Abstract
Objective
To investigate the association between serpin family E member 1 (SERPINE1) -844 A/G and -675 4G/5G polymorphisms and chronic obstructive pulmonary disease (COPD) in a Chinese Han population.
Method
SERPINE1 -844 A/G and -675 4G/5G polymorphisms were assessed by polymerase chain reaction-restriction fragment length polymorphism sequencing of genomic DNA from patients with COPD and healthy smoking controls.
Results
Out of 140 patients with COPD and 100 controls, all SERPINE1 -844 and -675 polymorphisms were in Hardy-Weinberg equilibrium. Differences in SERPINE1 -675 4G and 5G allele frequencies were statistically significant between the COPD and control groups (odds ratio [OR] 1.45, 95% confidence interval [CI] 1.00, 2.09), but there was no significant between-group difference in SERPINE1 -844 A and G allele frequencies. The SERPINE1 -675 4G/4G genotype was associated with COPD (OR 1.87, 95% CI 1.06, 3.32 [binary logistic regression]). Haplotype analysis showed that COPD was associated with SERPINE1 -844G/4G (OR 2.11, 95% CI 1.32, 3.38) and SERPINE1 -844G/5G (OR 0.66, 95% CI 0.45, 0.95).
Conclusion
The SERPINE1 -675 polymorphism, but not SERPINE1 -844 polymorphism, was associated with susceptibility to COPD in a Chinese Han population.
Keywords: Chronic obstructive pulmonary disease, plasminogen activator inhibitor-1, single-nucleotide polymorphism
Introduction
Chronic obstructive pulmonary disease (COPD) is a common respiratory disease characterized by abnormal inflammatory responses of the lung to noxious particles or gases.1 Persistent airway inflammation results in airway destruction and remodelling leading to progressive airway stenosis, airflow restriction and lung function impairment. Inhalation of cigarette smoke is the most significant environmental risk factor for the development of COPD, however, less than 20% of long-term cigarette smokers develop the disease,2 suggesting that genetic factors impact the effect of cigarette smoking on COPD pathogenesis.
Plasminogen activator inhibitor-1 (PAI-1) is a member of serine protease inhibitor family. PAI-1 blocks the conversion of plasminogen to plasmin through covalent binding to plasminogen activator, thereby inhibiting activation of matrix metalloproteinases. 3 PAI-1 participates in the process of inflammation through modulating cell migration and levels of cytokines, such as tumour necrosis factor-α, interleukin-6, and interferon-γ in the lung.4,5 Thus, PAI-1 may play an important role in the pathogenesis of COPD through its effect on extracellular matrix deposition and inflammation.
The serpin family E member 1 (SERPINE1) gene, which encodes the PAI-1 protein, is located in chromosome 7q21.3–q22 and contains nine exons. Several single-nucleotide polymorphisms (SNPs) have been described in SERPINE1, such as -844 Adenine (A)/Guanine (G), -675 4G/5G, 43G/A, 9785G/A and 11053 Thymine (T)/G,6,7 of which, the promoter region -844 A/G (rs2227631) and -675 4G/5G (rs1799889) polymorphisms have been shown to affect SERPINE1 expression. The A allele in the -844 A/G substitution polymorphism is associated with increased PAI-1 mRNA and protein levels.8,9 In terms of the SERPINE1 -675 4G/5G insertion/deletion polymorphism, the 4G allele has been shown to upregulate PAI-1 mRNA and protein levels,10,11,12 and populations with the 4G/4G genotype have higher plasma PAI-1 levels than those with the 4G/5G and 5G/5G genotypes.6 SERPINE1 -844 and -675 genetic polymorphisms have been found associated with several diseases, such as stroke, rheumatoid arthritis, acute coronary syndrome and metabolic syndrome, in different populations.6,7,9,13,14
Several studies have investigated the association between SERPINE1 -675 4G/5G polymorphism and chronic pulmonary diseases, including asthma and pulmonary fibrosis.10,15,16,17,18 The SERPINE1 -675 polymorphism is shown to be associated with risk and severity of asthma, 10,15,16 and to impact the effect of inhaled glucocorticosteroid.16 Frequency of the 4G allele is significantly higher than the 5G allele in patients with nonspecific interstitial pneumonia,17 and the SERPINE1 5G/5G genotype has a significant negative association with the development of idiopathic pulmonary fibrosis.18 Thus, it may be reasonable to expect a relationship between SERPINE1 polymorphisms and COPD. One study in Egyptian male patients, found an association between the development of COPD and the SERPINE1 -675 polymorphism 4G/4G genotype.19 The present study investigated whether the SERPINE1 -844 A/G and -675 4G/5G polymorphisms were associated with susceptibility to COPD in a Chinese Han population.
