Abstract
Objective
To describe the deletion patterns and distribution characteristics of the dystrophin gene in a Chinese population of patients with Duchenne muscular dystrophy (DMD) or Becker muscular dystrophy (BMD).
Methods
Patients with DMD/BMD were recruited. Deletions in 19 exons of the dystrophin gene were evaluated using accurate multiplex polymerase chain reaction (PCR).
Result
Multiplex PCR identified deletions in 238/401 (59.4%) patients with DMD/BMD. Of these, 196 (82.4%) were in the distal hotspot, 32 (13.4%) were in the proximal hotspot, five (2.1%) were in both regions and five (2.1%) were in neither hotspot. Deletions were classified into 54 patterns. Exon 49 was the most frequently deleted. The reading frame rule was upheld for 91.9% of cases.
Conclusion
Accurate multiplex PCR for 19 exons is an effective diagnostic tool.
Keywords: Duchenne muscular dystrophy, Becker muscular dystrophy, dystrophin gene, exon deletion, accurate multiplex polymerase chain reaction
Introduction
Duchenne and Becker muscular dystrophy (DMD and BMD) are the most common X-linked recessive neuromuscular disorders observed during childhood, with incidences of ∼1 in 3500 and 1 in 18500 live male births, respectively.1,2 DMD is clinically more severe than BMD; patients with DMD usually die from cardiac or respiratory failure in their second or third decade of life. Both DMD and BMD are caused by mutations in the dystrophin gene (GenBank accession number: NG_012232), which is located on Xp21.2. The largest known human gene dystrophin spans ∼2.4 Mb and contains 79 exons and eight tissue-specific promoters.3 Since the cloning of the dystrophin gene sequence in 1987,3 exon deletion has been determined to be the most common molecular defect underlying the disease, accounting for ∼60% of all cases of DMD and BMD.4,5 Intragenic deletions usually encompass one or more exons and are nonrandomly distributed, clustered in two specific hotspots. The distal hotspot is near the central part of the gene (around exons 44 to 53) and the proximal hotspot is located toward the 5′ end.3,6 Mutational analysis is complicated by the large size of the gene, although an accurate multiplex polymerase chain reaction (PCR) method using 19-exon primers can detect ∼98% of dystrophin deletions.7–9
The aim of the present study was to use accurate multiplex PCR to describe the deletion patterns and distribution characteristics of the dystrophin gene in a Chinese population of patients with DMD or BMD.
Patients and methods
Study population
Unrelated male patients with DMD or BMD were recruited from the Neuromuscular Clinic, The Affiliated Children’s Hospital, Capital Institute of Paediatrics, Beijing, China between January 2001 and December 2010. DMD and BMD were diagnosed according to clinical phenotype based on symptoms (elevated serum creatine kinase activity, age of onset, calf muscle hypertrophy, age at loss of ambulation, Gower’s sign, electromyography, family history, and/or dystrophic pattern at muscle biopsy). Male control volunteer subjects without any neuromuscular disease were recruited and used as positive controls for PCR.
The study was approved by the ethics committee of the Capital Institute of Paediatrics, Beijing, China, and all patients and/or their guardians provided written informed consent prior to enrolment.
Accurate multiplex PCR
Peripheral blood (5 ml) was taken from each patient using standard methods. Genomic DNA was isolated from leukocytes using the standard phenol–chloroform method.10 Deletion screening was performed with two sets of primers (which together cover 18 exons and the muscle-specific promoter [Pm]): Set I, exon 4, 8, 12, 17, 19, 44, 45, 48, 51; 7,8 Set II: exon 3, 6, 13, 43, 47, 49, 50, 52, 60 and the muscle-specific promoter.9 Primer sequences are shown in Table 1 (Life Technologies Co., Ltd; Shanghai, China). The reaction mix (25 µl) contained 100 ng genomic DNA, 1 × PCR buffer, 25 mmol/l each dNTP, 25 mmol/l magnesium chloride, 20 µmol/l each primer and 1.5 U Taq polymerase (Promega, Madison, WI, USA). Cycling conditions for Set I were 30 cycles of denaturation at 94℃ for 1 min, annealing at 55℃ for 1 min, and extension at 72℃ for 1 min. PCR conditions for Set II were 30 cycles of denaturation at 94℃ for 1 min, annealing at 60℃ for 1 min, extension at 72℃ for 1 min and a final extension at 72℃ for 5 min. All reactions were carried out using a Techne thermal cycler (Bibby Scientific Ltd, Staffordshire, UK). A normal genomic DNA sample (from control subjects) and a blank buffer served as the positive and negative controls, respectively. PCR products (5 µl) and a DNA size ladder were electrophoresed on 1.5% agarose gel stained with 0.5 µg/ml ethidium bromide, and visualized and photographed on an ultraviolet transilluminator.
