Abstract
Objective
To evaluate treatment response, survival, and the associations between KRAS mutation status and tumour expression levels of BRCA1, TYMS and SRC retrospectively in a cohort of patients with non-small cell lung cancer (NSCLC), treated exclusively with conjunctive platinum-based doublet chemotherapy.
Methods
KRAS mutation status was determined via amplification refractory mutation and multiple quantitative polymerase chain reaction (PCR) analysis. Tumour expression levels of BRCA1, TYMS and SRC were determined via real time quantitative PCR.
Results
Patients with KRAS mutations (n = 3) had significantly shorter survival duration than patients with wild type KRAS (n = 42). Tumour expression levels of BRCA1 and TYMS, but not SRC, were significantly lower in patients with, than in those without, KRAS mutations. Tumour expression level of BRCA1 was positively correlated with survival duration.
Conclusions
KRAS mutation status and BRCA1 tumour expression are potential biomarkers for tailoring chemotherapy and predicting clinical outcome.
Keywords: Nonsmall cell lung cancer (NSCLC), KRAS, BCA1, TYMS, platinum doublets, overall survival, drug response
Introduction
Non-small cell lung cancer (NSCLC) accounts for ∼80% of lung cancer cases and has a 5-year survival rate of 16%.1 A major obstacle to increasing overall survival is the poor response to available treatments.2 Research into the genetics and molecular biology of lung cancer has identified multiple mutations and molecular pathways that are involved in tumour pathogenesis and drug resistance,3 allowing the design of tailored treatment plans and the prediction of treatment outcomes in selected lung cancer patients, based on the results of molecular screening.4 These include EGFR mutations associated with improved overall response rate to EGFR tyrosine kinase inhibitors such as gefitinib and erlotinib,5,6 and ALK fusion gene mutations that improve response to ALK inhibitors such as crizotinib and ceritinib.7 Despite these advances, predicting treatment outcome and overall survival remains difficult in the majority of patients, and it is therefore important to identify additional relevant genetic mutations.
Oncogenes in the RAS family (HRAS, KRAS, and NRAS) encode small GTPases that act as intracellular transducer proteins and transmit extracellular signals through their receptors, such as EGFR, to downstream signalling transduction pathways.8 Proteins encoded by mutated RAS genes cannot hydrolyze GTP to GDP, leading to aberrant activities mediated by downstream signalling pathways.9 About 30% of patients with lung adenocarcinoma carry mutant KRAS,10 but it is unclear whether this mutation is a causative factor in poor survival and treatment resistance.11
Platinum-based chemotherapy remains popular for treating cancers, especially in patients with genetic or pathological profiles that respond poorly to targeted therapies. The platinum compound binds to DNA and forms platinum–DNA adducts that in turn cause the formation of crosslinks among DNA strands and the distortion of DNA conformation, leading to inhibition of DNA replication.12 DNA adduct formation and nucleotide excision repair (NER) of platinum–DNA adducts may influence clinical response and therapeutic outcome.13–15 The BRCA1 (breast cancer 1) and TYMS (thymidylate synthetase) genes encode proteins involved in adduct formation and NER, and have been evaluated as biomarkers for prognostic purposes and treatment selection.16–20 It is not clear whether the expression of these genes is affected by KRAS mutation status, however.
The aim of the present retrospective study was to evaluate the associations between KRAS mutation status and tumour expression levels of BRCA1, TYMS and SRC (a gene that is aberrantly overexpressed in lung cancer21) in a cohort of patients with NSCLC treated exclusively with conjunctive platinum-based doublet chemotherapy. The association between survival and BRCA1, TYMS and SRC expression or KRAS mutation status was investigated, in an attempt to identify additional molecular biomarkers for drug selection and outcome prediction.
Patients and methods
Study population
The study retrospectively recruited consecutive patients diagnosed with NSCLC at Liyang People’s Hospital, Liyang, Jiangsu, China, between February 2009 and December 2013. There were no age or sex restrictions. Tumour tissue samples were collected by surgical section or computed tomography-guided biopsy, processed immediately and preserved in formalin-fixed paraffin wax-embedded (FFPE) tissue blocks. Tumour staging was undertaken using the American Joint Committee on Cancer Staging Manual, 7th edition.22 Histology was determined according to World Health Organization criteria.23 Histopathological characteristics, including tumour subtype, were independently evaluated by two pathologists (including Y.D.), with disagreements resolved by a third pathologist.