Patients and methods
Study population
The present observational cohort study sequentially enrolled patients with COPD who were attending Shandong University Qilu Hospital, Jinan, China between May 2015 and December 2015 and who consented to participate in the study. COPD was diagnosed and graded according to the Global Initiative for Chronic Obstructive Lung Disease (GOLD) guidelines,1 and comprised medical history, physical examination and post-bronchodilator forced expiratory volume in one second (FEV1)/forced vital capacity (FVC) < 0.70. Patients with other respiratory diseases (e.g. asthma, lung cancer, pulmonary tuberculosis and bronchiectasis) were excluded. Healthy smokers > 45 years of age with smoking index (cigarette packs per day × number of smoking years) ≥ 10 pack-years who attended Shandong University Qilu Hospital for a routine examination were recruited as the control group. The study was approved by the Medical Ethics Committee of Shandong University and all participants provided written informed consent.
Lung function
Each participant completed a lung function test using a MasterScreen™ computerised spirometer (Jaeger Corp., Hoechberg, Germany) according to American Thoracic Society and European Respiratory Society recommendations.20 FEV1 % predicted (participant FEV1/predicted normal FEV1 value) and FVC % predicted (participant FVC/predicted normal FVC value) were calculated.
DNA extraction and genotyping
Peripheral venous blood samples (3 ml) were collected into tubes containing 1.8 mg/ml ethylenediaminetetra-acetic acid and stored at –80℃ prior to DNA extraction. Genomic DNA was isolated from peripheral blood mononuclear cells using a DNA extraction kit (Tiangen Biotech Co., Beijing, China) according to the manufacturer’s instructions, and stored at –20℃ prior to use in genotyping.
Serpin family E member 1 (SERPINE1) -844 and -675 polymorphisms were genotyped using polymerase chain reaction (PCR)-restriction fragment length polymorphism sequencing. Sense and antisense primer sequences were 5′-GGACCACTGCTCCACAGAAT-3′ and 5′-AACCTCCATCAAAACGTGGA-3′, respectively (Shanghai Sangon Biological Engineering Technology & Service, Shanghai, China), and produced an amplified fragment length of 591 bp. The PCR was performed in a 25 µl reaction volume containing 60 ng of genomic DNA, 3.2 pmol each of forward and reverse primers, 0.25 µl Taq DNA polymerase (Tiangen Biotech Co.), 200 µM dNTPs (Tiangen Biotech Co.), 2.5 µl 10 × PCR buffer (Tiangen Biotech Co.) and 17.25 µl double distilled H2O. The cycling programme involved preliminary denaturation at 95℃ for 5 min, followed by 40 cycles of denaturation at 95℃ for 30 s, annealing at 58℃ for 30 s, and elongation at 72℃ for 1 min, followed by a final elongation step at 72℃ for 5 min. The PCR products were purified and sequenced using a BigDye® Terminator v3.1 Cycle Sequencing Kit (ThermoFisher Scientific, Waltham, MA, USA) with the Applied Biosystems™ 3730xl DNA Analyzer (Applied Biosystems, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Statistical analyses
Statistical analyses were performed using SPSS software, version 17.0 (SPSS Inc., Chicago, IL, USA). Quantitative data are presented as mean ± SD and were analysed by Student’s t-test. Hardy-Weinberg equilibrium was assessed using χ2-test to evaluate the deviation of genotype distribution. Between-group differences in genotypes and alleles were analysed using χ2-test. Binary logistic regression was used to assess the association between susceptibility to COPD and SERPINE1 -844 and -675 genotypes. Odds ratios (ORs) for SERPINE1 -844 and -675 polymorphisms in recessive (wild type homozygous + heterozygous versus minor allele homozygous), dominant (wild type homozygous versus heterozygous + minor allele homozygous), codominant (heterozygous versus wild type homozygous, minor allele homozygous verse wild type homozygous) and overdominant (wild type homozygous + minor allele homozygous versus heterozygous) models were analysed. Haplotypes with frequencies > 3% were selected for analysis, and haplotype analysis was conducted using Haploview 4.2 software (Broad Institute of MIT and Harvard, Boston, MA, USA). A P value < 0.05 was considered statistically significant.
