DMEM in the absence (open bars) or presence (solid bars) of neutralizing IL-6 receptor antibodies (1 μg/ml). Cell lysates were prepared 48 h after transfection and were used to measure both luciferase and β-gal activity. The data shown are means ± SEM and are representative of similar experiments.
To verify that the effects of Rac V12 on STAT3 response gene expression are mediated by autocrine IL-6 activity, neutralizing IL-6 receptor antibodies were added to cotransfection assays. The STAT-responsive luciferase reporter gene, STAT3, and Rac V12 were cotransfected, and the transcriptional activation of STAT3 in response to Rac V12 was found to depend on signal transduction through the IL-6 receptor (Fig. 4C). To be sure these results were not due to overexpression of signaling mediators, we investigated endogenous STAT activity in HT1080 cells, a fibrosarcoma cell line that expresses constitutively active Ras (37). These cells exhibited high levels of the STAT-responsive luciferase gene in the absence of STAT3 or Ras overexpression, indicating the endogenous Ras mutation promotes STAT activation (Fig. 4C). Moreover, neutralizing IL-6 receptor antibodies effectively attenuated this STAT activity in HT1080 cells suggesting the involvement of autocrine IL-6 (Fig. 4C). Control antibodies had no effect (data not shown).
To confirm further the requirement of IL-6-mediated signal transduction for Rac1-induced activation of STAT3, a natural inhibitor of JAK activity was evaluated. A family of proteins has been identified that mediates the down-regulation of JAK/STAT signaling by binding to the tyrosine phosphorylated JAK proteins or the cytokine receptors. This family is known as suppressors of cytokine signaling (SOCS), and SOCS3 was identified to block IL-6 signal transduction specifically (25). For this reason we tested the effect of SOCS3 expression on Rac1-induced transcriptional activation of STAT3. SOCS3 expression dramatically inhibited STAT3 activation in response to Rac V12 expression as well as to exogenous IL-6 treatment (Fig. 5A). Similarly, SOCS3 expression inhibited Vav (Δ1–187)-induced STAT3 activation (data not shown). These data indicate that the JAK/STAT3 pathway is activated by IL-6 in response to Rac V12 expression.
Figure 5.
Effects of blocking IL-6 or NF-κB signaling. (A) Rat1 cells were cotransfected as described for Fig. 4C with pCDNA3 (control), STAT3, or Rac V12 in the absence or presence of SOCS-3. Cells were either untreated (open bars) or treated with IL-6 (20 ng/ml) (solid bars) for 6 h before harvest. (B) Rat1 cells were cotransfected with (NF-κB)5-Luc, SV40-LacZ, along with either control pCDNA3 plasmid (open bar) or increasing amounts of plasmid DNA encoding Rac V12 (solid bars) and Rac V12L37 (shaded bars). Cell lysates were prepared 48 h after transfection and were used to measure both luciferase and β-gal activity. The data shown are means ± SEM and are representative of two to four similar experiments. (C) Rat1 cells were cotransfected with STAT3-GFP alone (a), or with Rac V12 in the absence (b) or presence of IκBα S32AS36A (c). Cells were placed in serum-free DMEM for 24 h, fixed, and examined by fluorescent microscopy.