The institutional ethics committee of Liyang People’s Hospital, Liyang, Jiangsu, China, approved the study, and all patients provided written informed consent.
Treatment
All patients received conjunctive platinum-based doublet chemotherapy after surgery as first-line treatment, using cisplatin, carboplatin or nedaplatin in combination with gemcitabine, pemetrexed or docetaxel for between 4 and 6 weeks. Treatment was terminated if patients exhibited unacceptable toxic adverse events or progressive disease. All patients were followed up until September 2014 or death. Overall survival was calculated from the date of first chemotherapy to date of death or last visit.
Detection of KRAS mutations
Genomic DNA was extracted from tumour tissue via QIAamp® DNA FFPE kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s instructions. KRAS mutations were detected using an amplification refractory mutation system and multiple quantitative polymerase chain reaction (PCR) analysis (ARMS-multi-qPCR) with a Human KRAS and BRAF Mutation Detection Kit (YuanQi Bio-Pharmaceutical Co., Ltd., Shanghai, China). PCR cycling conditions were 94℃ for 3 min followed by 40 cycles of 94℃ for 15 s and 60℃ for 1 min, using an ABI 7500 Real Time PCR System (ThermoFisher Scientific, Waltham, MA, USA). Primer sequences were: forward 5′-TTTGTATTAAAAGGTACTGGTGG-′3, and reverse 5′-CCTCTATTGTTGGATCATATTCG-3′. Mutations were verified via direct sequencing using the following primer: 5′-TGTATTAAAAGGTACTGGTGGAG-3′.
RNA isolation and gene expression analysis
Total RNA was extracted from tumour tissue using an RNeasy FFPE Kit (Qiagen), as described.24 RNA was treated with DNase I (DNA-free; ThermoFisher Scientific) to remove any potential DNA contamination, quantified and quality-checked, and first strand cDNA was synthesized using a SuperScript® III First-Strand Synthesis System (ThermoFisher Scientific). The cDNA was used in real time quantitative PCR for BRCA1, SRC and TYMS, using the appropriate Tumour Gene Expression Analysis Kit (YuanQi Bio-Pharmaceutical Co., Ltd., Shanghai, China), according to the manufacturer’s instructions. The housekeeping gene ABL was used as an internal control. Cycling conditions for all genes were 94℃ for 5 min followed by 40 cycles of 94℃ for 15 s and 60℃ for 1 min, using an ABI 7500 Real Time PCR System (ThermoFisher Scientific).
Statistical analyses
Data were presented as mean ± SEM or n (%). Pairwise comparisons were performed using Student’s t test for normally distributed data and nonparametric tests for non-normally distributed data. Categorical data were analysed using χ2-test or Fisher’s exact test. Overall survival was plotted using Kaplan–Meier survival curves, and differences between patients with or without KRAS mutations were evaluated using log rank test with χ2. Pearson’s correlation analysis was performed to determine correlations among BRCA1, TYMS and SRC expression levels, and between overall survival and expression level of each gene. Statistical analyses were performed using GraphPad Prism version 6 (GraphPad Software, Inc., San Diego, CA, USA). P-values < 0.05 were considered statistically significant.
Results
The study enrolled 46 patients, one of whom was excluded from the final analysis due to lack of clinical data. The final study population included 45 patients (29 male/16 female; median age 62.4 years; age range 40 – 90 years). Demographic and clinical characteristics of the study population are given in Table 1. Of patients with advanced stage cancer (total n = 19; stage IIIb, n = 4; stage IV, n = 15), 14 had adenocarcinoma (n = 3 at stage IIIb; n = 11 at stage IV).
Table 1.
