Abstract
Host tRNAs cleaved near the anticodon occur specifically in T4-infected Escherichia coli prr strains which restrict polynucleotide kinase (pnk) or RNA ligase (rli) phage mutants. The cleavage products are transient with wt but accumulate in pnk- or rli- infections, implicating the affected enzymes in repair of the damaged tRNAs. Their roles in the pathway were elucidated by comparing the mutant infection intermediates with intact tRNA counterparts before or late in wt infection. Thus, the T4-induced anticodon nuclease cleaves lysine tRNA 5' to the wobble position, yielding 2':3'-P greater than and 5'-OH termini. Polynucleotide kinase converts them into a 3'-OH and 5' P pair joined in turn by RNA ligase. Presumably, lysine tRNA depletion, in the absence of polynucleotide kinase and RNA ligase mediated repair, underlies prr restriction. However, the nuclease, kinase and ligase may benefit T4 directly, by adapting levels or decoding specificities of host tRNAs to T4 codon usage.
Full text
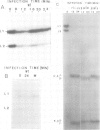
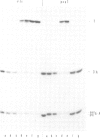
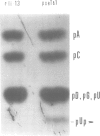
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker A., Hurwitz J. The enzymatic cleavage of phosphate termini from polynucleotides. J Biol Chem. 1967 Mar 10;242(5):936–950. [PubMed] [Google Scholar]
- Boguski M. S., Hieter P. A., Levy C. C. Identification of a cytidine-specific ribonuclease from chicken liver. J Biol Chem. 1980 Mar 10;255(5):2160–2163. [PubMed] [Google Scholar]
- Cameron V., Uhlenbeck O. C. 3'-Phosphatase activity in T4 polynucleotide kinase. Biochemistry. 1977 Nov 15;16(23):5120–5126. doi: 10.1021/bi00642a027. [DOI] [PubMed] [Google Scholar]
- Chakraburtty K., Steinschneider A., Case R. V., Mehler A. H. Primary structure of tRNA-Lys of E. coli B. Nucleic Acids Res. 1975 Nov;2(11):2069–2075. doi: 10.1093/nar/2.11.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M., Borasio G. D., Kaufmann G. Bacteriophage T4-induced anticodon-loop nuclease detected in a host strain restrictive to RNA ligase mutants. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7097–7101. doi: 10.1073/pnas.79.23.7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M., Borasio G. D., Kaufmann G. T4 bacteriophage-coded polynucleotide kinase and RNA ligase are involved in host tRNA alteration or repair. Virology. 1982 Dec;123(2):480–483. doi: 10.1016/0042-6822(82)90284-7. [DOI] [PubMed] [Google Scholar]
- David M., Vekstein R., Kaufmann G. RNA ligase reaction products in plasmolyzed Escherichia coli cells infected by T4 bacteriophage. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5430–5434. doi: 10.1073/pnas.76.11.5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew R. E., Cozzarelli N. R. Genetics and physiology of bacteriophage T4 3'-phosphatase: evidence for involvement of the enzyme in T4 DNA metabolism. J Virol. 1974 Apr;13(4):888–897. doi: 10.1128/jvi.13.4.888-897.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew R. E., Snopek T. J., Cozzarelli N. R. Characterization of a new class of deletions of the D region of the bacteriophage T4 genome. Virology. 1975 Mar;64(1):144–145. doi: 10.1016/0042-6822(75)90086-0. [DOI] [PubMed] [Google Scholar]
- Diamond A., Dudock B. Methods of RNA sequence analysis. Methods Enzymol. 1983;100:431–453. doi: 10.1016/0076-6879(83)00072-5. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry. 1978 May 30;17(11):2069–2076. doi: 10.1021/bi00604a008. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981 Feb 15;146(1):1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Jabbar M. A., Snyder L. Genetic and physiological studies of an Escherichia coli locus that restricts polynucleotide kinase- and RNA ligase-deficient mutants of bacteriophage T4. J Virol. 1984 Aug;51(2):522–529. doi: 10.1128/jvi.51.2.522-529.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano-Sueoka T., Sueoka N. Characterization of a modified leucyl-tRNA of Escherichia coli after bacteriophage T2 infection. J Mol Biol. 1968 Nov 14;37(3):475–491. doi: 10.1016/0022-2836(68)90116-2. [DOI] [PubMed] [Google Scholar]
