Abstract
Objective
A meta-analysis of the association between haplotypical variants of the apolipoprotein E (APOE) gene (ɛ2/ɛ3/ɛ4) and obstructive sleep apnoea (OSA) risk and changes in lipid profile.
Methods
Electronic databases were searched to retrieve articles that provided data on APOE gene ɛ2/ɛ3/ɛ4 variants in patients with OSA and healthy controls. Data were extracted from eligible articles and statistical analyses were performed.
Results
The meta-analysis included 14 articles involving 19 study populations (3198 patients and 6031 controls). There was no significant association between the presence of the ɛ4 allele and OSA risk. The presence of ɛ4 was associated with significantly increased total cholesterol and decreased high-density lipoprotein cholesterol, compared with ɛ4 allele negative individuals. There was a low probability of publication bias but significant heterogeneity.
Conclusions
There was no association between APOE ɛ2/ɛ3/ɛ4 and OSA susceptibility. The presence of APOE ɛ4 was associated with changes in lipid profile.
Keywords: Obstructive sleep apnoea, apolipoprotein E, variant, meta-analysis
Introduction
Obstructive sleep apnoea (OSA) is the most common form of apnoea, and is characterized by snoring, periodic apnoea, hypoxemia during sleep and daytime hypersomnolence.1 Despite several well-established modifiable risk factors such as obesity, compelling evidence supports a genetic component underlying the pathogenesis of OSA.2 As documented by family studies, individuals who had affected first-degree relatives were more likely to be at risk of OSA compared with those without an affected first-degree relative, and the risk increased in proportion to the number of affected relatives.3,4 It is estimated that up to 35% of the variability in OSA severity (as measured by apnoea–hypopnea index [AHI]) may be due to genetic determinants.5 Thus far, 85 genes have been listed as candidate OSA-susceptibility genes (hugenavigator.net/), with the gene encoding apolipoprotein E (APOE) ranked in the top three. Over the past decade, several association studies have independently assessed the relationship between OSA risk and a well-characterized haplotypical variant of the APOE gene (ɛ2/ɛ3/ɛ4; defined by the loci rs429358 and rs7412).6–8 These studies had poor reproducibility, possibly due to genetic heterogeneity across ethnic groups, methodological divergences and other confounding factors such as the coexistence of hypertension. To fully address this issue, this meta-analysis updates the findings of these analyses6–8 in order to re-evaluate the association between OSA risk and APOE ɛ2/ɛ3/ɛ4 alleles. In addition, we analysed changes in lipid profile and explored potential sources of heterogeneity.
Materials and methods
The implementation of this meta-analysis adheres to the protocols outlined in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (Supplementary PRISMA checklist).
Literature search
The electronic databases PubMed®, Web of Science™, Wanfang (Chinese) and CNKI (Chinese) were searched to retrieve potentially eligible articles that provided data on APOE ɛ2/ɛ3/ɛ4 in patients with OSA and healthy controls published up to and including 10 May 2015. The key words were ‘obstructive sleep apnoea’ or ‘sleep disorder’ or ‘breathing’ [Title] and ‘apolipoprotein E’ or ‘APOE’ or ‘APO E’ [Abstract], and ‘allele’ or ‘genotype’ or ‘polymorphism’ or ‘variant’ or ‘SNP’ [Abstract]. We additionally checked the reference list of each major article to ensure comprehensive coverage.
Eligibility criteria
Inclusion criteria were: (i) OSA as the clinical endpoint; (ii) case–control design; (iii) the genotype or allele counts of APOE gene ɛ2/ɛ3/ɛ4 or the counts of ɛ4 allele positive and negative individuals in patients and controls; (iv) effect-size estimates presented as odds ratio (OR) with 95% confidence interval (95% CI). In the case of sample duplication the study with the larger sample size and more complete information was retained. Articles written in languages other than English and/or Chinese were excluded.
The title and abstract of each article were assessed for primary eligibility by two investigators acting independently and in duplicate (Z.L. and X.W.). In the case of uncertainty, the full text was retrieved for further evaluation and disagreements were resolved by consensus.
Data retrieval
The following data were extracted independently and in duplicate by two investigators (Z.L. and X.W.): first author’s last name; year of publication; race; study design; source of controls; AHI; diagnostic method for OSA; sample size; genotype/allele counts/ORs and 95% CIs; mean body mass index (BMI); triglyceride; total cholesterol; high- and low-density lipoprotein cholesterol (HDLC and LDLC); age; sex; prevalence of smoking; duration of education; and prevalence of hypertension and diabetes mellitus. Any disagreements were resolved during data retrieval by consensus and review of the full text of the article in question.