Induction of IL-6 gene expression is known to depend on the activity of the transcription factor, nuclear factor-κB (NF-κB) (38), and Rac1 is known to stimulate NF-κB activation (39). NF-κB exists in a latent state in the cytoplasm complexed with an inhibitor protein IκB. NF-κB becomes activated and localizes to the nucleus after phosphorylation and subsequent dissociation of IκB (40). We measured NF-κB-dependent luciferase reporter gene activity in response to Rac V12 expression in Rat1 cells. Rac V12 stimulated NF-κB activity in a concentration-dependent manner (Fig. 5B). Because the Rac V12L37 mutant was deficient in rendering STAT3 transcriptionally active (Fig. 3B), we tested its ability to stimulate NF-κB activity. Consistent with previous reports (41), Rac V12L37 was significantly impaired in its ability to elicit NF-κB activation (Fig. 5B). The defect in NF-κB activation by Rac V12L37 correlates with its failure to stimulate STAT3 activation. If Rac1-induced NF-κB activation leads to IL-6 gene induction and consequential autocrine stimulation of the JAK/STAT3 pathway, inhibition of NF-κB should block STAT3 activation in response to Rac V12 expression. IκBα S32AS36A is a mutant form of the inhibitor protein that lacks specific phosphorylation target sites rendering it a constitutive inhibitor of NF-κB activation (24). IκBα S32AS36A was expressed in cells with STAT3-GFP and Rac V12, to determine whether NF-κB activation by Rac1 was required for Rac1-induced STAT3 activation and nuclear accumulation. IκBα S32AS36A expression was found to inhibit the ability of Rac V12 to induce STAT3 activation and nuclear accumulation (Fig. 5C), and to block NF-κB-dependent gene induction by Rac V12 completely (data not shown).
These results provide evidence that Rac1 stimulates STAT3 activation indirectly through the induction of an autocrine IL-6 feedback loop that leads to the activation of the JAK/STAT3 pathway. Our findings are particularly significant in cases where genetic mutations result in constitutive activity of small GTPases and aberrant growth. The persistent activity of Ras/Rho GTPases may stimulate STAT3, a known modulator of cell growth, through the induction and autocrine signaling of IL-6 through its receptor.
Discussion
Activation of specific JAK/STAT signal pathways can lead to a variety of biological responses. Recent studies have suggested that the abnormal regulation of STAT signaling may be involved in malignant transformation. Constitutive STAT3 activity has been reported in a growing number of cancers and has been identified as a potential oncogene (6, 8, 42). In this study we investigated effects of a gain-of-function mutation in small GTPases that promote the activation of STAT3. We presented evidence demonstrating that constitutively active forms of Ras and Rac1 specifically stimulate STAT3-dependent gene expression. The results suggest a mechanism in which GTPases activate NF-κB leading to transcriptional induction of the IL-6 gene. Consequently, IL-6 stimulates the JAK/STAT3 signal pathway in an autocrine fashion via the IL-6 receptor (Fig. 6). Although components of this signaling pathway have been established previously in the context of “outside-in” signaling events, our report links “inside-out” signaling events resulting from oncogenic/gain-of-function mutations.
Figure 6.
Proposed model for Rac1-mediated STAT3 activation. We show that constitutively active small GTPases such as Rac1 stimulate the JAK/STAT3 pathway from an “inside-out” signaling pathway involving the induction of an autocrine IL-6 activation loop. The left side of the diagram is a simple illustration of an oncogenic mutation rendering constitutively active Ras or Rac1 (Ras*/Rac1*) signaling to one of its known downstream targets, the NF-κB transcription factor. NF-κB activation leads to its translocation to the nucleus, DNA binding, and subsequent induction of NF-κB-dependent genes such as IL-6. After IL-6 binds to its receptor, the JAKs are activated and in turn tyrosine phosphorylate STAT3 (-pY). Signal transduction by the JAK/STAT pathway for STAT3 is depicted on the right side of the diagram. Phosphorylated STAT3 molecules form dimers and translocate to the nucleus to bind to promoter elements of responsive genes. Hence, gain-of-function mutations that render small GTPases constitutively active mediate STAT3 activation through the induction of an autocrine IL-6 activation loop.
Ras and Rac1 have been implicated recently in the resultant phosphorylation of STAT3 on Ser-727 by activation of either p38/JNK or an upstream kinase, MKK4 (10, 11). Our results demonstrate the ability of constitutively active forms of Ras or Rac1 alone to stimulate transcriptional activation by STAT3 independent of phosphorylation on Ser-727. Although a contribution of other serine or threonine residues in our studies cannot be excluded, our results are consistent with accumulating reports that suggest a role of serine phosphorylation in the transcriptional potential of STATs may be cell type, agonist, or target gene specific (43–46).
