Abstract
Objective
Cutaneous leishmaniasis (CL) is a significant disease in south-eastern Anatolia because it is prevalent among Syrian refugees. We identified the causative Leishmania species in CL patients using molecular methods.
Methods
Novy–MacNeal–Nicolle medium was inoculated with aspirated fluid from suspected CL lesions and tested for amastigotes with Giemsa staining. PCR amplified the internal transcribed spacer 1 (ITS1) of the Leishmania genome in cultures containing Leishmania promastigotes from 100 patients, which were genotyped with a restriction fragment length polymorphism (RFLP) analysis. A phylogenetic tree was constructed from ITS1 sequences of 95 culture fluid samples from these patients.
Results
Leishmania amastigotes were detected in 92% of cultures with growth. Leishmania promastigotes were typed as Leishmania tropica with both PCR–RFLP and sequencing.
Conclusions
Identification of L. tropica as the causative agent of CL in our region allows the clinical course to be predicted, and guides treatment decisions and preventive measures.
Keywords: Cutaneous leishmaniasis, culture, PCR–RFLP, sequence analysis, phylogenetic tree
Introduction
As in other parts of the world, cutaneous leishmaniasis (CL) is a major public health problem in Turkey. Anthroponotic CL caused by Leishmania tropica has been reported in Turkey since 1883, sometimes occurring as epidemics. Leishmaniasis is endemic to many countries in Africa, America, Asia, and Europe, but the majority of CL cases (> 90%) occur in Afghanistan, Brazil, Iran, Peru, Saudi Arabia, and Syria.1,2 Recently, millions of Syrians have entered Turkey in response to conflicts in the Syrian Arab Republic and are still residing in this country. Although refugee camps have been established in many cities, including Gaziantep, many Syrian refugees are also living in other parts of Turkey.3
The accurate diagnosis of CL, its treatment, disease prevention, strategies for its control, and management decisions require the identification of the causative species of the Leishmania parasite.4–6 The diagnosis of CL is based on clinical features and laboratory tests, including a direct parasitological examination and/or indirect testing with serology and molecular diagnostics.7 A universal PCR method targeting the internal transcribed spacer 1 (ITS1) region, which occurs between the genes encoding 18S rRNA and 5.8S rRNA, has proved useful in the direct diagnosis and identification of the Leishmania parasite because this region is highly conserved among species.8
Anthroponotic CL caused by L. tropica is highly endemic in south-eastern Anatolia and the eastern Mediterranean and Aegean regions of Turkey. In south Anatolia, both L. infantum and L. tropica have been reported as the causative agents of human CL. Leishmania major is known to be endemic in the countries bordering southern Turkey: Syria, Iraq, and Iran.9 In Syria, CL has two forms: zoonotic CL caused by L. major and anthroponotic CL caused by L. tropica.10
In this study, we identified the causative Leishmania species using PCR–restriction fragment length polymorphism (RFLP) and sequencing analyses of patients diagnosed with CL.
Materials and methods
Study population
Approval for this study was obtained from the Ethics Committee of Gaziantep University Faculty of Medicine (reference number: 221). In total, 458 patients were enrolled in the treatment program, including 433 patients of Syrian origin (94.5%) residing in refugee camps and 25 Turkish citizens (5.5%) with a diagnosis of CL, who were referred by primary and secondary health-care facilities in the city of Gaziantep to the Leishmaniasis Detection and Treatment Center established at the Department of Dermatology and Sexually Transmitted Diseases in Gaziantep University Sahinbey Research and Practice Hospital between April 2014 and April 2015. The diagnosis of CL was made based on clinical features and microscopic ± culture methods that detected the presence of amastigotes in suspected CL lesions of the patients.11,12 Of these patients, 100 (96 Syrian refugees and four Turkish citizens) in whom promastigotes were demonstrated in culture were included in the study to detect and type the Leishmania parasites. The mean age of our patients (62% males, 38% females) was 19.4 years. Culture fluids from these 100 patients were analysed with PCR–RFLP, and a sequencing analysis was also performed in 95 of these patients to type the Leishmania parasite.