- Kaufmann G., Amitsur M. Host transfer RNA cleavage and reunion in T4-infected Escherichia coli CTr5x. Nucleic Acids Res. 1985 Jun 25;13(12):4333–4341. doi: 10.1093/nar/13.12.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann G., David M., Borasio G. D., Teichmann A., Paz A., Amitsur M. Phage and host genetic determinants of the specific anticodon loop cleavages in bacteriophage T4-infected Escherichia coli CTr5X. J Mol Biol. 1986 Mar 5;188(1):15–22. doi: 10.1016/0022-2836(86)90476-6. [DOI] [PubMed] [Google Scholar]
- Kaufmann G., Kallenbach N. R. Determination of recognition sites of T4 RNA ligase on the 3'-OH and 5' -P termini of polyribonucleotide chains. Nature. 1975 Apr 3;254(5499):452–454. doi: 10.1038/254452a0. [DOI] [PubMed] [Google Scholar]
- Maruyama T., Gojobori T., Aota S., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1986;14 (Suppl):r151–r197. doi: 10.1093/nar/14.suppl.r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Guthrie C., Barrell B. G. Eight transfer RNAs induced by infection of Escherichia coli with bacteriophage T4. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3703–3707. doi: 10.1073/pnas.69.12.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nishimura S. Minor components in transfer RNA: their characterization, location, and function. Prog Nucleic Acid Res Mol Biol. 1972;12:49–85. [PubMed] [Google Scholar]
- Rand K. N., Gait M. J. Sequence and cloning of bacteriophage T4 gene 63 encoding RNA ligase and tail fibre attachment activities. EMBO J. 1984 Feb;3(2):397–402. doi: 10.1002/j.1460-2075.1984.tb01819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C. C. Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc Natl Acad Sci U S A. 1965 Jul;54(1):158–165. doi: 10.1073/pnas.54.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runnels J. M., Soltis D., Hey T., Snyder L. Genetic and physiological studies of the role of the RNA ligase of bacteriophage T4. J Mol Biol. 1982 Jan 15;154(2):273–286. doi: 10.1016/0022-2836(82)90064-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherberg N. H., Weiss S. B. T4 transfer RNAs: codon recognition and translational properties. Proc Natl Acad Sci U S A. 1972 May;69(5):1114–1118. doi: 10.1073/pnas.69.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber R., Malathi V. G., Hurwitz J. Purification and properties of bacteriophage T4-induced RNA ligase. Proc Natl Acad Sci U S A. 1972 Oct;69(10):3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- Sirotkin K., Cooley W., Runnels J., Snyder L. R. A role in true-late gene expression for the T4 bacteriophage 5' polynucleotide kinase 3' phosphatase. J Mol Biol. 1978 Aug 5;123(2):221–233. doi: 10.1016/0022-2836(78)90322-4. [DOI] [PubMed] [Google Scholar]
- Snopek T. J., Wood W. B., Conley M. P., Chen P., Cozzarelli N. R. Bacteriophage T4 RNA ligase is gene 63 product, the protein that promotes tail fiber attachment to the baseplate. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3355–3359. doi: 10.1073/pnas.74.8.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder L., Gold L., Kutter E. A gene of bacteriophage T4 whose product prevents true late transcription on cytosine-containing T4 DNA. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3098–3102. doi: 10.1073/pnas.73.9.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Moll J., Meissner F., Hartmann T. Compilation of tRNA sequences. Nucleic Acids Res. 1985;13 (Suppl):r1–49. doi: 10.1093/nar/13.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley J., Vassilenko S. A different approach to RNA sequencing. Nature. 1978 Jul 6;274(5666):87–89. doi: 10.1038/274087a0. [DOI] [PubMed] [Google Scholar]
- Sueoka N., Kano-Sueoka T. Transfer RNA and cell differentiation. Prog Nucleic Acid Res Mol Biol. 1970;10:23–55. doi: 10.1016/s0079-6603(08)60560-7. [DOI] [PubMed] [Google Scholar]
- Uemura H., Thorbjarnardóttir S., Gamulin V., Yano J., Andrésson O. S., Söll D., Eggertsson G. supN ochre suppressor gene in Escherichia coli codes for tRNALys. J Bacteriol. 1985 Sep;163(3):1288–1289. doi: 10.1128/jb.163.3.1288-1289.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura M., Inokuchi H., Ozeki H. Identification of transfer RNA suppressors in Escherichia coli. IV. Amber suppressor Su+6 a double mutant of a new species of leucine tRNA. J Mol Biol. 1984 Aug 25;177(4):627–644. doi: 10.1016/0022-2836(84)90041-x. [DOI] [PubMed] [Google Scholar]
- Yudelevich A. Specific cleavage of an Escherichia coli leucine transfer RNA following bacteriophage T4 infection. J Mol Biol. 1971 Aug 28;60(1):21–29. doi: 10.1016/0022-2836(71)90444-x. [DOI] [PubMed] [Google Scholar]