Statistical analyses
The DerSimonian and Laird method and a random-effects model were used to pool individual effect-size estimates for the association between APOE ɛ2/ɛ3/ɛ4 and OSA susceptibility.9 Differences in BMI, triglyceride, total cholesterol, HDLC and LDLC between ɛ4 allele positive and negative carriers were expressed as weighted mean difference (WMD) with 95% CI.
Heterogeneity was judged by the inconsistency index (I2) statistic, with statistically significant heterogeneity indicated by I2 > 50%. Sources of heterogeneity were evaluated by stratified analysis of categorical variables (study design, source of controls, AHI cut off, sample size) and by meta-regression analysis of continuous variables (age, sex, BMI, smoking, education, hypertension and diabetes mellitus). The probability of publication bias was visually inspected using Begg’s funnel plots and statistically assessed with Egger’s test (significance level 10%), using the trim-and-fill method to impute the presence of missing studies to yield an unbiased pooled estimate.
All statistical analyses were completed with Stata® software version 12.0 (StataCorp, College Station, TX, USA) for Windows®. Unless otherwise indicated, P-values < 0.05 were considered statistically significant.
Results
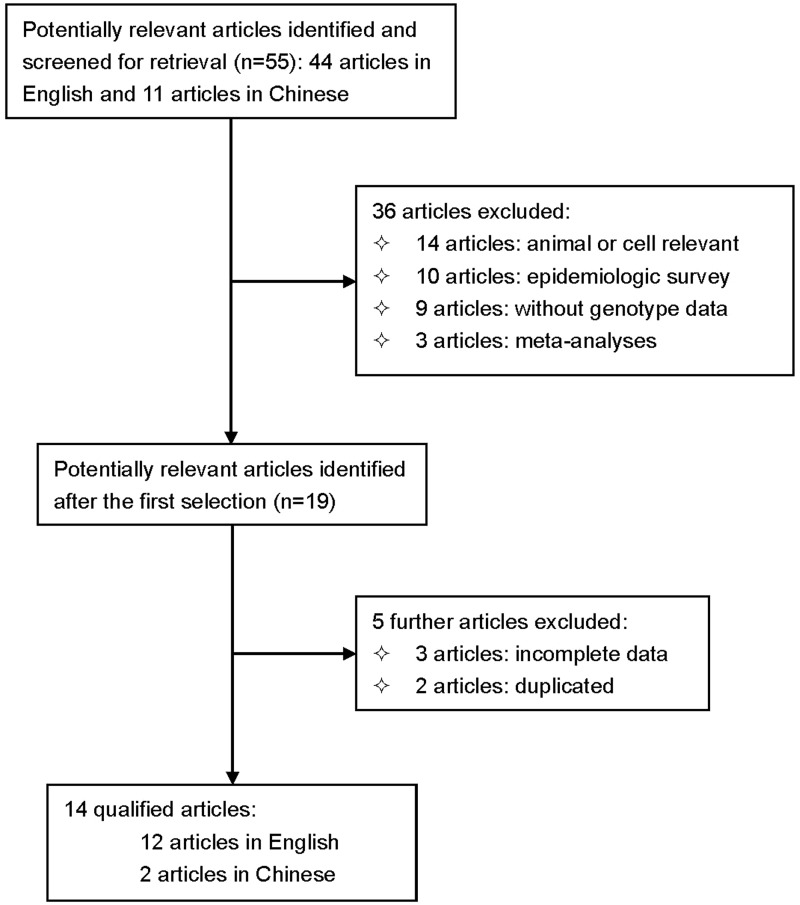
The initial literature search identified 55 potential articles. After exclusions, data were extracted from 14 articles (12 in English and two in Chinese) that fulfilled the predetermined eligibility criteria.10–23 Figure 1 presents a flow diagram of search strategy and study selection; Table 1 shows the characteristics of all study populations. A total of 19 study populations were available from the 14 included studies, with 3198 patients and 6031 controls. There were no statistically significant between-group differences in age, smoking, hypertension and diabetes mellitus. Patients were significantly more likely to be obese (P = 0.0009) and male (P = 0.0016) than controls (Table 1).
Figure 1.
Flow diagram of search strategy and study selection for a meta-analysis evaluating the association between obstructive sleep apnoea (OSA) risk and the apolipoprotein gene (APOE) ɛ2/ɛ3/ɛ4 alleles.
Table 1.