Sampling
To collect aspiration fluid from the lesion sites, 0.5 ml of a sterile 0.9% sodium chloride (NaCl) solution was drawn into a syringe. After the lesion site was cleaned with 70% alcohol, the lesion was penetrated with the syringe through the margin of the lesion close to the intact skin. Sterile NaCl solution was delivered into the lesion and the intra-lesional fluid was collected into the syringe. The aspiration fluid was used to inoculate Now–McNeal–Nicolle culture medium (Besimik Ltd, Sti, Turkey), and a portion of the collected sample was transferred onto a clean slide to prepare a smear. The smear was then stained with Giemsa staining solution and examined under a light microscope (Olympus CX31, Tokyo, Japan) with a 100× objective lens. Preparations showing amastigotes were considered to be positive (+) for Leishmania spp. and preparations with no amastigotes were considered negative (–) for Leishmania spp. All results were recorded. The inoculated culture tubes were stored at 23–25℃ and monitored every second day for 1 month for the presence of promastigotes. Promastigotes were detected with a direct microscopic examination and Giemsa staining of the culture fluid.11 The samples of culture fluid containing promastigotes were collected in sterile Eppendorf tubes and stored at −80℃ until typing.
Isolation of Leishmania DNA
Leishmania spp. DNA was isolated from the culture fluids with the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) and stored at −20℃.
PCR–RFLP analysis of ITS1 region
The forward primer (LITSR) 5′-CTGGATCATTTTCCGATG-3′ and reverse primer (L5.8S) 5′-TGATACCACTTATCGCACTT-3′, specific to the ribosomal ITS1 region that occurs between the genes encoding the small subunit (18S) ribosomal RNA and 5.8S rRNA, were used in this study.13,14 A 50 µl reaction mix was prepared in a PCR tube by adding 1 µl of 10 nmol forward primer, 1 µl of 10 nmol reverse primer, 25 µl of the Taq 2× master mix (25 mM Taq 2× master mix; New England Biolabs Inc., UK), 13 µl of nuclease-free water, and 10 µl of template DNA. The 50 µl reaction mix was processed in a thermal cycler (Explera, Olly 96, Italy) using the programmed PCR protocol: one cycle at 95℃ for 30 s; 30 cycles of 20 s at 95℃, 40 s at 56℃, and 60 s at 68℃; one cycle at 68℃ for 5 min; and hold at 4℃. A 1.5% agarose gel was prepared containing 3.75 g of agarose (AB Analitica, Italy), 250 ml of 10× TBE solution (Bio Basic Canada Inc., Canada), and 25 µl of ethidium bromide (Amresco Inc., OH, USA). A 10 µl aliquot of the PCR product was loaded onto the agarose gel together with 10-bp and 100-bp markers and the DNA of L. infantum, L. major, and L. tropica reference strains (as the positive controls), after combination with 3 µl of loading dye. The samples were run on the agarose gel for 1–1.5 h at 80 mA and 130 V, and visualized with a UV transilluminator (UVP Upland, USA). The DNA of reference strains L. infantum MHOM/TN/1980/IPT1, L. major MHOM/IR/173, and L. tropica MHOM/AZ/1974/SAF-K27 was used in the analysis.
RFLP
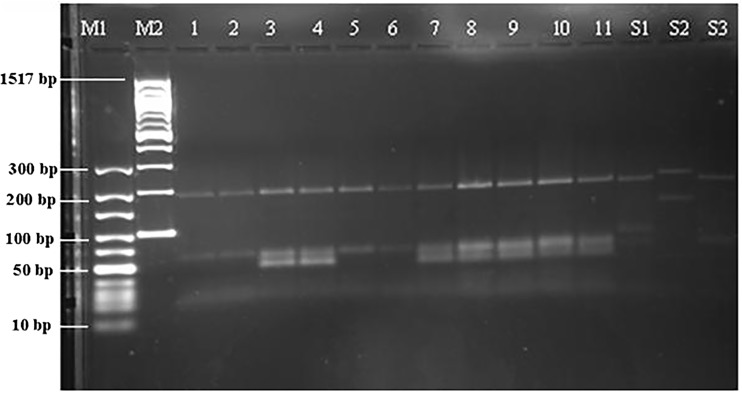
The RFLP procedure was performed with the HaeIII restriction enzyme (New England BioLabs, Inc.).13 In this step, 2.5 µl of 10× buffer, 1.5 µl of H2O, 1 µl of HaeIII restriction enzyme, and 15 µl of PCR product were added to a PCR tube and incubated at 37℃ for 2 h. A 2% agarose gel was prepared containing 5 g of Metaphor agarose (BioShop Canada Inc., Canada), 250 ml of 10× TBE solution (Bio Basic Canada Inc.), and 25 µl of ethidium bromide (Amresco Inc.). The PCR products digested with the restriction enzyme were loaded onto the gel together with 10-bp and 100-bp markers and the DNA of the L. infantum, L. major, and L. tropica reference strains (as positive controls), after combination with 3 µl of loading dye. The samples were run for 3–4 h at 80 mA and 130 V, and visualized with a UV transilluminator (UVP Inc., Upland, CA, USA). The samples were evaluated by comparing them with the markers and positive controls (Figure 1).