Because Rac1 has been reported to mediate some of its specific effects through a direct physical association with effector molecules, we investigated whether Rac1 associated directly with STAT3 in our system (14). A recent report proposed a direct physical association between Rac1 and STAT3 that led to STAT3 regulation; however, the mechanism or specificity of this proposed mode of activation was not described (13). Our studies did not demonstrate any specific physical association in vivo by using coimmunoprecipitation techniques. STAT3 antibodies and the appropriate control antibodies generated indistinguishable levels of Rac1 associated with the immunocomplexes (data not shown). Hence, we have no evidence of a physical association between Rac1 and STAT3 in our system.
Mutations of Rac1 that alter their ability to stimulate downstream effectors were evaluated to discern a pathway of STAT3 activation. The Rac V12H40 mutant, although deficient in JNK activation, still led to STAT3-driven gene expression. Hence, the kinase activity of JNK seems not to be required for Rac1-mediated effects on STAT3. These results are consistent with another study that indicated that IL-6-mediated STAT3 activation occurred in the absence of activated JNK (11). The Rac V12L37 mutant failed to mediate STAT3 activation. This deficiency may reflect what we and others have shown, an impaired ability of Rac V12L37 to elicit NF-κB-dependent gene induction (41).
To determine whether gene induction by STAT3 was an indirect effect of NF-κB activation, we tested the effect of an NF-κB inhibitor, IκBα S32AS36A. Inhibition of NF-κB activity by IκBα S32AS36A expression correlated with a lack of STAT3 nuclear accumulation in response to Rac1 activity. These observations lend support to the hypothesis that Rac1-induced STAT3 activation occurs through the NF-κB-dependent induction of IL-6. The fact that neutralizing IL-6 receptor antibodies inhibited Rac1-induced STAT3 activation further demonstrates the requirement of an autocrine IL-6 activation loop for Rac1-mediated STAT3 activation.
In view of our results, we propose a model of STAT3 activation that may play a role during aberrant cell growth in which oncogenic mutations can result in constitutive small GTPase activity (Fig. 6). GTPases are normally regulated by the activity of GEFs and GTPase activating proteins, but mutations resulting in constitutive activity contribute to oncogenesis (15, 20, 47, 48). Persistently active Rac1 results in signaling pathways that lead to NF-κB activation and in turn IL-6 gene induction. An autocrine IL-6 activation loop is thereby initiated. Receptor-mediated IL-6 signal transduction results in the activation of JAKs and subsequent tyrosine phosphorylation and activation of STAT3. Tyrosine phosphorylated STAT3 forms dimers that translocate to the nucleus to bind DNA target sites in responsive genes. Because constitutive STAT3 activity has been shown to result in oncogenic transformation (6), this autocrine loop may play a role in the transforming potential of Ras/Rho GTPases.
Acknowledgments
We thank Michael Hayman, Patrick Hearing, Todd Miller, and our laboratory colleagues for their helpful discussions. We also thank James Darnell, Jr., (The Rockefeller University) for STAT3 cDNA, Douglas Hilton (University of Melbourne) for SOCS-3 cDNA, Dean Ballard (Vanderbilt University) for IκBα S32AS36A cDNA, Steve McKnight (University of Texas Southwestern Medical Center) for STAT5a cDNA, and Rolf de Groot (University Hospital Utrecht) for STAT3β cDNA. This work was supported by Grants PO1CA28146 (to D.B. and N.C.R.) and RO1CA50773 from the National Institutes of Health and in part by a Carol M. Baldwin Breast Cancer Award (to N.C.R.) and by National Research Service Award funding (5T32DK07521) (to T.R.F.).