Demographic and clinical characteristics of patients with nonsmall cell lung cancer included in a study to evaluate the associations between KRAS mutation status and tumour expression levels of BRCA1, TYMS and SRC (n = 45).
| Characteristic | n (%) |
|---|---|
| Sex, male/female | 29/17 (64.4/37.8) |
| Age, years | |
| 40–49 | 4 (8.9) |
| 50–59 | 10 (22.2) |
| 60–69 | 21 (46.7) |
| ≥70 | 10 (22.2) |
| Smoking history | |
| Nonsmoker | 16 (35.6) |
| Smoker | 28 (62.2) |
| Unknown | 1 (2.2) |
| Family history of cancer | |
| No | 35 (77.8) |
| Yes | 6 (13.3) |
| Unknown | 4 (8.9) |
| Metastasis | |
| No | 15 (33.3) |
| Yes | 30 (66.7) |
| Tumour stage | |
| I/II | 20 (44.4) |
| IIIa | 6 (13.3) |
| IIIb | 4 (8.9) |
| IV | 15 (33.3) |
| Histology | |
| Adenocarcinoma | 34 (75.6) |
| Squamous | 9 (20.0) |
| Other | 2 (4.4) |
| Response* | |
| CR/PR | 16 (41.0) |
| SD | 9 (23.1) |
| PD | 14 (35.9) |
Data presented as n (%) of patients.
Excluding six uncharacterized cases; n = 39.
A total of three patients were found to have KRAS mutations Table 2, and all had adenocarcinoma. Patients with KRAS mutations were significantly more likely to have stage IV disease than patients without mutations (P < 0.03). There was no significant between-group difference in the presence of metastasis. The prevalence of KRAS mutation was 8.8% in adenocarcinoma (three of 34) and 21.4% (three of 14) in advanced stage adenocarcinoma (IIIb and IV). None of the patients carrying KRAS mutations survived to the end of the investigation (mean survival 4.5 ± 1.5 months).
Table 2.
Clinical characteristics of patients with nonsmall cell lung cancer and KRAS mutations.
| Case ID | Sex | Age, years | Histology | Smoking status | Family history | Metastasis | Tumour stage | Survival |
|---|---|---|---|---|---|---|---|---|
| 1300636 | M | 70 | Ad | Yes | No | Yes | IV | No |
| 1307722 | F | 58 | Ad | No | No | Yes | IV | No |
| 1308892 | M | 61 | Ad | Yes | Yes | Yes | IV | No |
Ad, adenocarcinoma.
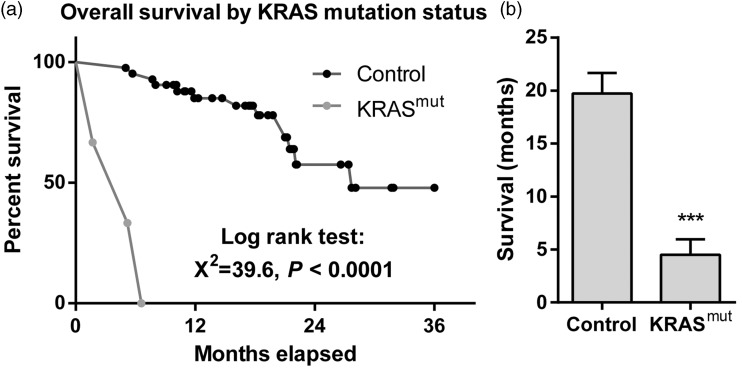
The mean duration of survival was significantly shorter in patients with KRAS mutations than in those without (P < 0.001; Figure 1).
Figure 1.
Overall survival of patients with nonsmall cell lung cancer, stratified by the presence of KRAS mutations. (a) Kaplan–Meier survival curve. (b) Mean ± SEM survival of patients without (control; n = 42) or with (n = 3) KRAS mutations. ***P < 0.001; Student’s t test.