Figure 1.
PCR-RFLP image. M1:10 bp marker, M2: 100 bp marker, Patient no: 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, S1: L. infantum MHOM/TN/1980/IPT1, S2: L. major MHOM/IR/173, S3: L. tropica MHOM/AZ/1974/SAF-K27.
Sequencing the ITS1 region
The DNA extracted from the Leishmania samples was analysed in the laboratories of Iontek Anonim Sirketi (Iontec A.S., Istanbul, Turkey). The extracted DNA was amplified by PCR using primers specific for Leishmania. The primers used for sequencing (LITSR 5′-CTGGATCATTTTCCGATG-3′ and L5.8S 5′-TGATACCACTTATCGCACTT-3′) targeted the ITS1 region. After the PCR procedure, the PCR product was run on an agarose gel together with a marker of known size and concentration to check whether the PCR reaction had occurred. The samples visualized on the gel were purified and then sequenced with an ABI Prism 310 Genetic Analyzer using an ABI PRISM® BigDye Terminator Cycle Sequencing Kit. A BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was performed for all the sequences and the sequences were typed.
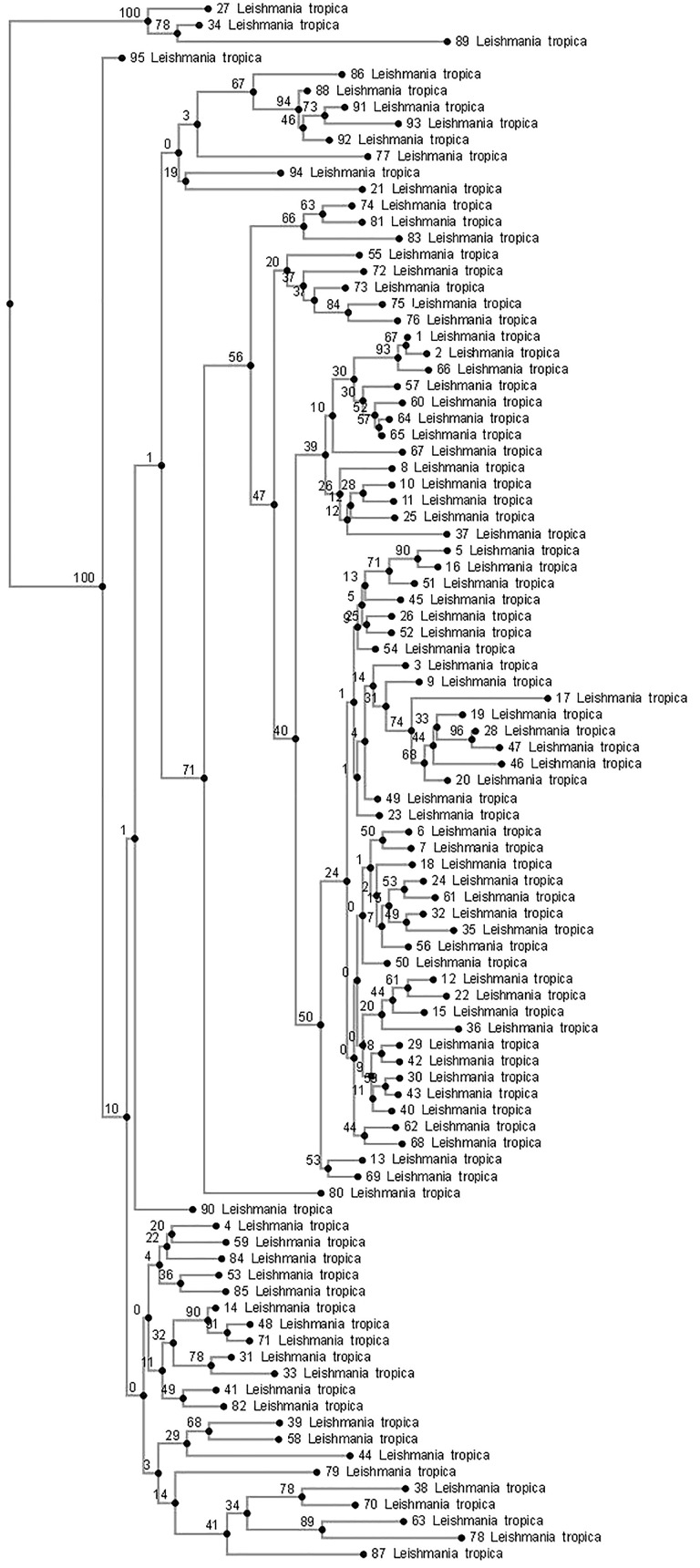
A phylogenetic tree was constructed by the Iontek Anonim Sirketi laboratory (Iontec A.S.). The phylogenetic relationships were inferred from an ITS1 nucleotide sequence alignment produced with the MAFFT multiple alignment program using a combination of the E-INS-i alignment options.15 ‘Neighbor joining method conserved sites’ was chosen as the method of phylogenetic tree construction. For the bootstrap analysis, 1000 re-samplings was assigned in the program code.
Results
The 100 patient samples that showed growth of Leishmania promastigotes in culture were included in the study and genotyped. A light microscopic examination of the aspirated samples stained with Giemsa staining solution showed the presence of Leishmania amastigotes in 92 (92%) patients and an absence of amastigotes in the remaining eight patients (8%). A PCR–RFLP analysis of the DNA from the Leishmania promastigotes grown in culture identified all of them as L. tropica (Figure 1). A sequencing analysis of 95 of the Leishmania DNA samples confirmed the presence of L. tropica. A phylogenetic tree constructed from these sequences and the genetic heterogeneity of the L. tropica parasite are shown in Figure 2.
Figure 2.
Numbers on the branches of the phylogenetic tree indicate bootstrap values and numbers given next to L. tropica indicate the number of study patients. The phylogenetic tree shows L. tropica genotypes isolated from Turkish patients (no. 12, 37, 58 and 68) and Syrian refugees.
Discussion