Results
A total of 140 patients with COPD were included (69 patients with COPD grades 1 and 2 and 71 patients with COPD grades 3 and 4, according to GOLD guidelines),1 and a total of 100 healthy smokers were included as the control group (Table 1). There were no statistically significant differences in sex, age and smoking index between the COPD and control groups. Statistically significant between-group differences were observed in terms of FEV1 % predicted and FVC % predicted (P < 0.01).
Table 1.
Characteristics of Chinese Han patients with chronic obstructive pulmonary disease (COPD) and healthy smokers (controls).
| Study group |
|||
|---|---|---|---|
| Characteristic | COPD (n = 140) | Control (n = 100) | Statistical significance |
| Sex, male/female | 101/39 | 70/30 | NS |
| Age, years | 66.0 ± 9.1 | 62.5 ± 11.2 | NS |
| Smoking index, pack-years | 35.2 ± 21.6 | 30.2 ± 19.2 | NS |
| FEV1 % predicted | 51.3 ± 18.6 | 95.3 ± 13.1 | P < 0.01 |
| FVC % predicted | 77.4 ± 17.1 | 97.2 ± 12.6 | P < 0.01 |
Data presented as n participant incidence or mean ± SD.
FEV1 % predicted, participant FEV1/predicted normal FEV1 value; FVC % predicted, participant FVC/predicted normal FVC value.
NS, no statistically significant between-group difference (P > 0.05; Student’s t-test).
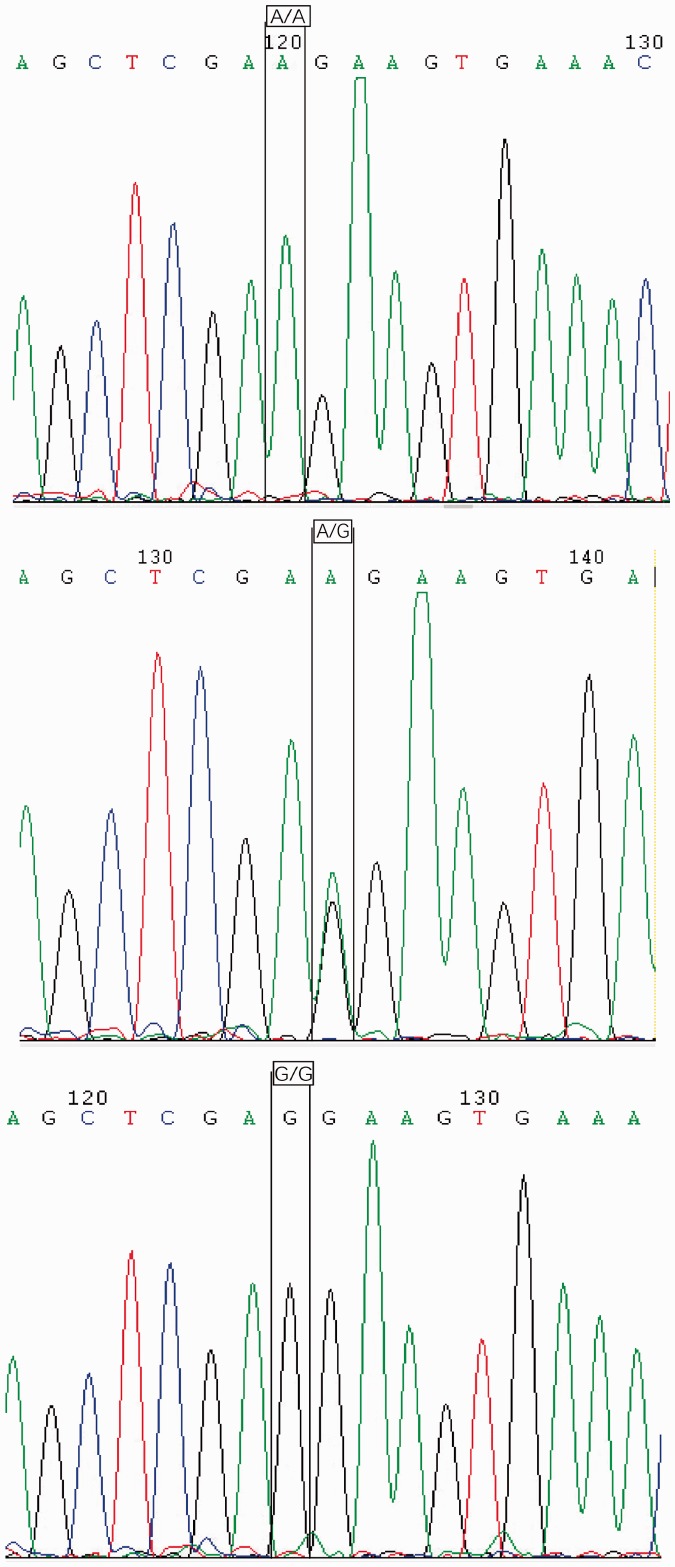
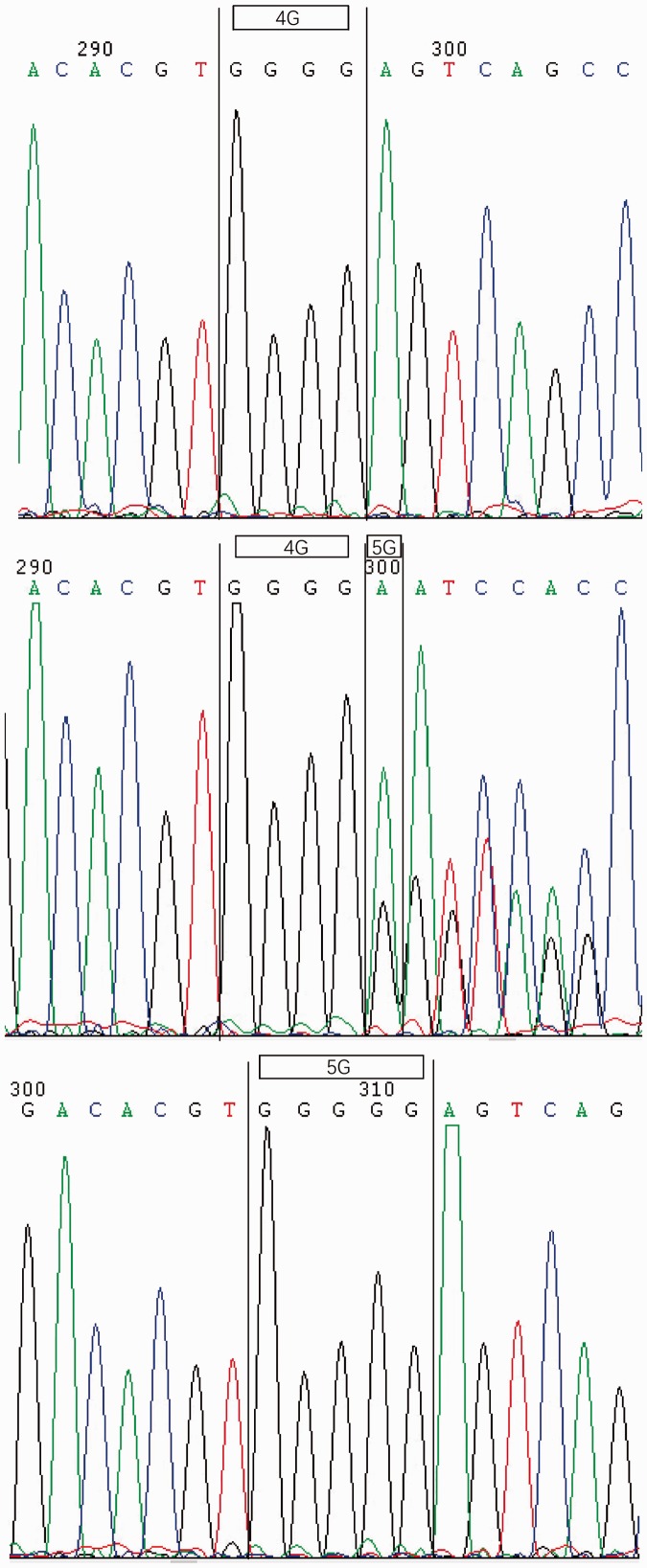
Representative electropherograms showing sequence analyses of SERPINE1 -844 A/A, A/G and G/G and SERPINE1 -675 4G/4G, 4G/5G and 5G/5G genotypes are presented in Figures 1 and 2, respectively. SERPINE1 -844 A/G and -675 4G/5G polymorphisms in the COPD and control groups were all found to be in Hardy-Weinberg equilibrium (Table 2). No statistically significant difference in genotype distribution was found between the COPD and control groups (Table 3). There was no statistically significant difference in frequency of SERPINE1 -844 polymorphism A and G alleles between the COPD and control groups, however, the frequency of SERPINE1 -675 polymorphism 4G and 5G alleles was significantly different between patients with COPD and controls (P = 0.04; OR 1.45, 95% confidence interval [CI] 1.00, 2.09; Table 4).
