Abstract
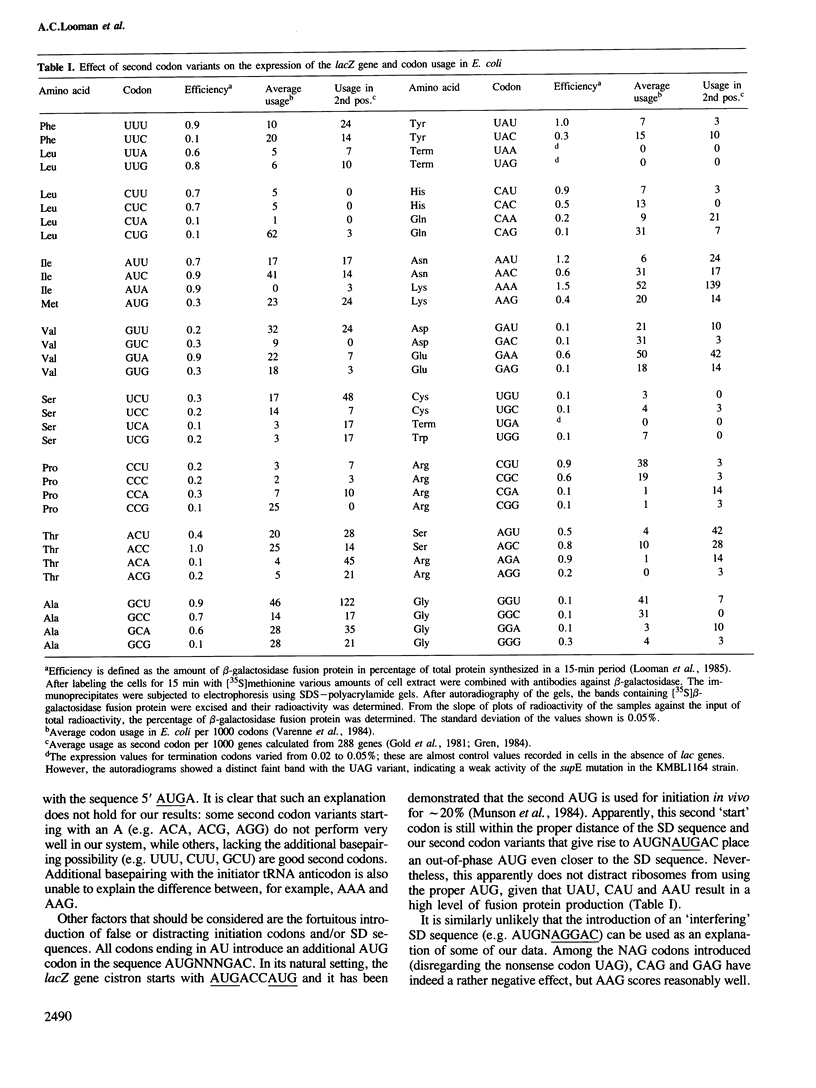
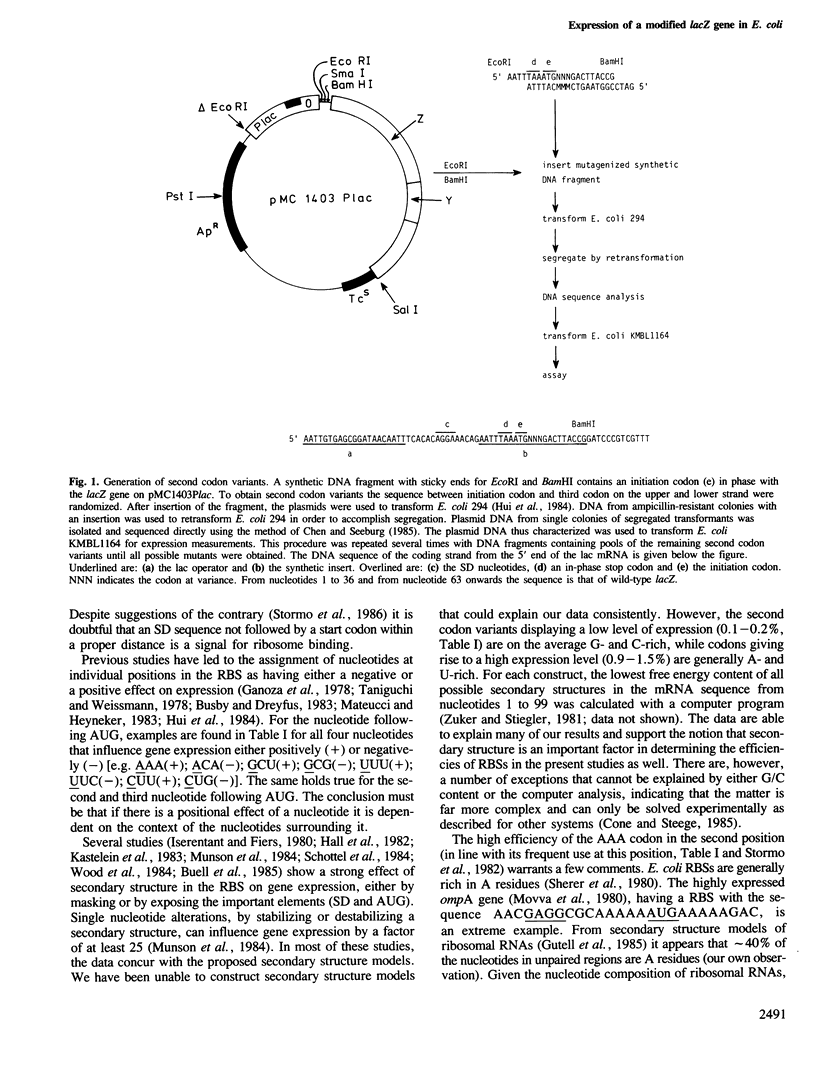
In a lacZ expression vector (pMC1403Plac), all 64 codons were introduced immediately 3' from the AUG initiation codon. The expression of the second codon variants was measured by immunoprecipitation of the plasmid-coded fusion proteins. A 15-fold difference in expression was found among the codon variants. No distinct correlation could be made with the level of tRNA corresponding to the codons and large differences were observed between synonymous codons that use the same tRNA. Therefore the effect of the second codon is likely to be due to the influence of its composing nucleotides, presumably on the structure of the ribosomal binding site. An analysis of the known sequences of a large number of Escherichia coli genes shows that the use of codons in the second position deviates strongly from the overall codon usage in E. coli. It is proposed that codon selection at the second position is not based on requirements of the gene product (a protein) but is determined by factors governing gene regulation at the initiation step of translation.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berkhout B., van Duin J. Mechanism of translational coupling between coat protein and replicase genes of RNA bacteriophage MS2. Nucleic Acids Res. 1985 Oct 11;13(19):6955–6967. doi: 10.1093/nar/13.19.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell G., Schulz M. F., Selzer G., Chollet A., Movva N. R., Semon D., Escanez S., Kawashima E. Optimizing the expression in E. coli of a synthetic gene encoding somatomedin-C (IGF-I). Nucleic Acids Res. 1985 Mar 25;13(6):1923–1938. doi: 10.1093/nar/13.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby S., Dreyfus M. Segment-specific mutagenesis of the regulatory region in the Escherichia coli galactose operon: isolation of mutations reducing the initiation of transcription and translation. Gene. 1983 Jan-Feb;21(1-2):121–131. doi: 10.1016/0378-1119(83)90154-3. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Cone K. C., Steege D. A. Messenger RNA conformation and ribosome selection of translational reinitiation sites in the lac repressor mRNA. J Mol Biol. 1985 Dec 20;186(4):725–732. doi: 10.1016/0022-2836(85)90392-4. [DOI] [PubMed] [Google Scholar]