Abbreviations
- STAT
signal transducer and activator of transcription
- JAK
Janus kinase
- GEF
guanine nucleotide exchange factor
- SOCS
suppressor of cytokine signaling
- GFP
green fluorescent protein
- SV40
simian virus 40
- JNK
c-Jun terminal kinase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- β-gal
β-galactosidase
References
- 1.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Schindler C, Darnell J E., Jr Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 3.Ihle J N. Nature (London) 1995;377:591–594. doi: 10.1038/377591a0. [DOI] [PubMed] [Google Scholar]
- 4.Leonard W J, O'Shea J J. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 5.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 6.Bromberg J F, Wrzeszczynska M H, Devgan G, Zhao Y, Pestell R G, Albanese C, Darnell J E., Jr Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 7.Yu C L, Meyer D J, Campbell G S, Larner A C, Carter-Su C, Schwartz J, Jove R. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 8.Garcia R, Jove R. J Biomed Sci. 1998;5:79–85. doi: 10.1007/BF02258360. [DOI] [PubMed] [Google Scholar]
- 9.Besser D, Bromberg J F, Darnell J E, Jr, Hanafusa H. Mol Cell Biol. 1999;19:1401–1409. doi: 10.1128/mcb.19.2.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turkson J, Bowman T, Adnane J, Zhang Y, Djeu J Y, Sekharam M, Frank D A, Holzman L B, Wu J, Sebti S, Jove R. Mol Cell Biol. 1999;19:7519–7528. doi: 10.1128/mcb.19.11.7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuringa J J, Jonk L J, Dokter W H, Vellenga E, Kruijer W. Biochem J. 2000;347:89–96. [PMC free article] [PubMed] [Google Scholar]
- 12.Uddin S, Lekmine F, Sharma N, Majchrzak B, Mayer I, Young P R, Bokoch G M, Fish E N, Platanias L C. J Biol Chem. 2000;275:27634–27640. doi: 10.1074/jbc.M003170200. [DOI] [PubMed] [Google Scholar]
- 13.Simon A R, Vikis H G, Stewart S, Fanburg B L, Cochran B H, Guan K L. Science. 2000;290:144–147. doi: 10.1126/science.290.5489.144. [DOI] [PubMed] [Google Scholar]
- 14.Van Aelst L, D'Souza-Schorey C. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 15.Bos J L. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 16.Qiu R G, Chen J, Kirn D, McCormick F, Symons M. Nature (London) 1995;374:457–459. doi: 10.1038/374457a0. [DOI] [PubMed] [Google Scholar]
- 17.Joneson T, McDonough M, Bar-Sagi D, Van Aelst L. Science. 1996;274:1374–1376. doi: 10.1126/science.274.5291.1374. [DOI] [PubMed] [Google Scholar]
- 18.Qiu R G, Abo A, McCormick F, Symons M. Mol Cell Biol. 1997;17:3449–3458. doi: 10.1128/mcb.17.6.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roux P, Gauthier-Rouviere C, Doucet-Brutin S, Fort P. Curr Biol. 1997;7:629–637. doi: 10.1016/s0960-9822(06)00289-2. [DOI] [PubMed] [Google Scholar]
- 20.Katzav S, Martin-Zanca D, Barbacid M. EMBO J. 1989;8:2283–2290. doi: 10.1002/j.1460-2075.1989.tb08354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong Z, Wen Z, Darnell J E., Jr Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 22.Heinrich P C, Behrmann I, Muller-Newen G, Schaper F, Graeve L. Biochem J. 1998;334:297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coso O A, Chiariello M, Yu J C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 24.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Starr R, Willson T A, Viney E M, Murray L J, Rayner J R, Jenkins B J, Gonda T J, Alexander W S, Metcalf D, Nicola N A, Hilton D J. Nature (London) 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 26.Wigler M, Pellicer A, Silverstein S, Axel R, Urlaub G, Chasin L. Proc Natl Acad Sci USA. 1979;76:1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotanides H, Moczygemba M, White M F, Reich N C. J Biol Chem. 1995;270:19481–19486. doi: 10.1074/jbc.270.33.19481. [DOI] [PubMed] [Google Scholar]