Table 1.
Primer sequences for polymerase chain reaction of the human dystrophin gene.
| Exon | Primer | Sequence (5′–3′) |
|---|---|---|
| Pm | Forward | GAA GAT CTA GAC AGT GGA TAC ATA ACA AAT GCA TG |
| Reverse | TTC TCC GAA GGT AAT TGC CTC CCA GAT CTG AGT CC | |
| 3 | Forward | TCA TCC ATC ATC TTC GGC AGA TTA A |
| Reverse | CAG GCG GTA GAG TAT GCC AAA TGA AAA TCA | |
| 4 | Forward | TTG TCG GTC TCT CTG CTG GTC AGT G |
| Reverse | CAA AGC CCT CAC TCA AAC ATG AAG C | |
| 6 | Forward | CCA CAT GTA GGT CAA AAA TGT AAT GAA |
| Reverse | GTC TCA GTA ATC TTC TTA CCT ATG ACT ATG G | |
| 8 | Forward | GTC CTT TAC ACA CTT TAC CTG TTG AG |
| Reverse | GGC CTC ATT CTC ATG TTC TAA TTA G | |
| 12 | Forward | GAT AGT GGG CTT TAC TTA CAT CCT TC |
| Reverse | GAA AGC ACG CAA CAT AAG ATA CAC CT | |
| 13 | Forward | AAT AGG AGT ACC TGA GAT GTA GCA GAA AT |
| Reverse | CTG ACC TTA AGT TGT TCT TCC AAA GCA G | |
| 17 | Forward | GAC TTT CGA TGT TGA GAT TAC TTT CCC |
| Reverse | AAG CTT GAG ATG CTC TCA CCT TTT CC | |
| 19 | Forward | TTC TAC CAC ATC CCA TTT TCT TCC A |
| Reverse | GAT GGC AAA AGT GTT GAG AAA AAG TC | |
| 43 | Forward | GAA CAT GTC AAA GTC ACT GGA CTT CAT GG |
| Reverse | ATA TAT GTG TTA CCT ACC CTT GTC GGT CC | |
| 44 | Forward | CTT GAT CCA TAT GCT TTT ACC TGC A |
| Reverse | TCC ATC ACC CTT CAG AAC CTG ATC T | |
| 45 | Forward | AAA CAT GGA ACA TCC TTG TGG GGA C |
| Reverse | CAT TCC TAT TAG ATC TGT CGC CCT AC | |
| 47 | Forward | CGT TGT TGC ATT TGT CTG TTT CAG TTA C |
| Reverse | GTC TAA CCT TTA TCC ACT GGA GAT TTG | |
| 48 | Forward | TTG AAT ACA TTG GTT AAA TCC CAA CAT G |
| Reverse | CCT GAA TAA AGT CTT CCT TAC CAC AC | |
| 49 | Forward | GTG CCC TTA TGT ACC AGG CAG AAA TTG |
| Reverse | GCA ATG ACT CGT TAA TAG CCT TAA GAT C | |
| 50 | Forward | CAC CAA ATG GAT TAA GAT GTT CAT GAA T |
| Reverse | TCT CTC TCA CCC AGT CAT CAC TTC ATA G | |
| 51 | Forward | GAA ATT GGC TCT TTA GCT TGT GTT TC |
| Reverse | GGA GAG TAA AGT GAT TGG TGG AAA ATC | |
| 52 | Forward | AAT GCA GGA TTT GGA ACA GAG GCG TCC |
| Reverse | TTC GAT CCG TAA TGA TTG TTC TAG CCT C | |
| 60 | Forward | AGG AGA AAT TGC GCC TCT GAA AGA GAA CG |
| Reverse | CTG CAG AAG CTT CCA TCT GGT GTT CAG G |
Pm, muscle-specific promoter.