- Ganoza M. C., Fraser A. R., Neilson T. Nucleotides contiguous to AUG affect translational initiation. Biochemistry. 1978 Jul 11;17(14):2769–2775. doi: 10.1021/bi00607a011. [DOI] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Gren E. J. Recognition of messenger RNA during translational initiation in Escherichia coli. Biochimie. 1984 Jan;66(1):1–29. doi: 10.1016/0300-9084(84)90188-3. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Gryczan T. J., Grandi G., Hahn J., Grandi R., Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 1980 Dec 20;8(24):6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Weiser B., Woese C. R., Noller H. F. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Gabay J., Débarbouillé M., Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982 Feb 18;295(5850):616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui A., Hayflick J., Dinkelspiel K., de Boer H. A. Mutagenesis of the three bases preceding the start codon of the beta-galactosidase mRNA and its effect on translation in Escherichia coli. EMBO J. 1984 Mar;3(3):623–629. doi: 10.1002/j.1460-2075.1984.tb01858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Iserentant D., Fiers W. Secondary structure of mRNA and efficiency of translation initiation. Gene. 1980 Apr;9(1-2):1–12. doi: 10.1016/0378-1119(80)90163-8. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looman A. C., Bodlaender J., de Gruyter M., Vogelaar A., van Knippenberg P. H. Secondary structure as primary determinant of the efficiency of ribosomal binding sites in Escherichia coli. Nucleic Acids Res. 1986 Jul 11;14(13):5481–5497. doi: 10.1093/nar/14.13.5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looman A. C., de Gruyter M., Vogelaar A., van Knippenberg P. H. Effects of heterologous ribosomal binding sites on the transcription and translation of the lacZ gene of Escherichia coli. Gene. 1985;37(1-3):145–154. doi: 10.1016/0378-1119(85)90267-7. [DOI] [PubMed] [Google Scholar]