Leishmaniasis is endemic in more than 80 countries. Its overall global prevalence is estimated to be around 12 million cases and this figure increases by 1.5–2 million each year.16 According to data for 2008 from the World Health Organization (WHO), CL occurs in 82 countries and 1.5 million new cases are recorded every year.17 Migration, travel, and ecological changes contribute to this increase. Host, parasite, and vector characteristics determine the occurrence and course of the disease.2 CL is highly prevalent in our neighbouring countries of Iraq, Iran, and Syria and there are massive migratory flows into Turkey from these countries, both controlled and uncontrolled. According to a report issued by the United Nations High Commissioner for Refugees, 2,750,000 Syrian refugees are currently living in Turkey. Some Syrians with manifest CL have come to Turkey and live in camps or various cities.3 According to a WHO Report of 2010, Syria is one of the countries most affected by CL, with more than 25,000 cases per year.18 CL has been endemic in the Aleppo region of Syria for a very long time. However, there has been a gradual increase in the number of cases reported since the 1990s, with a maximum number of 58,156 cases reported in 2011 in the Idlib, Hamah, and Halab Provinces.19 An outbreak has occurred with the current war in Syria and measures to combat the disease are lacking, particularly in the besieged and medically underserved areas. New publications from the Turkish Ministry of Health reveal shocking statistics. An incidence rate of 53,000 cases was observed in 2012, and 41,000 cases were reported in the first 6 months of 2013.20
The identification of the different species of the Leishmania parasite is based on an isoenzyme analysis or molecular biological methods.21 In the present study, Leishmania promastigotes isolated from patient samples were typed with PCR–RFLP and DNA sequencing using primers specific for the ITS1 region. The ITS1 fragment that were similar to those obtained with standard Leishmania strains was chosen as the target for the diagnostic PCR analysis,22 because ITS1 of the ribosomal DNA repeat unit has previously been used to distinguish Old World Leishmania species using RFLP and DNA sequencing.22,23 The sensitivity of the ITS1 analysis has been demonstrated and PCR–RFLP with HaeIII is an efficient technique for the identification of Leishmania species.22
In patients with CL, the lesions differ throughout the clinical course of the disease and in response to treatment, depending on the causative agent. In the present study, the Leishmania species isolated from samples from Turkish citizens was identified as L. tropica. Leishmania tropica is the predominant cause of CL in the south-eastern Anatolia region,2 although in recent years, L. major and L. donovani have also been shown to be causative agents of CL in Turkey.24 Toz et al.23 examined the parasites in samples obtained from humans and dogs living in 31 cities in Turkey, with the majority of samples originating from Izmir, Aydın, Hatay, and Sanliurfa, and identified 80.43% of the human and canine visceral leishmaniasis samples as L. infantum and 6.52% as L. tropica, and 52.46% of the CL samples as L. infantum and 26.90% as L. tropica.