The size of each fragment was evaluated by comparison with the DNA size ladder. Deletions were diagnosed if PCR fragments were clearly absent in the patient sample compared with the normal male control sample. PCR results showing the absence of exons were confirmed by repeat PCR in addition to simultaneous amplification of each suspected missing exon, and a specifically designed control exon in a duplex PCR (for patients and normal male controls).
Reading frame analysis
The genotypes and phenotypes of patients with deletion patterns where the start and end points were clearly visible were analysed using an online DMD exonic deletion reading frame checker (version 1.9; available at: http://www.dmd.nl).
Statistical analyses
Data were presented as n (%) and mean. Differences between Hebei and Henan districts in the distribution of DMD gene deletions were evaluated using χ2-test or Fisher’s exact test (when t < 5 or n < 40) for categorical variables. Statistical analyses were performed using SPSS® version 16.0 (SPSS Inc., Chicago, IL, USA) for Windows®. P-values < 0.05 were considered statistically significant.
Results
The study recruited 401 male patients with DMD or BMD (mean age 6.6 ± 2.9 years; age range 3 months–14 years). Of these, multiplex PCR identified deletions in 238 patients (59.4%). A total of 854 exon deletions were detected (mean 3.59 per patient). Deletions were in the distal hotspot in 196 patients (82.4%) and the proximal hotspot in 32 patients (13.4%); five patients (2.1%) had deletions in both regions. The deletions of a further five patients were located outside the hotspot regions.
Single exon deletions were present in 67/238 patients (28.2%), with the most common in exon 45 (n = 20; 29.9%). There were no single exon deletions detected in exons Pm, 4, 6, 12, 13, 17, 48, 49 or 50. Multiple exon deletions were found in 171/238 patients (71.8%), the most common of which was exon 45–52 (n = 23; 13.5%). Nine patients (3.8%) had deletions of more than 10 exons.
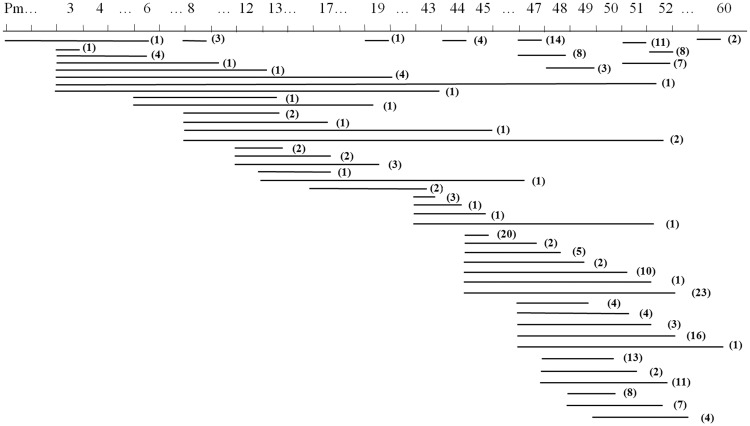
Deletions could be classified into 54 patterns (Figure 1), 22 of which were located in the proximal hotspot and 30 in the distal hotspot. Of these deletion patterns, 13/22 (59.1%) in the proximal hotspot and 7/30 (23.3%) in the distal hotspot occurred once only.
Figure 1.
Distribution of dystrophin gene deletions in 238 Chinese male patients with Duchenne or Becker muscular dystrophy. The top numbers represent the amplified exons, horizontal lines represent different deletion patterns, and the number in parentheses represents the number of cases with the same deletion. Pm, muscle-specific promoter.