Characteristics of studies included in a meta-analysis evaluating the association between obstructive sleep apnoea risk and the apolipoprotein gene (APOE) ɛ2/ɛ3/ɛ4 alleles.
|
n
|
Age, years | Male sex, % | |||||||||||
| Author, year |
Country | Design | Source | AHI cut off | Method | Cases | Controls | Cases | Controls | Cases | Controls | ||
| Uyrum, 201510 | Turkey | Pro | Hosp | ≥5 | PSG | 42 | 31 | 54 | 44 | 59.5 | 38.8 | ||
| Tisko (mild), 201411 | Slovakia | Retro | Hosp | >5–<15 | PSG | 126 | 128 | 49.5 | 47.8 | 70.6 | 53.1 | ||
| Tisko (moderate), 201411 | Slovakia | Retro | Hosp | > 15–<30 | PSG | 66 | 128 | 51.6 | 47.8 | 68.2 | 53.1 | ||
| Tisko (severe), 201411 | Slovakia | Retro | Hosp | ≥30 | PSG | 199 | 128 | 51.2 | 47.8 | 83.9 | 53.1 | ||
| Osorio (mild), 201412 | USA | Pro | Pop | >5–15 | PSG | 52 | 25 | 67.8 | 65.3 | 41.2 | 32.0 | ||
| Osorio (moderate/ severe), 201412 | USA | Pro | Pop | ≥15 | PSG | 19 | 25 | 70.1 | 65.3 | 42.1 | 32.0 | ||
| Nikodemova (mild), 201313 | USA | Pro | Pop | >5–<15 | PSG | 399 | 1146 | 56.4 | 52.1 | 62.2 | 54.1 | ||
| Nikodemova (moderate/ severe), 201313 | USA | Pro | Pop | ≥15 | PSG | 298 | 1146 | 56.6 | 52.1 | 71.5 | 54.1 | ||
| Cosentino, 200814 | Italy | Retro | Pop | ≥15 | PSG | 123 | 121 | 58.6 | 57.9 | 66.7 | 64.5 | ||
| Sheng, 200815 | China | Retro | Pop | ≥5 | PSG | 84 | 106 | 48.6 | 49.8 | 86.9 | 86.8 | ||
| Zheng, 200716 | China | Retro | Hosp | ≥5 | PSG | 50 | 40 | 39 | 44.5 | 100 | 100 | ||
| Gozal, 200717 | USA | Retro | Pop | >1 | PSG | 112 | 146 | 6.3 | 6.4 | 54.1 | 55.4 | ||
| Craig, 200618 | UK | Retro | Hosp | Other | NPI-D | 217 | 185 | 78 | 78 | 40.0 | 33.0 | ||
| Larkin (white), 200619 | USA | Pro | Pop | ≥15 | PSG | 218 | 796 | 40 | 38.7 | 48.2 | 45.7 | ||
| Larkin (black), 200619 | USA | Pro | Pop | ≥15 | PSG | 197 | 796 | 37.1 | 38.7 | 42.8 | 45.7 | ||
| Gottlieb, 200420 | USA | Pro | Pop | ≥15 | PSG | 337 | 1438 | 71 | 71 | 45.0 | 45.0 | ||
| Kadotani, 200121 | USA | Pro | Pop | ≥15 | PSG | 66 | 725 | 49 | 49 | 58.3 | 58.3 | ||
| Foley, 200122 | USA | Pro | Pop | ≥15 | PSG | 302 | 416 | NA | NA | 100 | 100 | ||
| Saarelainen, 199823 | Finland | Retro | Pop | ≥5 | PSG | 291 | 728 | 53.3 | 53.7 | 90.7 | 77.6 | ||
| BMI, kg/m2 |
Smoking, % | Education, years | AHI | Hypertension, % | Diabetes mellitus, % | ||||||||
| Cases |
Controls |
Cases |
Controls |
Cases |
Controls |
Cases |
Controls |
Cases |
Controls |
Cases |
Controls |
OR; 95% CI |
Adjusted |
| 35 | 31.8 | NA | NA | NA | NA | 31.3 | 2.2 | NA | NA | NA | NA | 2.90; 0.56, 15.05 | No |