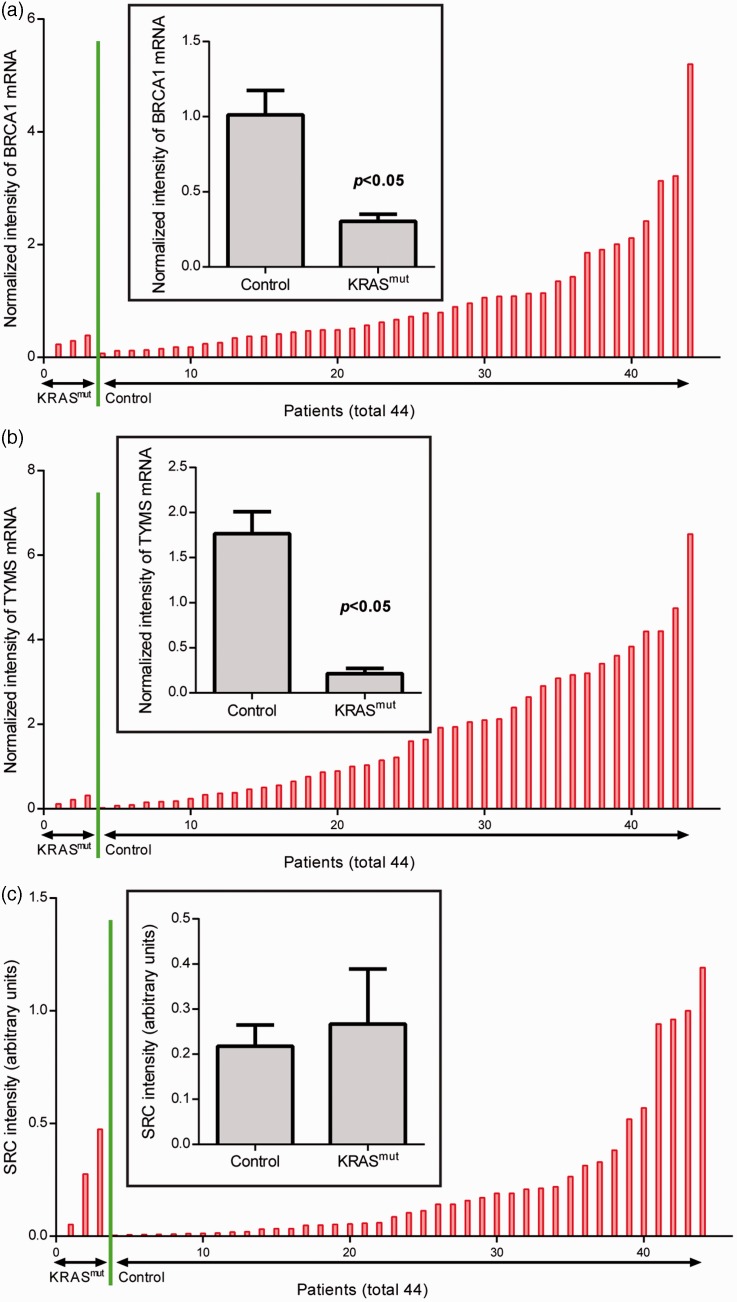
Tumour expression levels of BRCA1 and TYMS were significantly lower in patients with KRAS mutations compared with those without (P < 0.05; Figure 2a and b). There was no significant between-group difference in SRC expression (Figure 2c).
Figure 2.
Tumour levels of (a) BRCA1, (b) TYMS and (c) SRC mRNA in patients with nonsmall cell lung cancer, stratified by the presence (n = 3) or absence (control; n = 42) of KRAS mutations. Data presented as mean ± SEM; Student’s t-test.
In the total study population, there were significant positive correlations between overall survival duration and BRCA1 expression level (r = 0.3015; P < 0.02), and between BRCA1 and TYMS expression levels (r = 0.4749; P < 0.001). There were no significant correlations between survival duration and TYMS or SRC expression level.
Discussion
Resistance to platinum-based chemotherapy is a major impediment to successful treatment of lung cancer. In our cohort of 46 patients with NSCLC treated with platinum-based chemotherapy, we found that KRAS mutations were associated with shorter survival and thus poor treatment response. In addition, patients with KRAS mutations had significantly lower tumour expression of both BRCA1 and TYMS, but not SRC, compared with patients without such mutations. Our finding that BRCA1 levels alone (not TYMS or SRC) were correlated with overall survival duration underscores the complex pathology of NSCLC.