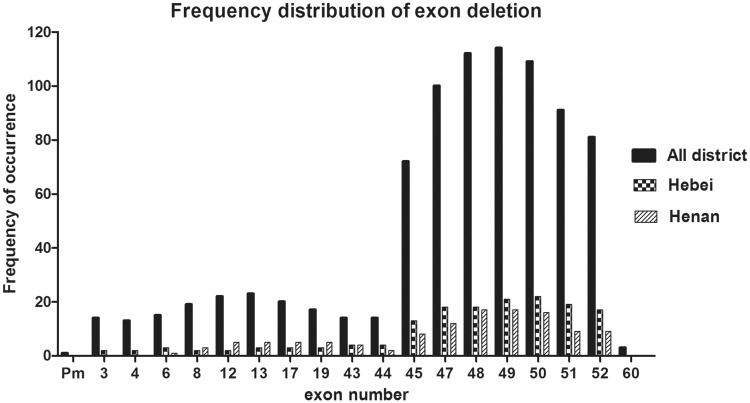
Deletion frequencies are shown in Figure 2. In the total patient group (n = 238), the most frequent deletion in the proximal hotspot was exon 13 (n = 23; 9.7%), followed by exon 12 (n = 22; 9.2%), exon 17 (n = 20; 8.4%), and exon 8 (n = 19; 8.0%). In the distal hotspot, exon 49 was the most common deletion (n = 114; 47.9%), followed by exon 48 (n = 112; 47.1%), exon 50 (n = 109; 45.8%) and exon 47 (n = 100; 42%).
Figure 2.
Frequency of individual exon deletions in the dystrophin gene in 238 Chinese male patients with Duchenne or Becker muscular dystrophy, stratified by geographical district.
Deletion frequencies were stratified by location (Figure 2). In Hebei district, there were 41 patients with deletions, of whom 36 (87.8%) showed deletions in the distal hotspot, three (7.3%) in the proximal hotspot, one (2.4%) in both regions and one (2.4%) in neither hotspot. The highest deletion frequency was in exon 50 (n = 22; 53.7%), followed by exon 49 (n = 21; 51.2%) and 51 (n = 19; 46.3%). In Henan district, there were 28 patients with deletions, 21 (75.0%) of which were in the distal hotspot, five (17.9%) in the proximal hotspot and two (7.1%) in both regions. Exons 48 and 49 had the highest deletion frequency (both n = 17; 60.7%), followed by exon 50 (n = 16; 57.1%) and exon 47 (n = 12; 42.9%). There were no statistically significant between-region differences in deletion distribution.
The genotypes and phenotypes of 62 patients were analysed with the exonic deletion reading frame checker. The reading frame rule explained the relationship between genotype and phenotype in 57 cases (91.9%; Table 2). Reading frame results were not in accordance with clinical diagnosis in the remaining five cases (Table 3).
Table 2.
Deletions in the dystrophin gene conforming to the reading frame rule (correlation between genotype and phenotype) in Chinese male patients with Duchenne or Becker muscular dystrophy (n = 62).
| Exon(s) deleted | Reading frame | Phenotype |
|
|---|---|---|---|
| DMD | BMD | ||
| 44 | Out | 4 | 0 |
| 44–50 | Out | 1 | 0 |
| 45–47 | In | 0 | 2 |
| 45–48 | In | 1 | 4 |
| 45–49 | In | 0 | 2 |
| 45–50 | Out | 10 | 0 |
| 45–51 | In | 0 | 1 |
| 48–49 | In | 1 | 2 |
| 48–50 | Out | 13 | 0 |
| 48–51 | In | 2 | 0 |
| 49–50 | Out | 7 | 1 |
| 51 | Out | 11 | 0 |
Data presented as n.
DMD, Duchenne muscular dystrophy; BMD, Becker muscular dystrophy.
Table 3.
Clinical characteristics of patients with Duchenne or Becker muscular dystrophy who do not conform to the reading frame rule (correlation between genotype and phenotype).
| Case no. | Age at diagnosis, years | Age at onset, years | Exon deletion pattern | CK, U/l | Clinical information | Clinical diagnosis | Reading frame |
|---|---|---|---|---|---|---|---|
| 1 | 3 | 1.5 | 45–48 | 14 230 | Pseudohypertrophy of calf muscles, MD, difficulty in climbing stairs, falling, positive Gower’s sign | DMD | In |
| 2 | 7.5 | 1.5 | 48–49 | 7 190 | Pseudohypertrophy of calf muscles, MD, difficulty in walking, positive Gower’s sign, positive family history | DMD | In |
| 3 | 2.8 | 2.75 | 48–51 | 12 300 | Pseudohypertrophy of calf muscles, MD, difficulty in climbing stairs, unable to jump, falling, positive Gower’s sign | DMD | In |
| 4 | 5.5 | 4 | 48–51 | 15 060 | Pseudohypertrophy of calf muscles, MD, difficulty in climbing stairs, waddling gait, positive Gower’s sign | DMD | In |
| 5 | 10 | 9 | 49–50 | 17 597 | Pseudohypertrophy of calf muscles, fatigability | BMD | Out |
CK, creatinine kinase; MD, myogenic damage; DMD, Duchenne muscular dystrophy; BMD, Becker muscular dystrophy.