| 29.6 | 28.4 | 34.9 | 34.4 | NA | NA | 9.4 | 2.3 | 33.3 | 45.3 | 6.3 | 3.1 | 0.71; 0.40, 1.24 | No |
| 31.1 | 28.4 | 30.3 | 34.4 | NA | NA | 20.8 | 2.3 | 56.1 | 45.3 | 10.6 | 3.1 | 0.58; 0.28, 1.19 | No |
| 33.9 | 28.4 | 45.7 | 34.4 | NA | NA | 60.4 | 2.3 | 62.3 | 45.3 | 17.6 | 3.1 | 0.77; 0.47, 1.27 | No |
| 25.5 | 24.2 | NA | NA | 17.2 | 16.2 | 8.3 | 2.3 | 29.4 | 24.0 | 7.8 | 8.0 | 1.18; 0.41, 3.37 | No |
| 28.9 | 24.2 | NA | NA | 16.3 | 16.2 | 30.7 | 2.3 | 31.6 | 24.0 | 5.3 | 8.0 | 1.19; 0.32, 4.37 | No |
| 32.5 | 28.9 | 12.0 | 14.5 | 14.2 | 14.7 | 8.7 | 1.4 | 34.8 | 20.5 | NA | NA | 0.81; 0.62, 1.06 | No |
| 36.6 | 28.9 | 11.4 | 14.5 | 14.0 | 14.7 | 29.4 | 1.4 | 51.3 | 20.5 | NA | NA | 1.14; 0.86, 1.50 | No |
| 36.1 | 30.2 | 45.5 | 20.0 | 8.1 | 7.4 | NA | NA | 61.8 | 57.1 | 18.7 | 8.6 | 1.22; 0.64, 2.31 | No |
| 29.58 | 24.71 | NA | NA | NA | NA | NA | NA | NA | 0.0 | NA | 0.0 | 7.12; 3.41, 14.89 | No |
| NA | NA | NA | NA | NA | NA | NA | NA | 0.0 | 0.0 | 0.0 | 0.0 | 3.50; 1.05, 11.66 | No |
| 17 | 16.9 | NA | NA | NA | NA | 8.6 | 0.8 | NA | NA | NA | NA | 4.47; 1.27, 15.75 | No |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.03; 0.69, 1.53 | No |
| 29.6 | 30.3 | NA | NA | NA | NA | NA | NA | 23.6 | 28.7 | NA | NA | 0.85; 0.56, 1.00 | Yes |
| 31.1 | 30.3 | NA | NA | NA | NA | NA | NA | 35.0 | 28.7 | NA | NA | 0.64; 0.42, 0.98 | Yes |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.41; 1.06, 1.87 | Yes |
| 30 | 30 | 16.4 | 16.4 | NA | NA | NA | NA | 33.0 | 33.0 | NA | NA | 2.00; 1.20, 3.50 | Yes |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.77; 0.52, 1.14 | Yes |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 1.00; 0.75, 1.33 | No |
AHI, apnoea–hypopnea index; BMI, body mass index; OR, odds ratio; 95% CI, 95% confidence interval. Pro, prospective; Retro, retrospective; Hosp, hospital; Pop, population; PSG, polysomnography; NPI-D, neuropsychiatric inventory with caregiver distress; NA, not available;
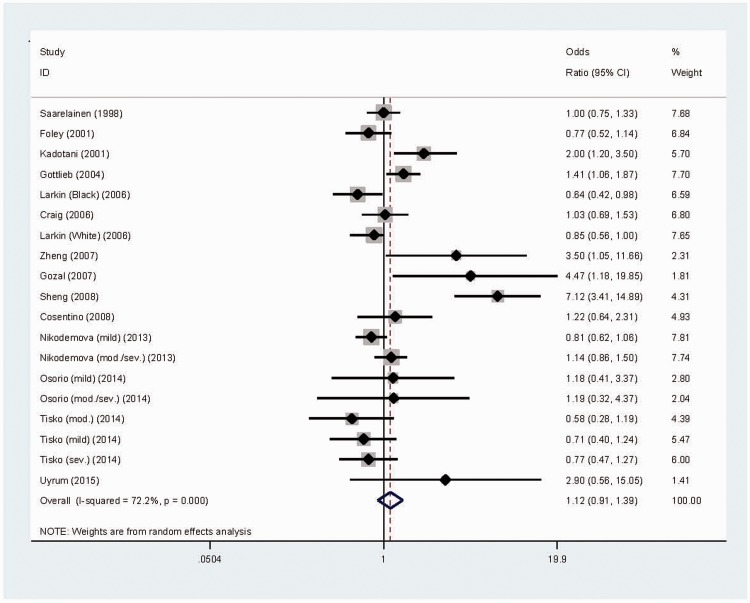
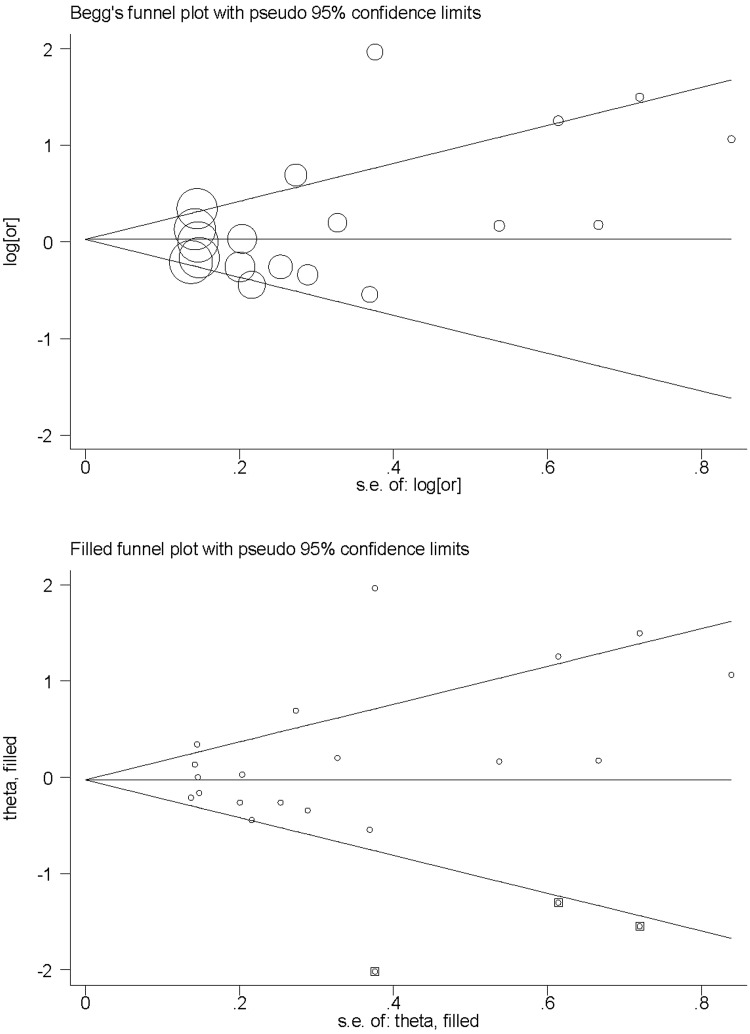
There was no significant association between APOE ɛ4-positivity and OSA risk in the pooled study population. There was significant heterogeneity (I2 = 72.2%; P < 0.0005; Figure 2) and a low probability of publication bias for this comparison, as illustrated by Begg’s funnel plot (Figure 3) and Egger’s test. Trim-and-fill analysis suggested that three studies were missing to general a symmetrical filled funnel plot (Figure 3). After adjusting for the three missing studies, the presence of ɛ4 allele was associated with a nonsignificant 2% reduction in OSA risk (95% CI 0.77, 1.25).