Proteins of the KRAS family transduce extracellular signals through membrane-bound receptors, such as from EGFR to downstream intracellular (RAS–RAF–MEK) pathways.25 KRAS mutations are mutually exclusive with EGFR mutations.26 Patients with EGFR mutations respond well to EGFR tyrosine kinase inhibitor-based drugs, including gefitinib and erlotinib, but these drugs are no more effective than platinum-based or other traditional therapies in patients carrying KRAS mutations.27 This poor response from KRAS mutation-harbouring tumours is likely due to aberrant activation of the RAS–RAF–MEK–ERK signalling pathway, which is downstream to EGFR and independent of EGFR tyrosine kinase activity inhibition.11 Although it is the most common mutation in lung adenocarcinoma, KRAS mutation remains an intriguing therapeutic target, as there has been little success in attempts to directly inhibit KRAS activity.11
It has been shown that KRAS mutations are present in up to 30% of lung adenocarcinomas, but are rarely found in squamous cell lung cancers.28 This is in accordance with our current finding that no squamous carcinomas were positive for KRAS mutations. The frequency of KRAS mutation in adenocarcinoma was 8.8% in the present study, consistent with studies indicating that the prevalence of KRAS mutation in adenocarcinomas is much lower in Asians than in Caucasians.29 All KRAS mutations in the present study were found in stage IV metastatic tumours. This is in line with the hypothesis that KRAS mutations play a critical role in NSCLC tumourigenesis and are more common in advanced lung cancers.11
An accumulating pool of evidence suggests that KRAS mutations are associated with poor overall survival,30–33 although some studies have reported inconclusive results.11,34 Patients with KRAS mutations had a significantly shorter survival time than patients with wild type KRAS in the present study, supporting the notion that KRAS mutation is associated with poor overall survival in NSCLC. Interestingly, we also found that KRAS mutation was associated with low tumour expression of BRCA1, which encodes a tumour suppressor protein involved in repairing damaged DNA.35 BRCA1 mutation has been linked to increased risk of breast and ovarian cancers,35,36 and its expression level has been identified as a potential biomarker for prognostic purpose in several cancers.16,17,37–44 In support of this hypothesis, we found that low BRCA1 expression was associated with poor survival in our study. It is possible that oncogenic KRAS mutations may lead to decreased BRCA1 expression, which in turn may lead to resistance to platinum-based doublet chemotherapy and poor overall survival.
Mutations in KRAS were associated with low TYMS expression in the present study, and there was a positive correlation between TYMS and BRCA1 expression levels. Others have suggested that TYMS is associated with treatment outcome and survival after fluorouracil (5-FU)/oxaliplatin chemotherapy in metastatic colorectal18,19 and gastric cancer,20 but there was no association between TYMS expression level and overall survival in the present study. This may reflect the heterogeneity and complexity of NSCLC and the functional differences in DNA repair between TYMS and BRCA1 or other DNA repair enzymes.18
The gene SRC is aberrantly activated and overexpressed in a wide range of human cancers,45 including lung cancers.21 Aberrant KRAS activation has been shown to increase SRC expression and activity, driving metastatic growth and therapy resistance in pancreatic cancer.46 There was no association between KRAS mutations and SRC expression level in the present study.
Our study is limited by the small cohort; the resulting low frequency of KRAS mutations may limit the power of statistical analyses.
In conclusion, KRAS mutations are associated with poor overall survival and resistance to platinum-based doublet chemotherapy in patients with NSCLC. In addition, KRAS mutations are associated with low tumour expression of BRCA1 and TYMS, but not SRC. KRAS mutation status and tumour expression of BRCA1 are potential biomarkers for tailoring chemotherapy and predicting clinical outcome.