Discussion
Detection of dystrophin deletion mutations is important in the diagnosis of DMD and BMD. Deletions tend to occur in hotspots, thus the analysis of this limited number of exons can detect 98% of dystrophin deletions. In the present study, accurate multiplex PCR was adopted to analyse 19 exons of the dystrophin gene in 401 Chinese patients and detected 59.4% of deletions.
Reported deletion detection rates vary widely, ranging between 31% and 73.86%.11–24 These differences may be caused by many factors such as race, sample size and inclusion criteria. It has been suggested that the relatively low deletion detection rate in some studies is caused by the inclusion of other similar muscle diseases, because diagnosis was based mainly on clinical symptoms and did not require muscle biopsy.17,21 Further, detailed studies are needed to verify the relationship between deletion frequency and population.
In accordance with our findings, others have shown that ∼20–30% of detected deletions cluster in the proximal hotspot and ∼70–80% in the distal hotspot.25 There were no differences in the deletion distribution for patients in Hebei and Henan regions in the present study, despite variations in customs and environment. This may be due to the small sample size or the fact that the residents of both districts are from the Han Chinese population, in which there is little genetic variation.
The most common deletion was in exon 49 in the present study; a finding that is consistent with a large-scale study in a Chinese population.26 We have identified several other high-frequency deletions, which may be a useful target for prenatal screening.
Our data showed that deletion segments were smaller and considerably less heterogeneous in the distal hotspot than those in the proximal hotspot. As others have suggested, distal deletions are therefore more suitable than proximal lesions for comparing populations.27,28
The frequency of specific single-exon deletions differs among populations. The most common single exon deletion in an Indian population was exon 50, with a frequency of 16.1% of all single exon deletions,29 but this deletion was not found in any case in the present study. Exon 44 deletion occurred at a considerably higher rate in a Turkish population27 compared with patients from India,29 USA30 or China (present study). In contrast, the single exon deletion of exon 51 had a similar frequency in all four populations. These data confirm the hypothesis, first suggested by Danieli et al31 and supported by others,12,20,27 that different populations possess specific differences in a certain intron sequence of the dystrophin gene. This sequence predisposes this locus to an increased frequency of breakpoint deletions. Population-based differences can therefore be ascribed to genetic drift.
The reading frame rule was first described by Monaco et al.32, who found that the severity of disease is not directly associated with deletion size but largely depends on whether or not the reading frame is disrupted. Mutations that disrupt the reading frame (out-of-frame) produce unstable RNA and nearly undetectable levels of truncated proteins, resulting in the DMD phenotype. Conversely, mutations that maintain the reading frame (in-frame) result in abnormal but partially functional dystrophin, leading to the BMD phenotype. The reading-frame hypothesis holds for >90% of cases and can guide the early-stage clinical evaluation of DMD and BMD patients. In the present study, 91.9% of cases were consistent with the reading frame rule, in agreement with others.5,33 We identified five patients with inconsistent genotype–phenotype (Table 3). Deletions in exons 45–48, 48–49, and 49–50 are known to occur in both DMD and BMD,34,35 possibly due to variation in the exact location of the breakpoints within the intron. Patients with apparently similar deletion mutations are likely to have lost different gene regions because of such breakpoint variation. Different intron sequences may contain motifs that affect gene splicing, and exon skipping events may be affected by different deletion breakpoints.36 Similar to another case report,37 two patients with in-frame deletion of exon 48–51 presented the DMD phenotype in the present study. It is possible that these deletions may be located in critical protein regions or lead to the production of unstable protein.
The present study was limited by evaluation of mutation at the DNA level only. Future studies should investigate muscle RNA or protein level detection. The accurate multiplex PCR technique is useful in the initial step of molecular diagnosis of DMD and BMD, but is unable to detect intragenic duplication mutations or female carriers.
In conclusion, the accurate multiplex PCR method for 19 exons is an effective diagnostic tool. The distribution of dystrophin gene deletions in the Chinese population differs from other populations. These population-based differences may be caused by genetic drift.