- Matteucci M. D., Heyneker H. L. Targeted random mutagenesis: the use of ambiguously synthesized oligonucleotides to mutagenize sequences immediately 5' of an ATG initiation codon. Nucleic Acids Res. 1983 May 25;11(10):3113–3121. doi: 10.1093/nar/11.10.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movva N. R., Nakamura K., Inouye M. Amino acid sequence of the signal peptide of ompA protein, a major outer membrane protein of Escherichia coli. J Biol Chem. 1980 Jan 10;255(1):27–29. [PubMed] [Google Scholar]
- Munson L. M., Stormo G. D., Niece R. L., Reznikoff W. S. lacZ translation initiation mutations. J Mol Biol. 1984 Aug 25;177(4):663–683. doi: 10.1016/0022-2836(84)90043-3. [DOI] [PubMed] [Google Scholar]
- Scherer G. F., Walkinshaw M. D., Arnott S., Morré D. J. The ribosome binding sites recognized by E. coli ribosomes have regions with signal character in both the leader and protein coding segments. Nucleic Acids Res. 1980 Sep 11;8(17):3895–3907. doi: 10.1093/nar/8.17.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottel J. L., Sninsky J. J., Cohen S. N. Effects of alterations in the translation control region on bacterial gene expression: use of cat gene constructs transcribed from the lac promoter as a model system. Gene. 1984 May;28(2):177–193. doi: 10.1016/0378-1119(84)90255-5. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. Quantitative analysis of the relationship between nucleotide sequence and functional activity. Nucleic Acids Res. 1986 Aug 26;14(16):6661–6679. doi: 10.1093/nar/14.16.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., Weissmann C. Inhibition of Qbeta RNA 70S ribosome initiation complex formation by an oligonucleotide complementary to the 3' terminal region of E. coli 16S ribosomal RNA. Nature. 1978 Oct 26;275(5682):770–772. doi: 10.1038/275770a0. [DOI] [PubMed] [Google Scholar]
- Varenne S., Buc J., Lloubes R., Lazdunski C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol. 1984 Dec 15;180(3):549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- Wood C. R., Boss M. A., Patel T. P., Emtage J. S. The influence of messenger RNA secondary structure on expression of an immunoglobulin heavy chain in Escherichia coli. Nucleic Acids Res. 1984 May 11;12(9):3937–3950. doi: 10.1093/nar/12.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]