In the present study, the Leishmania parasite isolated from lesions of Syrian refugees was L. tropica and no other species was identified. Several studies have reported that the most prevalent causative agents of CL in Syria are L. tropica8,25 and L. major.26 Consistent with our findings, Yehia et al.27 established L. tropica as the causative Leishmania species in biopsy samples isolated from lesions of Lebanese, Syrian, and Saudi Arabian patients with suspected Leishmaniasis, using PCR–RFLP. In Lebanon, Saroufim et al.28 identified L. tropica in 85% and L. major in 15% of Leishmania isolated from the lesions of Syrian refugees with CL.
Significant heterogeneity was observed in the genetic structure of the L. tropica parasite that was analysed phylogenetically in the present study. Leishmania tropica is a very heterogeneous species and a high degree of intra-species polymorphism has been described based on an isoenzyme analysis and other molecular methods.29,30 This genetic variation may produce different phenotypes that are associated with a diversity of clinical manifestations and geographic distributions.31 In their study, Ajaoud et al.32 reported that an analysis of the ITS1–5.8S rRNA gene–ITS2 sequence in 31 specimens of the Phlebotomus sergenti vector showed great heterogeneity among L. tropica, segregating them into 16 haplotypes and demonstrating their phylogenetic relatedness to Indian strains and one Moroccan strain isolated from a CL patient. The heterozygosity of L. tropica isolates has also been suggested in a previous study.33 Schwenkenbecher et al.34 developed 16 polymorphic microsatellite markers for the phylogenetic analysis of L. tropica. They found a high degree of allelic heterozygosity among the strains, which suggested sexual recombination within the species. The variations reported in the Old World Leishmania species using ITS1 PCR–RFLP analyses range from highest to lowest in the order: L. tropica > L. aethiopica > L. major > L. donovani.9 Eroglu et al.35 reported that DNA sequencing revealed genetic variation in both L. infantum (variants 1–3) and L. tropica (variants 1–5), and suggested that the increase in disease occurrence may be attributable to the geographic expansion of the disease, changing patterns of international travel, population migrations, and the entry of non-immune people into endemic regions and of infected people into non-endemic regions.
Conclusion
There are many diseases prevalent among the Syrian and Iraqi refugees fleeing the civil wars in their countries, primarily CL. The risk of epidemics is greater for anthroponotic infections because of the transmission cycle from infected human to vector to human. Infected migrants may carry the disease to our region, particularly Nizip and its surrounds, where environmental/climatic conditions favour the vector. There is also a risk that the disease will spread to other regions. The identification of the causative Leishmania parasite in the cases of CL detected in our region will guide treatment decisions and the implementation of the necessary preventive measures and vector control strategies.