Figure 1.
Representative electropherogram showing sequencing analysis of serpin family E member 1 -844 polymorphisms: homozygous A/A, heterozygous A/G and homozygous G/G genotypes.
Figure 2.
Representative electropherogram showing sequencing analysis of serpin family E member 1 -675 polymorphisms: homozygous 4G/4G, heterozygous 4G/5G and homozygous 5G/5G genotypes.
Table 2.
Hardy-Weinberg equilibrium of serpin family E member 1 (SERPINE1) -844 and -675 polymorphisms in patients with chronic obstructive pulmonary disease (COPD; n = 140) and healthy smoking controls (n = 100).
| Study group |
SERPINE1 -844 A/G |
χ2 | Statistical significance |
SERPINE1 -675 4G/5G |
χ2 | Statistical significance | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| A/A | A/G | G/G | 4G/4G | 4G/5G | 5G/5G | |||||
| COPD | 18 | 69 | 53 | 0.37 | NS | 55 | 65 | 20 | 0.01 | NS |
| Control | 13 | 51 | 36 | 0.59 | NS | 26 | 55 | 19 | 1.11 | NS |
Data presented as n prevalence.
NS, no statistically significant between-group difference (P > 0.05; χ2-test).
Table 3.
Genotype frequencies of serpin family E member 1 (SERPINE1) -844 and -675 polymorphisms in patients with chronic obstructive pulmonary disease (COPD; n = 140) and healthy smoking controls (n = 100).
| Study group |
SERPINE1 -844 A/G |
SERPINE1 -675 4G/5G |
||||
|---|---|---|---|---|---|---|
| A/A | A/G | G/G | 4G/4G | 4G/5G | 5G/5G | |
| COPD | 18 (12.9) | 69 (49.3) | 53 (37.9) | 55 (39.3) | 65 (46.4) | 20 (14.3) |
| Control | 13 (13.0) | 51 (51.0) | 36 (36.0) | 26 (26.0) | 55 (55.0) | 19 (19.0) |
| Statistical significance | χ2 = 0.09 | NS | χ2 = 4.71 | NS | ||
Data presented as n (%) prevalence.
NS, no statistically significant between-group difference (P > 0.05; χ2-test).
Table 4.
Allele frequencies of serpin family E member 1 (SERPINE1) -844 and -675 polymorphisms in patients with chronic obstructive pulmonary disease (COPD, n = 140) and healthy smoking controls (n = 100).
| Study group |
Statistical significance | ||||||
|---|---|---|---|---|---|---|---|
| Polymorphism | Allele | COPD | Control | χ 2 | OR | 95% CI | |
| SERPINE1 -844 | A | 105 (37.5) | 77 (38.5) | 0.05 | NS | 0.96 | 0.66, 1.39 |
| G | 175 (62.5) | 123 (61.5) | |||||
| SERPINE1 -675 | 4G | 175 (62.5) | 107 (53.5) | 3.90 | P = 0.04 | 1.45 | 1.00, 2.09 |
| 5G | 105 (37.5) | 93 (46.5) | |||||
Data presented as n (%) prevalence.
OR, odds ratio; CI, confidence interval.
NS, no statistically significant between-group difference (P > 0.05; χ2-test).
Binary logistic regression analyses of the whole study population showed that only the SERPINE1 -675 4G/4G genotype was associated with susceptibility to COPD (P = 0.03; OR 1.87, 95% CI 1.06, 3.32). Other genotypes failed to show any association with susceptibility to COPD (P > 0.05; Table 5).
Table 5.