Declaration of conflicting interest
The authors declare that there are no conflicts of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2.Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 2002; 346: 92–98. [DOI] [PubMed] [Google Scholar]
- 3.Cheng L, Alexander RE, Maclennan GT, et al. Molecular pathology of lung cancer: key to personalized medicine. Mod Pathol 2012; 25: 347–369. [DOI] [PubMed] [Google Scholar]
- 4.Buettner R, Wolf J, Thomas RK. Lessons learned from lung cancer genomics: the emerging concept of individualized diagnostics and treatment. J Clin Oncol 2013; 31: 1858–1865. [DOI] [PubMed] [Google Scholar]
- 5.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004; 350: 2129–2139. [DOI] [PubMed] [Google Scholar]
- 6.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304: 1497–1500. [DOI] [PubMed] [Google Scholar]
- 7.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007; 448: 561–566. [DOI] [PubMed] [Google Scholar]
- 8.Bos JL. Ras oncogenes in human cancer: a review. Cancer Res 1989; 49: 4682–4689. [PubMed] [Google Scholar]
- 9.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol 2008; 9: 517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slebos RJ, Kibbelaar RE, Dalesio O, et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med 1990; 323: 561–565. [DOI] [PubMed] [Google Scholar]
- 11.Roberts PJ, Stinchcombe TE. KRAS mutation: should we test for it, and does it matter? J Clin Oncol 2013; 31: 1112–1121. [DOI] [PubMed] [Google Scholar]
- 12.Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev 2007; 107: 1387–1407. [DOI] [PubMed] [Google Scholar]
- 13.van de Vaart PJ, Belderbos J, de Jong D, et al. DNA-adduct levels as a predictor of outcome for NSCLC patients receiving daily cisplatin and radiotherapy. Int J Cancer 2000; 89: 160–166. [PubMed] [Google Scholar]
- 14.Jordan P, Carmo-Fonseca M. Molecular mechanisms involved in cisplatin cytotoxicity. Cell Mol Life Sci 2000; 57: 1229–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Y, Geacintov NE, Broyde S. Nucleotide excision repair efficiencies of bulky carcinogen-DNA adducts are governed by a balance between stabilizing and destabilizing interactions. Biochemistry 2012; 51: 1486–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennedy RD, Quinn JE, Johnston PG, et al. BRCA1: mechanisms of inactivation and implications for management of patients. Lancet 2002; 360: 1007–1014. [DOI] [PubMed] [Google Scholar]
- 17.Taron M, Rosell R, Felip E, et al. BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum Mol Genet 2004; 13: 2443–2449. [DOI] [PubMed] [Google Scholar]
- 18.Shirota Y, Stoehlmacher J, Brabender J, et al. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J Clin Oncol 2001; 19: 4298–4304. [DOI] [PubMed] [Google Scholar]
- 19.Salonga D, Danenberg KD, Johnson M, et al. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res 2000; 6: 1322–1327. [PubMed] [Google Scholar]
- 20.Lenz HJ, Leichman CG, Danenberg KD, et al. Thymidylate synthase mRNA level in adenocarcinoma of the stomach: a predictor for primary tumor response and overall survival. J Clin Oncol 1996; 14: 176–182. [DOI] [PubMed] [Google Scholar]
- 21.Mazurenko NN, Kogan EA, Zborovskaya IB, et al. Expression of pp60c-src in human small cell and non-small cell lung carcinomas. Eur J Cancer 1992; 28: 372–377. [DOI] [PubMed] [Google Scholar]
- 22.Edge S, Byrd DR, Compton CC, et al (eds). AJCC Cancer Staging Manual. 7th ed. Berlin: Springer, 2010.