Acknowledgements
This study was supported by the Gaziantep University Scientific Research Projects Management Unit (project number TF.14.05). The DNA of the L. infantum MHOM/TN/1980/IPT1, L. major MHOM/IR/173, and L. tropica MHOM/AZ/1974/SAF-K27 reference strains used in this study was provided by Prof. Dr Ahmet Ozbilgin, founder of the Parasite and DNA Bank at Celal Bayar University, to whom we are grateful.
Declaration of conflicting interest
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1.Özbel Y, Töz SÖ. Leishmaniosis. In: Özcel MA. (ed). Tıbbi parazit hastaliklari 1, Baskı, İzmir: Mete Basım Matbaacılık Hizmetleri, 2007, pp. 198–230. [Google Scholar]
- 2.Gürel MS, Yeş ilova Y, Olgen MK, et al. Cutaneous leishmaniasis in Turkey. Turkiye Parazitol Derg 2012; 36: 121–129. [in Turkish, English Abstract]. [DOI] [PubMed] [Google Scholar]
- 3.UNHCR - BM Mülteciler Yüksek Komiserliği (www.unhcr.org.tr).
- 4.Wortmann G, Hochberg L, Houng HH, et al. Rapid identification of Leishmania complexes by a real-time PCR assay. Am J Trop Med Hyg 2005; 73: 999–1004. [PubMed] [Google Scholar]
- 5.Ben Abda I, de Monbrison F, Bousslimi N, et al. Advantages and limits of real-time PCR assay and PCR-restriction fragment length polymorphism for the identification of cutaneous Leishmania species in Tunisia. Trans R Soc Trop Med Hyg 2011; 105: 17–22. [DOI] [PubMed] [Google Scholar]
- 6.Jirkù M, Zemanová E, Al-Jawabreh A, et al. Development of a direct species-specific PCR assay for differential diagnosis of Leishmania tropica. Diagn Microbiol Infect Dis 2006; 55: 75–79. [DOI] [PubMed] [Google Scholar]
- 7.de Viries HJ, Reedijk SH, Schallig HD. Cutaneous leishmaniasis: recent developments in diagnosis and management. Am J Clin Dermatol 2015; 16: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Nahhas SA, Kaldas RM. Characterization of leishmania species isolated from cutaneous human samples from central region of Syria by RFLP analysis. ISRN Parasitol 2013; 2013: 308726–308726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toz SO, Nasereddin A, Ozbel Y, et al. Leishmaniasis in Turkey: molecular characterization of Leishmania from human and canine clinical samples. Trop Med Int Health 2009; 14: 1401–1406. [DOI] [PubMed] [Google Scholar]
- 10.Knio K, Baydoun E, Tawk R, et al. Isoenzyme characterization of Leishmania isolates from Lebanon and Syria. Am J Trop Med Hyg 2000; 63: 43–47. [DOI] [PubMed] [Google Scholar]
- 11.Özbel Y and Töz SO. Leishmaniasis. In: Korkmaz M and Ok ÜZ (ed) Parazitolojide laboratuar. Türkiye Parazitoloji Derneği, Yayın No:23, 2011, pp.307–320.
- 12.Handler MZ, Patel PA, Kapila R, et al. Cutaneous and mucocutaneous leishmaniasis: differential diagnosis, diagnosis, histopathology, and management. J Am Acad Dermatol 2014; 73: 911–926. [DOI] [PubMed] [Google Scholar]
- 13.Manual molecular procedures. Training course Molecular Epidemiology Leishmaniasis. Rio de Janeiro Brazil: Instituto Oswaldo Cruz, 2009, pp. 27–32. Available at: http://clioc.fiocruz.br/documents/mmp.pdf.
- 14.el Tai NO, Osman OF, el Fari M, et al. Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans R Soc Trop Med Hyg 2000; 94: 575–579. [DOI] [PubMed] [Google Scholar]
- 15.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 2008; 9: 286–298. [DOI] [PubMed] [Google Scholar]
- 16.Ameen M. Cutaneous leishmaniasis: advances in disease pathogenesis, diagnostics and therapeutics. Clin Exp Dermatol 2010; 35: 699–705. [DOI] [PubMed] [Google Scholar]
- 17.WHO. Report of the consultative meeting on cutaneous leishmaniasis Geneva, WHO Headquarters, 30 April to 2 May 2007; WHO/HTM/NTD/IDM/2008.7 http://www.who.int/leishmaniasis/resources/Cutaneous_leish_cm_2008.pdf.