Binary logistic regression analysis showing association between serpin family E member 1 (SERPINE1) polymorphisms and susceptibility to chronic obstructive pulmonary disease (COPD) in patients with COPD (n = 140) and healthy smoking controls (n = 100).
| Genotype | Statistical significance | OR | 95% CI |
|---|---|---|---|
| SERPINE1 -844 recessive | NS | 1.08 | 0.64, 1.84 |
| dominant | NS | 0.99 | 0.46, 2.12 |
| codominanta | NS | 1.02 | 0.46, 2.28 |
| codominantb | NS | 0.94 | 0.41, 2.15 |
| overdominant | NS | 0.93 | 0.56, 1.56 |
| SERPINE1 -675 recessive | NS | 0.84 | 0.60, 1.19 |
| dominant | P = 0.03 | 1.87 | 1.06, 3.32 |
| codominanta | NS | 1.12 | 0.54, 2.31 |
| codominantb | NS | 2.01 | 0.92, 4.39 |
| overdominant | NS | 1.41 | 0.84, 2.36 |
OR, odds ratio; CI, confidence interval.
Heterozygous versus wild type homozygous; bminor allele homozygous verse wild type homozygous.
NS, no statistically significant correlation between SERPINE1 polymorphism and susceptibility to COPD (P > 0.05).
In terms of haplotype analyses of the whole study population, SERPINE1 -844G/4G showed a significantly positive association with susceptibility to COPD (P = 0.001; OR 2.11, 95% CI 1.32, 3.38) and SERPINE1 -844G/5G showed a statistically significant inverse association with susceptibility to COPD (P = 0.03; OR 0.66, 95% CI 0.45, 0.95; Table 6).
Table 6.
Haplotype analysis of serpin family E member 1 -844 and -675 polymorphisms and association with susceptibility to chronic obstructive pulmonary disease (COPD) in patients with COPD (n = 140) and healthy smoking controls (n = 100).
| Study group |
|||||
|---|---|---|---|---|---|
| Haplotype | COPD | Control | Statistical significance | OR | 95% CI |
| −844A/4G | 100 (35.7) | 77 (38.5) | NS | 0.91 | 0.62, 1.33 |
| −844G/4G | 75 (26.8) | 30 (15.0) | P = 0.001 | 2.11 | 1.32, 3.38 |
| −844G/5G | 100 (35.7) | 93 (46.5) | P = 0.03 | 0.66 | 0.45, 0.95 |
Data presented as n (%) prevalence.
OR, odds ratio; CI, confidence interval.
NS, no statistically significant correlation between haplotype and susceptibility to COPD (P > 0.05).
Discussion
The present study investigated the association between SERPINE1 -844 and -675 polymorphisms and susceptibility to COPD in patients with COPD and healthy smokers in a Chinese Han population. In agreement with a published study conducted in Egypt,19 the SERPINE1 -675 4G/5G polymorphism was found to be associated with susceptibility to COPD in the present study. The SERPINE1 -844 polymorphism was not found to be associated with susceptibility to COPD.
In the present study, frequencies of SERPINE1 -844 A/A genotype and A allele in healthy controls were 13.0% and 38.5%, respectively, and were similar to those previously described in a Chinese study population of 19.3% and 43.8%, respectively.21 Frequencies of SERPINE1 -675 4G/4G genotype and 4G allele in healthy controls (26% and 53.5%, respectively) were also similar to previously published frequencies in Chinese (28% and 54.1%, respectively) and Polish (29.4% and 51.25%, respectively) populations.14,22
COPD is a prevalent chronic airway inflammatory disease associated with persisting airway inflammation and remodelling that results in airway obstruction during disease progression. Besides environmental factors, genetic factors are reported to affect the development of COPD.2,23,24
Levels of PAI-1 are increased in the lung tissues of patients with COPD, and are associated with alveolar epithelial cell apoptosis and exacerbation of lung inflammation in mice with passive cigarette smoke exposure.21 PAI-1 levels in the induced sputum of patients with COPD are significantly elevated and negatively related to FEV1 % predicted,25 and correlate with sputum malondialdehyde (MDA) in patients with COPD.26 In addition, a positive correlation has been reported between the amount of collagen and the percentage of PAI-1-positive macrophages in pulmonary specimens from patients with COPD.27 PAI-1 affects the expression of cytokines (interleukin-8 and leukotriene B4) and monocyte migration in the inflammation induced by cigarette smoke extract.5 As an important factor which impacts airway inflammation and remodelling, PAI-1 appears to play an essential role in the development of asthma and COPD. The present results suggest an association between PAI-1 and COPD from the aspect of genetic polymorphisms, in that there was a significant difference between the SERPINE1 -675 polymorphism 4G and 5G allele between patients with COPD and healthy smoking controls. The carriers of SERPINE1 4G/4G genotype were shown to have increased odds of COPD compared with the carriers of other genotypes. It was also apparent in haplotype analysis that the SERPINE1 -844G/4G haplotype was a risk factor for COPD and -844G/5G haplotype was a protective factor for COPD. It may be the case that the 4G allele upregulates expression of SERPINE1 compared with the 5G allele, and it may further promote airway remodelling and inflammation.