- 23.Beasley MB, Brambilla E, Travis WD. The 2004 World Health Organization classification of lung tumors. Semin Roentgenol 2005; 40: 90–97. [DOI] [PubMed] [Google Scholar]
- 24.Guo Y, Ma J, Lyu X, et al. Non-small cell lung cancer with EML4-ALK translocation in Chinese male never-smokers is characterized with early-onset. BMC Cancer 2014; 14: 834–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahearn IM, Haigis K, Bar-Sagi D, et al. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol 2011; 13: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karachaliou N, Mayo C, Costa C, et al. KRAS mutations in lung cancer. Clin Lung Cancer 2013; 14: 205–214. [DOI] [PubMed] [Google Scholar]
- 27.Mao C, Qiu LX, Liao RY, et al. KRAS mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: a meta-analysis of 22 studies. Lung Cancer 2010; 69: 272–278. [DOI] [PubMed] [Google Scholar]
- 28.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res 2012; 72: 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korpanty GJ, Graham DM, Vincent MD, et al. Biomarkers that currently effect clinical practice in lung cancer: EGFR, ALK, MET, ROS-1 and KRAS. Front Oncol 2014; 4: 204–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 2011; 22: 2616–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson ML, Sima CS, Chaft J, et al. Association of KRAS and EGFR mutations with survival in patients with advanced lung adenocarcinomas. Cancer 2013; 119: 356–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paik PK, Johnson ML, D’Angelo SP, et al. Driver mutations determine survival in smokers and never-smokers with stage IIIB/IV lung adenocarcinomas. Cancer 2012; 118: 5840–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metro G, Chiari R, Bennati C, et al. Clinical outcome with platinum-based chemotherapy in patients with advanced nonsquamous EGFR wild-type non–small-cell lung cancer segregated according to KRAS mutation status. Clin Lung Cancer 2014; 15: 86–92. [DOI] [PubMed] [Google Scholar]
- 34.Macerelli M, Caramella C, Faivre L, et al. Does KRAS mutational status predict chemoresistance in advanced non-small cell lung cancer (NSCLC)? Lung Cancer 2014; 83: 383–388. [DOI] [PubMed] [Google Scholar]
- 35.Campeau PM, Foulkes WD, Tischkowitz MD. Hereditary breast cancer: new genetic developments, new therapeutic avenues. Hum Genet 2008; 124: 31–42. [DOI] [PubMed] [Google Scholar]
- 36.Pal T, Permuth-Wey J, Betts JA, et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer 2005; 104: 2807–2816. [DOI] [PubMed] [Google Scholar]
- 37.James CR, Quinn JE, Mullan PB, et al. BRCA1, a potential predictive biomarker in the treatment of breast cancer. Oncologist 2007; 12: 142–150. [DOI] [PubMed] [Google Scholar]
- 38.Carser JE, Quinn JE, Michie CO, et al. BRCA1 is both a prognostic and predictive biomarker of response to chemotherapy in sporadic epithelial ovarian cancer. Gynecol Oncol 2011; 123: 492–498. [DOI] [PubMed] [Google Scholar]
- 39.Karachaliou N, Papadaki C, Lagoudaki E, et al. Predictive value of BRCA1, ERCC1, ATP7B, PKM2, TOPOI, TOPΟ-IIA, TOPOIIB and C-MYC genes in patients with small cell lung cancer (SCLC) who received first line therapy with cisplatin and etoposide. PLoS ONE 2013; 8: e74611–e74611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang JG, Jin ZY, Gao XD, et al. Predictive role of RRM1 and BRCA1 mRNA expression on the clinical outcome of advanced non-small cell lung cancer. Genet Mol Res 2014; 13: 5292–5298. [DOI] [PubMed] [Google Scholar]
- 41.Wang TB, Zhang NL, Wang SH, et al. Expression of ERCC1 and BRCA1 predict the clinical outcome of non-small cell lung cancer in patients receiving platinum-based chemotherapy. Genet Mol Res 2014; 13: 3704–3710. [DOI] [PubMed] [Google Scholar]
- 42.Han Y, Wang XB, Xiao N, et al. mRNA expression and clinical significance of ERCC1, BRCA1, RRM1, TYMS and TUBB3 in postoperative patients with non-small cell lung cancer. Asian Pac J Cancer Prev 2013; 14: 2987–2990. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Yang H, Qiu Y, et al. Association between epidermal growth factor receptor gene copy number and ERCC1, BRCA1 protein expression in Chinese patients with non-small cell lung cancer. Med Oncol 2014; 31: 803–803. [DOI] [PubMed] [Google Scholar]
- 44.Qin X, Yao W, Li W, et al. ERCC1 and BRCA1 mRNA expressions are associated with clinical outcome of non-small cell lung cancer treated with platinum-based chemotherapy. Tumour Biol 2014; 35: 4697–4704. [DOI] [PubMed] [Google Scholar]
- 45.Irby RB, Yeatman TJ. Role of Src expression and activation in human cancer. Oncogene 2000; 19: 5636–5642. [DOI] [PubMed] [Google Scholar]
- 46.Kelber JA, Reno T, Kaushal S, et al. KRas induces a Src/PEAK1/ErbB2 kinase amplification loop that drives metastatic growth and therapy resistance in pancreatic cancer. Cancer Res 2012; 72: 2554–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]