- 18.Anonymous, WHO Working to overcome the global impact of neglected tropical diseases. First WHO report on neglected tropical diseases, 2010, pp.91–96.
- 19.Salam N, Al-Shaqha WM, Azzi A. Leishmaniasis in the middle East: incidence and epidemiology. PLOS Negl TropDis 2014; 8: e3208–e3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National reports for the Ministry of Health, Syrian Arab Republic. (2012, 2013) http://www.moh.gov.sy.
- 21.Klaus SN, Frankenburg S, Ingber A. Epidemiology of cutaneous leishmaniasis. Clin Dermatol 1999; 17: 257–260. [DOI] [PubMed] [Google Scholar]
- 22.Schönian G, Nasereddin A, Dinse N, et al. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis 2003; 47: 349–358. [DOI] [PubMed] [Google Scholar]
- 23.Toz SO, Culha G, Zeyrek FY, et al. A real-time ITS1-PCR based method in the diagnosis and species identification of Leishmania parasite from human and dog clinical samples in Turkey. PLOS Negl Trop Dis 2013; 7: e2205–e2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koltas IS, Eroglu F, Alabaz D, et al. The emergence of Leishmania major and Leishmania donovani in southern Turkey. Trans R Soc Trop Med Hyg 2014; 108: 154–158. [DOI] [PubMed] [Google Scholar]
- 25.Haddad N, Saliba H, Altawil A, et al. Cutaneous leishmaniasis in the central provinces of Hama and Edlib in Syria: vector identification and parasite typing. Parasit Vectors 2015; 8: 524–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization. Leishmaniasis, http://www.who.int/leishmaniasis/resources/SYRIAN_ARAB_REPUBLIC.pdf (Accessed 15 September 2014).
- 27.Yehia L, Adib-Houreih M, Raslan WF, et al. Molecular diagnosis of cutaneous leishmaniasis and species identification: analysis of 122 biopsies with varied parasite index. J Cutan Pathol 2012; 39: 347–355. [DOI] [PubMed] [Google Scholar]
- 28.Saroufim M, Charafeddine K, Issa G, et al. Ongoing epidemic of cutaneous leishmaniasis among Syrian refugees, Lebanon. Emerg Infect Dis 2014; 20: 1712–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azmi K, Schnur L, Schonian G, et al. Genetic, serological and biochemical characterization of Leishmania tropica from foci in northern Palestine and discovery of zymodeme MON-307. Parasit Vectors 2012; 5: 121–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schönian G, Schnur L, el Fari M, et al. Genetic heterogeneity in the species Leishmania tropica revealed by different PCR-based methods. Trans R Soc Trop Med Hyg 2001; 95: 217–224. [DOI] [PubMed] [Google Scholar]
- 31.Mahnaz T, Al-jawabreh A, Kuhis K, et al. Multilocus microsatellite typing shows three different genetic clusters of Leishmania major in Iran. Microbes Infect 2011; 13: 937–942. [DOI] [PubMed] [Google Scholar]
- 32.Ajaoud M, Es-Sette N, Charrel RN, et al. Phlebotomus sergenti in a cutaneous leishmaniasis focus in Azilal province (High Atlas, Morocco): molecular detection and genotyping of Leishmania tropica, and feeding behavio. PLOS Negl Trop Dis 2015. doi:10.1371/journal.pntd.000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Odiwuor S, Muia A, Magiri C, et al. Identification of Leishmania tropica from micro-foci of cutaneous leishmaniasis in the Kenyan Rift valley. Pathog Glob Health 2012; 106: 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwenkenbecher JM, Fröhlich C, Gehre F, et al. Evolution and conservation of microsatellite markers for Leishmania tropica. Infect Genet Evol 2004; 4: 99–105. [DOI] [PubMed] [Google Scholar]
- 35.Eroglu F, Koltas IS, Alabaz D, et al. Clinical manifestations and genetic variation of Leishmania infantum and Leishmania tropica in Southern Turkey. Exp Parasitol 2015; 154: 67–74. [DOI] [PubMed] [Google Scholar]