Figure 2.
Forest plot of a meta-analysis of the association between obstructive sleep apnoea (OSA) risk and the apolipoprotein gene (APOE) ɛ4 allele.10–23 The colour version of this figure is available at: http://imr.sagepub.com.
Figure 3.
Begg’s and Filled funnel plots for a meta-analysis of the association between obstructive sleep apnoea (OSA) risk and the apolipoprotein gene (APOE) ɛ4 allele.10–23
There was no significant association between APOE ɛ4-positivity and OSA risk in adults10–17,19–23 or when analysis was limited to study populations with adjusted effect-size estimates.19–22
Data regarding APOE gene ɛ2/ɛ3/ɛ4 alleles were provided in 11 study populations.10–12,14–16,18,23 When using the ɛ3 allele as a reference, there was no significant association between either ɛ2 or ɛ4 and OSA risk. There was significant heterogeneity for this comparison (I2 = 66.2%; P = 0.001).
Data regarding BMI and lipid parameters were provided by four studies.15,16,20,21 Total cholesterol was significantly higher (P = 0.007) and HDLC was significantly lower (P = 0.040) in ɛ4-positive individuals than ɛ4-negative individuals (Table 2).
Table 2.
Body mass index and lipid parameters in apolipoprotein gene (APOE) ɛ4-positive and negative individuals.
| Parameter | Studies, n |
n
|
WMD | 95% CI | Statistical significance | Heterogeneity | |
|---|---|---|---|---|---|---|---|
| ɛ4+ | ɛ4− | ||||||
| BMI, kg/m2 | 4 | 713 | 2017 | 0.027 | −0.817, 0.871 | NS | I2 = 56.4% |
| TG, mmol/l | 4 | 713 | 2017 | 0.203 | −0.085, 0.491 | NS | I2 = 77.4% |
| TC, mmol/l | 4 | 713 | 2017 | 0.342 | 0.095, 0.590 | P = 0.007 | I2 = 78.7% |
| HDLC, mmol/l | 4 | 713 | 2017 | −0.052 | −0.103, −0.002 | P = 0.040 | I2 = 31.5% |
| LDLC, mmol/l | 3 | 274 | 681 | 0.197 | −0.097, 0.491 | NS | I2 = 64.4% |
WMD, weighted mean difference; CI, confidence interval; BMI, body mass index; NS, not statistically significant (P ≥ 0.05; random effects model); TG, triglyceride; TC, total cholesterol; HDLC, high-density lipoprotein cholesterol; LDLC, low-density lipoprotein cholesterol.
Stratified analyses revealed no effect of study design (prospective vs retrospective), source of controls (population-based vs hospital-based), AHI cut off (≥15 vs >5–<15) and sample size (≥500 vs <500) on heterogeneity (Table 3). The presence of ɛ4 was significantly associated with OSA risk in studies including only Chinese individuals (OR 5.87; 95% CI 3.13, 11.00).15,16
Table 3.