Declaration of conflicting interest
The authors declare that there are no conflicts of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- 1.Emery AE. Population frequencies of inherited neuromuscular diseases–a world survey. Neuromuscul Disord 1991; 1: 19–29. [DOI] [PubMed] [Google Scholar]
- 2.Bushby KM, Thambyayah M, Gardner-Medwin D. Prevalence and incidence of Becker muscular dystrophy. Lancet 1991; 337: 1022–1024. [DOI] [PubMed] [Google Scholar]
- 3.Koenig M, Hoffman EP, Bertelson CJ, et al. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 1987; 50: 509–517. [DOI] [PubMed] [Google Scholar]
- 4.Read AP, Mountford RC, Forrest SM, et al. Patterns of exon deletions in Duchenne and Becker muscular dystrophy. Hum Genet 1988; 80: 152–156. [DOI] [PubMed] [Google Scholar]
- 5.Zimowski JG, Massalska D, Holding M, et al. MLPA based detection of mutations in the dystrophin gene of 180 polish families with Duchenne/Becker muscular dystrophy. Neurol Neurochir Pol 2014; 48: 416–422. [DOI] [PubMed] [Google Scholar]
- 6.Forrest SM, Cross GS, Flint T, et al. Further studies of gene deletions that cause Duchenne and Becker muscular dystrophies. Genomics 1988; 2: 109–114. [DOI] [PubMed] [Google Scholar]
- 7.Chamberlain JS, Gibbs RA, Ranier JE, et al. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res 1988; 16: 11141–11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamberlain JS, Gibbs RA, Ranier JE, et al. Multiplex PCR for the diagnosis of Duchenne muscular dystrophy. In: Innis MA, Gelfand DH, Sninsky JJ, White T. (eds). PCR protocols: a guide to methods and applications, New York, NY: Academic Press, 1990, pp. 272–281. [Google Scholar]
- 9.Beggs AH, Koenig M, Boyce FM, et al. Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum Genet 1990; 86: 45–48. [DOI] [PubMed] [Google Scholar]
- 10.Sambrook J, Russell DW. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2001. [Google Scholar]
- 11.Effat LK, El-Harouni AA, Amr KS, et al. Screening of dystrophin gene deletions in Egyptian patients with DMD/BMD muscular dystrophies. Dis Markers 2000; 16: 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hrdlicka I, Zadina J, Krejci R, et al. Patterns of deletions and the distribution of breakpoints in the dystrophin gene in Czech patients with Duchenne and Becker muscular dystrophy (statistical comparison with results from several other countries). Folia Biol (Praha) 2001; 47: 81–87. [DOI] [PubMed] [Google Scholar]
- 13.Sbiti A, El Kerch F, Sefiani A. Analysis of dystrophin gene deletions by multiplex PCR in Moroccan patients. J Biomed Biotechnol 2002; 2: 158–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Jumah M, Majumdar R, Al-Rajeh S, et al. Deletion mutations in the dystrophin gene of Saudi patients with Duchenne and Becker muscular dystrophy. Saudi Med J 2002; 23: 1478–1482. [PubMed] [Google Scholar]
- 15.Hallwirth Pillay KD, Bill PL, Madurai S, et al. Molecular deletion patterns in Duchenne and Becker muscular dystrophy patients from KwaZulu Natal. J Neurol Sci 2007; 252: 1–3. [DOI] [PubMed] [Google Scholar]
- 16.Hassan MJ, Mahmood S, Ali G, et al. Intragenic deletions in the dystrophin gene in 211 Pakistani Duchenne muscular dystrophy patients. Pediatr Int 2008; 50: 162–166. [DOI] [PubMed] [Google Scholar]
- 17.Sura T, Eu-ahsunthornwattana J, Pingsuthiwong S, et al. Sensitivity and frequencies of dystrophin gene mutations in Thai DMD/BMD patients as detected by multiplex PCR. Dis Markers 2008; 25: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basak J, Dasgupta UB, Mukherjee SC, et al. Deletional mutations of dystrophin gene and carrier detection in eastern India. Indian J Pediatr 2009; 76: 1007–1012. [DOI] [PubMed] [Google Scholar]