- 28.Joneson T, White M A, Wigler M H, Bar-Sagi D. Science. 1996;271:810–812. doi: 10.1126/science.271.5250.810. [DOI] [PubMed] [Google Scholar]
- 29.Stokoe D, Macdonald S G, Cadwallader K, Symons M, Hancock J F. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 30.Bustelo X R. Mol Cell Biol. 2000;20:1461–1477. doi: 10.1128/mcb.20.5.1461-1477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuebel K E, Movilla N, Rosa J L, Bustelo X R. EMBO J. 1998;17:6608–6621. doi: 10.1093/emboj/17.22.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaptein A, Paillard V, Saunders M. J Biol Chem. 1996;271:5961–5964. doi: 10.1074/jbc.271.11.5961. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Blenis J, Li H C, Schindler C, Chen-Kiang S. Science. 1995;267:1990–1994. doi: 10.1126/science.7701321. [DOI] [PubMed] [Google Scholar]
- 34.Wen Z, Zhong Z, Darnell J E., Jr Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 35.Caldenhoven E, van Dijk T B, Solari R, Armstrong J, Raaijmakers J A M, Lammers J W J, Koenderman L, de Groot R P. J Biol Chem. 1996;271:13221–13227. doi: 10.1074/jbc.271.22.13221. [DOI] [PubMed] [Google Scholar]
- 36.Song J S, Haleem-Smith H, Arudchandran R, Gomez J, Scott P M, Mill J F, Tan T H, Rivera J. J Immunol. 1999;163:802–810. [PubMed] [Google Scholar]
- 37.Brown R, Marshall C J, Pennie S G, Hall A. EMBO J. 1984;3:1321–1326. doi: 10.1002/j.1460-2075.1984.tb01970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Libermann T A, Baltimore D. Mol Cell Biol. 1990;10:2327–2334. doi: 10.1128/mcb.10.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sulciner D J, Irani K, Yu Z X, Ferrans V J, Goldschmidt-Clermont P, Finkel T. Mol Cell Biol. 1996;16:7115–7121. doi: 10.1128/mcb.16.12.7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baeuerle P A. Cell. 1998;95:729–731. doi: 10.1016/s0092-8674(00)81694-3. [DOI] [PubMed] [Google Scholar]
- 41.Joyce D, Bouzahzah B, Fu M, Albanese C, D'Amico M, Steer J, Klein J U, Lee R J, Segall J E, Westwick J K, et al. J Biol Chem. 1999;274:25245–25249. doi: 10.1074/jbc.274.36.25245. [DOI] [PubMed] [Google Scholar]
- 42.Grandis J R, Drenning S D, Zeng Q, Watkins S C, Melhem M F, Endo S, Johnson D E, Huang L, He Y, Kim J D. Proc Natl Acad Sci USA. 2000;97:4227–4232. doi: 10.1073/pnas.97.8.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung J, Uchida E, Grammer T C, Blenis J. Mol Cell Biol. 1997;17:6508–6516. doi: 10.1128/mcb.17.11.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wen Z, Darnell J E., Jr Nucleic Acids Res. 1997;25:2062–2067. doi: 10.1093/nar/25.11.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaefer L K, Wang S, Schaefer T S. Biochem Biophys Res Commun. 1999;266:481–487. doi: 10.1006/bbrc.1999.1853. [DOI] [PubMed] [Google Scholar]
- 46.Kovarik P, Mangold M, Ramsauer K, Heidari H, Steinborn R, Zotter A, Levy D E, Muller M, Decker T. EMBO J. 2001;20:91–100. doi: 10.1093/emboj/20.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boguski M S, McCormick F. Nature (London) 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 48.Aronheim A, Engelberg D, Li N, al-Alawi N, Schlessinger J, Karin M. Cell. 1994;78:949–961. doi: 10.1016/0092-8674(94)90271-2. [DOI] [PubMed] [Google Scholar]