Stratified analyses of the association between obstructive sleep apnoea (OSA) risk and the apolipoprotein gene (APOE) ɛ4 allele.
| Subgroups | Studies, n |
n
|
OR | 95% CI | Heterogeneity | |
|---|---|---|---|---|---|---|
| Patients | Controls | |||||
| Study design | ||||||
| Prospective | 10 | 1930 | 6544 | 1.02 | 0.82, 1.28 | I2 = 62.9% |
| Retrospective | 9 | 1268 | 1710 | 1.33 | 0.86, 2.05 | I2 = 80.0% |
| Source of controls | ||||||
| Population-based | 13 | 2498 | 7614 | 1.20 | 0.93,1.55 | I2 = 77.7% |
| Hospital-based | 6 | 700 | 640 | 0.64, 1.38 | I2 = 48.7% | |
| AHI cut off | ||||||
| >5–<15 | 3 | 577 | 1299 | 0.88 | 0.64, 1.02 | I2 = 0.0% |
| ≥15 | 10 | 1825 | 5719 | 0.99 | 0.79, 1.24 | I2 = 62.1% |
| ≥5 | 4 | 467 | 905 | 2.82 | 0.84, 9.45 | I2 = 88.9% |
| Total sample size | ||||||
| <500 | 11 | 1090 | 1063 | 1.45 | 0.91, 2.31 | I2 = 75.9% |
| ≥500 | 8 | 2108 | 7191 | 1.00 | 0.81, 1.22 | I2 = 68.8% |
| Chinese subjects | 2 | 134 | 146 | 5.87 | 3.13, 11.00 | I2 = 0.0% |
OR, odds ratio; CI, confidence interval; AHI, apnoea–hypopnea index.
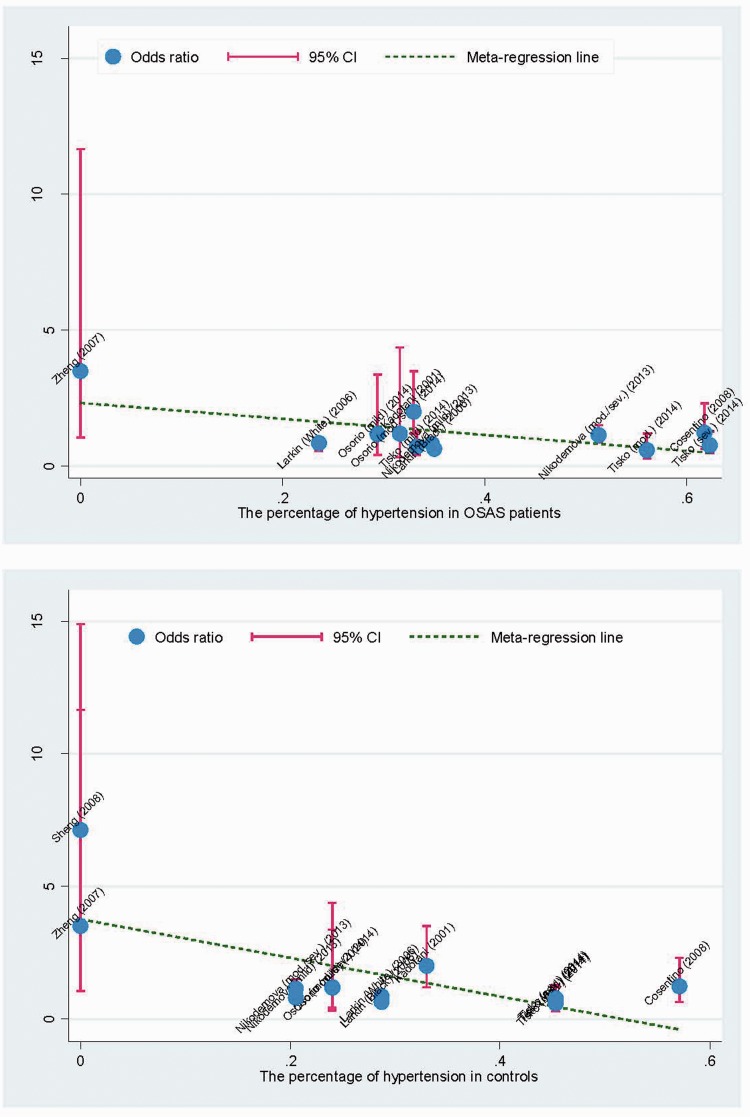
Meta-regression analysis found that hypertension was significantly correlated with OSA risk in both patients (r = −0.64; P = 0.024) and controls (r = −0.68; P = 0.01; Figure 4). There was no significant association between OSA risk and age, sex, BMI, smoking, duration of education, or diabetes mellitus.
Figure 4.
Meta-regression analysis of the association between hypertension and risk of obstructive sleep apnoea (OSA). The colour version of this figure is available at: http://imr.sagepub.com.