- 19.Madania A, Zarzour H, Jarjour RA, et al. Combination of conventional multiplex PCR and quantitative real-time PCR detects large rearrangements in the dystrophin gene in 59% of Syrian DMD/BMD patients. Clin Biochem 2010; 43: 836–842. [DOI] [PubMed] [Google Scholar]
- 20.Basumatary LJ, Das M, Goswami M, et al. Deletion pattern in the dystrophin gene in Duchenne muscular dystrophy patients in northeast India. J Neurosci Rural Pract 2013; 4: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran VK, Ta VT, Vu DC, et al. Exon deletion patterns of the dystrophin gene in 82 Vietnamese Duchenne/Becker muscular dystrophy patients. J Neurogenet 2013; 27: 170–175. [DOI] [PubMed] [Google Scholar]
- 22.Kerr R, Robinson C, Essop FB, et al. Genetic testing for Duchenne/Becker muscular dystrophy in Johannesburg, South Africa. S Afr Med J 2013; 103(12 Suppl 1): 999–1004. [DOI] [PubMed] [Google Scholar]
- 23.Nouri N, Fazel-Najafabadi E, Salehi M, et al. Evaluation of multiplex ligation-dependent probe amplification analysis versus multiplex polymerase chain reaction assays in the detection of dystrophin gene rearrangements in an Iranian population subset. Adv Biomed Res 2014; 3: 72–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao MV, Sindhav GM, Mehta JJ. Duchenne/Becker muscular dystrophy: a report on clinical, biochemical, and genetic study in Gujarat population, India. Ann Indian Acad Neurol 2014; 17: 303–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbs S, Yau SC, Clark S, et al. A convenient multiplex PCR system for the detection of dystrophin gene deletions: a comparative analysis with cDNA hybridisation shows mistypings by both methods. J Med Genet 1991; 28: 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji X, Zhang J, Xu Y, et al. MLPA application in clinical diagnosis of DMD/BMD in Shanghai. J Clin Lab Anal 2014; 29: 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Onengüt S, Kavaslar GN, Battaloğlu E, et al. Deletion pattern in the dystrophin gene in Turks and a comparison with Europeans and Indians. Ann Hum Genet 2000; 64(pt 1): 33–40. [DOI] [PubMed] [Google Scholar]
- 28.Lai PS, Takeshima Y, Adachi K, et al. Comparative study on deletions of the dystrophin gene in three Asian populations. J Hum Genet 2002; 47: 552–555. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee M, Verma IC. Are there ethnic differences in deletions in the dystrophin gene? Am J Med Genet 1997; 68: 152–157. [PubMed] [Google Scholar]
- 30.Cunniff C, Andrews J, Meaney FJ, et al. Mutation analysis in a population-based cohort of boys with Duchenne or Becker muscular dystrophy. J Child Neurol 2009; 24: 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danieli GA, Mioni F, Müller CR, et al. Patterns of deletions of the dystrophin gene in different European populations. Hum Genet 1993; 91: 342–346. [DOI] [PubMed] [Google Scholar]
- 32.Monaco AP, Bertelson CJ, Liechti-Gallati S, et al. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 1988; 2: 90–95. [DOI] [PubMed] [Google Scholar]
- 33.Takeshima Y, Yagi M, Okizuka Y, et al. Mutation spectrum of the dystrophin gene in 442 Duchenne/Becker muscular dystrophy cases from one Japanese referral center. J Hum Genet 2010; 55: 379–388. [DOI] [PubMed] [Google Scholar]
- 34.Aartsma-Rus A, Van Deutekom JC, Fokkema IF, et al. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve 2006; 34: 135–144. [DOI] [PubMed] [Google Scholar]
- 35.Guo R, Zhu G, Zhu H, et al. DMD mutation spectrum analysis in 613 Chinese patients with dystrophinopathy. J Hum Genet 2015; 60: 435–442. [DOI] [PubMed] [Google Scholar]
- 36.Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol 2003; 2: 731–740. [DOI] [PubMed] [Google Scholar]
- 37.Magri F, Govoni A, D’Angelo MG, et al. Genotype and phenotype characterization in a large dystrophinopathic cohort with extended follow-up. J Neurol 2011; 258: 1610–1623. [DOI] [PubMed] [Google Scholar]