The present results may be limited by the fact that this was a single-centre study, the study population was relatively small, and only SERPINE1 -844 and -675 polymorphisms were observed. Future studies to identify the role of SERPINE1 polymorphisms in the development of COPD should include a larger study population and a larger selection of SERPINE1 polymorphisms in addition to -844 and -675.
In conclusion, SERPINE1 -675 4G/5G polymorphism was associated with susceptibility to COPD in the present Chinese Han population, however, SERPINE1 -844 polymorphism did not appear to impact COPD development.
Acknowledgment
The authors are grateful to Professor Qiji Liu of Department of Medical Genetics, Shandong University, for his assistance on statistical analyses.
Declaration of conflicting interests
The authors declare that there is no conflict of interest.
Funding
This work was supported by Grants from Qilu Hospital (No. 2015QLQN04).
References
- 1.Vestbo J, Hurd SS, Agustí AG, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med 2013; 187: 347–365. [DOI] [PubMed] [Google Scholar]
- 2.Lomas DA, Silverman EK. The genetics of chronic obstructive pulmonary disease. Respir Res 2001; 2: 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmeliet P, Moons L, Lijnen R, et al. Urokinase-generated plasmin activates matrix metalloproteinases during aneurysm formation. Nat Genet 1997; 17: 439–444. [DOI] [PubMed] [Google Scholar]
- 4.Renckens R, Roelofs JJ, Bonta PI, et al. Plasminogen activator inhibitor type 1 is protective during severe gram-negative pneumonia. Blood 2007; 109: 1593–1601. [DOI] [PubMed] [Google Scholar]
- 5.Xu X, Wang H, Wang Z, et al. Plasminogen activator inhibitor-1 promotes inflammatory process induced by cigarette smoke extraction or lipopolysaccharides in alveolar epithelial cells. Exp Lung Res 2009; 35: 795–805. [DOI] [PubMed] [Google Scholar]
- 6.Morange PE, Saut N, Alessi MC, et al. Association of plasminogen activator inhibitor (PAI)-1 (SERPINE1) SNPs with myocardial infarction, plasma PAI-1, and metabolic parameters: the HIFMECH study. Arterioscler Thromb Vasc Biol 2007; 27: 2250–2257. [DOI] [PubMed] [Google Scholar]
- 7.Abboud N, Ghazouani L, Saidi S, et al. Association of PAI-1 4G/5G and -844G/A gene polymorphisms and changes in PAI-1/tissue plasminogen activator levels in myocardial infarction: a case-control study. Genet Test Mol Biomarkers 2010; 14: 23–27. [DOI] [PubMed] [Google Scholar]
- 8.Ma Z, Jhun B, Jung SY, et al. Binding of upstream stimulatory factor 1 to the E-box regulates the 4G/5G polymorphism-dependent plasminogen activator inhibitor 1 expression in mast cells. J Allergy Clin Immunol 2008; 121: 1006–1012. [DOI] [PubMed] [Google Scholar]
- 9.Ma Z, Paek D, Oh CK. Plasminogen activator inhibitor-1 and asthma: role in the pathogenesis and molecular regulation. Clin Exp Allergy 2009; 39: 1136–1144. [DOI] [PubMed] [Google Scholar]
- 10.Nie W, Li B, Xiu QY. The −675 4G/5G polymorphism in plasminogen activator inhibitor-1 gene is associated with risk of asthma: a meta-analysis. PLoS One 2012; 7: e34385–e34385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burzotta F, Di Castelnuovo A, Amore C, et al. 4G/5G promoter PAI-1 gene polymorphism is associated with plasmatic PAI-1 activity in Italians: a model of gene-environment interaction. Thromb Haemost 1998; 79: 354–358. [PubMed] [Google Scholar]
- 12.Dawson SJ, Wiman B, Hamsten A, et al. The two allele sequences of a common polymorphism in the promoter of the plasminogen activator inhibitor-1 (PAI-1) gene respond differently to interleukin-1 in HepG2 cells. J Biol Chem 1993; 268: 10739–10745. [PubMed] [Google Scholar]