Discussion
In accordance with the findings of others,6–8 the present meta-analysis of 14 articles and 9229 study subjects found no association between OSA risk and APOE ɛ2/ɛ3/ɛ4 positivity. The presence of the ɛ4 allele was significantly correlated with increased total cholesterol and decreased HDLC, however.
There is a growing recognition that pathophysiological mechanisms involving dysregulated lipid metabolism underlie OSA.11,24 APOE is a lipid transport and signalling protein with a key role in lipid metabolism,25 and its function is determined by the presence of three common alleles (ɛ2, ɛ3, ɛ4).26 Generally, a particular genetic variant could alter disease risk through its effects on either circulating concentrations or physiological function of a particular protein. The present analysis confirms the observation of others6–8 that individuals with different APOE ɛ2/ɛ3/ɛ4 genotypes show statistically significant differences in their circulating total cholesterol and HDLC levels. The absence of an association between ɛ2/ɛ3/ɛ4 alleles and OSA risk in the present analysis suggest that the principal differences in lipid profile driven by these variants relate to protein concentrations rather than function. The ɛ2/ɛ3/ɛ4 alleles appear to play a significant role in cholesterol regulation, although this is not strong enough to predict individual differences in OSA susceptibility.
Genetic epidemiological studies have shown varying and often nonreproducible findings regarding the association between APOE ɛ2/ɛ3/ɛ4 alleles and OSA susceptibility across ethnic groups. For example, the presence of the ɛ4 allele conferred a reduced risk for OSA in one study from the USA19 but an increased risk in another,21 and seemed to be neutral in a UK population.18 This lack of significance may be due to heterogeneity of effect associated with the presence of hypertension, as reflected in our meta-regression analysis. It is worth noting that the presence of hypertension might neutralize the contributory role of the ɛ4 allele in the pathogenesis of OSA, since ɛ4 was strongly associated with OSA risk after restricting analysis to the two studies of Chinese ancestry with normotensive controls.15,16 This finding may be too underpowered to be generalizable to a general population and other ethnic groups. On the other hand, OSA is an established risk factor for arterial hypertension,27 and the severity of hypertension is reported to be in proportion to that of OSA.28 Analyses stratified by OSA severity were still not significant in this meta-analysis, however. In view of the lack of necessary information, we agree that further adjustment for the severity of hypertension is critical to quantify reliably the association between APOE ɛ2/ɛ3/ɛ4 and OSA susceptibility.
The present analysis has several limitations. First, OSA is a polygenic disease, and it is not possible to unravel its genetic underpinnings by evaluating APOE ɛ2/ɛ3/ɛ4 alone. Secondly, all studies included in this meta-analysis were case–control in design. Thirdly, there was a very high level of heterogeneity between studies, but the level of publication bias was low. Finally, the limited sample sizes (especially in some stratified analyses) underline the requirement for large-scale, prospective studies.
In conclusion, this meta-analysis of 14 articles and 9229 study subjects failed to identify any association between APOE ɛ2/ɛ3/ɛ4 and OSA susceptibility. The presence of APOE ɛ4 was associated with changes in lipid profile. Importantly, hypertension was identified as a plausible source of heterogeneity between studies, and further studies incorporating information on the severity of hypertension are required to elucidate its role in OSA.
Declaration of conflicting interest
The authors declare that there are no conflicts of interest.
Funding
This analysis was supported by a grant from the “Miao Pu” Scientific Research Foundation of Fujian Medical University (2014MP030).
References
- 1.Eckert DJ, Jordan AS, Merchia P, et al. Central sleep apnea: pathophysiology and treatment. Chest 2007; 131: 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckert DJ, Malhotra A. Pathophysiology of adult obstructive sleep apnea. Proc Am Thorac Soc 2008; 5: 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Redline S, Tishler PV, Tosteson TD, et al. The familial aggregation of obstructive sleep apnea. Am J Respir Crit Care Med 1995; 151(3 pt 1): 682–687. [DOI] [PubMed] [Google Scholar]
- 4.Redline S, Tishler PV. The genetics of sleep apnea. Sleep Med Rev 2000; 4: 583–602. [DOI] [PubMed] [Google Scholar]
- 5.Buxbaum SG, Elston RC, Tishler PV, et al. Genetics of the apnea hypopnea index in Caucasians and African Americans: I. Segregation analysis. Genet Epidemiol 2002; 22: 243–253. [DOI] [PubMed] [Google Scholar]
- 6.Thakre TP, Mamtani MR, Kulkarni H. Lack of association of the APOE epsilon 4 allele with the risk of obstructive sleep apnea: meta-analysis and meta-regression. Sleep 2009; 32: 1507–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varvarigou V, Dahabreh IJ, Malhotra A, et al. A review of genetic association studies of obstructive sleep apnea: field synopsis and meta-analysis. Sleep 2011; 34: 1461–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H, Qian Y, Guan J, et al. No association between the ApoE ɛ2 and ɛ4 alleles and the risk of obstructive sleep apnea: a systematic review and meta-analysis. Biomed Rep 2015; 3: 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003; 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uyrum E, Balbay O, Annakkaya AN, et al. The relationship between obstructive sleep apnea syndrome and apolipoprotein E genetic variants. Respiration 2015; 89: 195–200. [DOI] [PubMed] [Google Scholar]
- 11.Tisko R, Sopkova Z, Habalova V, et al. Effects of apolipoprotein E genotype on serum lipids in obstructive sleep apnoea. Eur Respir J 2014; 43: 1097–1105. [DOI] [PubMed] [Google Scholar]
- 12.Osorio RS, Ayappa I, Mantua J, et al. Interaction between sleep-disordered breathing and apolipoprotein E genotype on cerebrospinal fluid biomarkers for Alzheimer’s disease in cognitively normal elderly individuals. Neurobiol Aging 2014; 35: 1318–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikodemova M, Finn L, Mignot E, et al. Association of sleep disordered breathing and cognitive deficit in APOE ɛ4 carriers. Sleep 2013; 36: 873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cosentino FI, Bosco P, Drago V, et al. The APOE epsilon4 allele increases the risk of impaired spatial working memory in obstructive sleep apnea. Sleep Med 2008; 9: 831–839. [DOI] [PubMed] [Google Scholar]
- 15.Sheng Y, Yu Q. Relationship between apolipoprotein E gene polymorphism and obstructive sleep apnea hypopnea syndrome. Int J Respir 2008; 28: 659–662. [Google Scholar]
- 16.Zheng H, Chang Z-w. Association of obstructive sleep apnea-hypopnea syndrome with gene polymorphism of apolipoprotein E. Clinical Focus 2007; 22: 1752–1756. [Google Scholar]
- 17.Gozal D, Capdevila OS, Kheirandish-Gozal L, et al. APOE epsilon 4 allele, cognitive dysfunction, and obstructive sleep apnea in children. Neurology 2007; 69: 243–249. [DOI] [PubMed] [Google Scholar]
- 18.Craig D, Hart DJ, Passmore AP. Genetically increased risk of sleep disruption in Alzheimer’s disease. Sleep 2006; 29: 1003–1007. [DOI] [PubMed] [Google Scholar]
- 19.Larkin EK, Patel SR, Redline S, et al. Apolipoprotein E and obstructive sleep apnea: evaluating whether a candidate gene explains a linkage peak. Genet Epidemiol 2006; 30: 101–110. [DOI] [PubMed] [Google Scholar]
- 20.Gottlieb DJ, DeStefano AL, Foley DJ, et al. APOE epsilon4 is associated with obstructive sleep apnea/hypopnea: the sleep heart health study. Neurology 2004; 63: 664–668. [DOI] [PubMed] [Google Scholar]
- 21.Kadotani H, Kadotani T, Young T, et al. Association between apolipoprotein E epsilon4 and sleep-disordered breathing in adults. JAMA 2001; 285: 2888–2890. [DOI] [PubMed] [Google Scholar]
- 22.Foley DJ, Masaki K, White L, et al. Relationship between apolipoprotein E epsilon4 and sleep-disordered breathing at different ages. JAMA 2001; 286: 1447–1448. [DOI] [PubMed] [Google Scholar]
- 23.Saarelainen S, Lehtimaki T, Kallonen E, et al. No relation between apolipoprotein E alleles and obstructive sleep apnea. Clin Genet 1998; 53: 147–148. [DOI] [PubMed] [Google Scholar]
- 24.Kheirandish L, Row BW, Li RC, et al. Apolipoprotein E-deficient mice exhibit increased vulnerability to intermittent hypoxia-induced spatial learning deficits. Sleep 2005; 28: 1412–1417. [DOI] [PubMed] [Google Scholar]
- 25.Huang Y, Mahley RW. Apolipoprotein E: structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol Dis 2014; 72 Pt A: 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svobodova H, Kucera F, Stulc T, et al. Apolipoprotein E gene polymorphism in the Mongolian population. Folia Biol (Praha) 2007; 53: 138–142. [PubMed] [Google Scholar]
- 27.Schulz R, Murzabekova G, Egemnazarov B, et al. Arterial hypertension in a murine model of sleep apnea: role of NADPH oxidase 2. J Hypertens 2014; 32: 300–305. [DOI] [PubMed] [Google Scholar]
- 28.Khan A, Patel NK, O’Hearn DJ, et al. Resistant hypertension and obstructive sleep apnea. Int J Hypertens 2013; 2013: 193010–193010. [DOI] [PMC free article] [PubMed] [Google Scholar]