- 13.Torres-Carrillo NM, Torres-Carrillo N, Vázquez-Del Mercado M, et al. The −844 G/A PAI-1 polymorphism is associated with mRNA expression in rheumatoid arthritis. Rheumatol Int 2008; 28: 355–360. [DOI] [PubMed] [Google Scholar]
- 14.Hu X, Zan X, Xie Z, et al. Association between plasminogen activator inhibitor-1 genetic polymorphisms and stroke susceptibility. Mol Neurobiol 2016. doi:10.1007/s12035-015-9549-8. [DOI] [PubMed] [Google Scholar]
- 15.Kowal K, Bodzenta-Lukaszyk A, Pampuch A, et al. Analysis of -675 4 g/5 G SERPINE1 and C-159T CD14 polymorphisms in house dust mite-allergic asthma patients. J Investig Allergol Clin Immunol 2008; 18: 284–292. [PubMed] [Google Scholar]
- 16.Dijkstra A, Postma DS, Bruinenberg M, et al. SERPINE1 -675 4G/5G polymorphism is associated with asthma severity and inhaled corticosteroid response. Eur Respir J 2011; 38: 1036–1043. [DOI] [PubMed] [Google Scholar]
- 17.Kim KK, Flaherty KR, Long Q, et al. A plasminogen activator inhibitor-1 promoter polymorphism and idiopathic interstitial pneumonia. Mol Med 2003; 9: 52–56. [PMC free article] [PubMed] [Google Scholar]
- 18.Li XX, Li N, Ban CJ, et al. Idiopathic pulmonary fibrosis in relation to gene polymorphisms of transforming growth factor-β1 and plasminogen activator inhibitor 1. Chin Med J (Engl) 2011; 124: 1923–1927. [PubMed] [Google Scholar]
- 19.Essa ES, El Wahsh RA. Association between plasminogen activator inhibitor-1-675 4G/5G insertion/deletion polymorphism and chronic obstructive pulmonary disease. COPD. Epub ahead of print April 2016: 1–4. DOI: 10.3109/15412555.2016.1168392. [DOI] [PubMed] [Google Scholar]
- 20.Hanania NA, Marciniuk DD. A unified front against COPD: clinical practice guidelines from the American College of Physicians, the American College of Chest Physicians, the American Thoracic Society, and the European Respiratory Society. Chest 2011; 140: 565–566. [DOI] [PubMed] [Google Scholar]
- 21.Liu SF, Chen YC, Wang CC, et al. IL13 promoter (-1055) polymorphisms associated with chronic obstructive pulmonary disease in Taiwanese. Exp Lung Res 2009; 35: 807–816. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Long J, Wang X, et al. Association of the plasminogen activator inhibitor-1 (PAI-1) Gene -675 4G/5G and -844 A/G promoter polymorphism with risk of keloid in a Chinese Han population. Med Sci Monit 2014; 20: 2069–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhandary YP, Shetty SK, Marudamuthu AS, et al. Plasminogen activator inhibitor-1 in cigarette smoke exposure and influenza A virus infection-induced lung injury. PLoS One 2015; 10: e0123187–e0123187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayada C, Toru Ü, Genç O, et al. Angiotensinogen gene M235T and angiontensin II-type 1 receptor gene A/C1166 polymorphisms in chronic obstructive pulmonary disease. Int J Clin Exp Med 2015; 8: 4521–4526. [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao W, Hsu YP, Ishizaka A, et al. Sputum cathelicidin, urokinase plasminogen activation system components, and cytokines discriminate cystic fibrosis, COPD and asthma inflammation. Chest 2005; 128: 2316–2326. [DOI] [PubMed] [Google Scholar]
- 26.To M, Takagi D, Akashi K, et al. Sputum plasminogen activator inhibitor-1 elevation by oxidative stress-dependent nuclear factor-κB activation in COPD. Chest 2013; 144: 515–521. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Xiao W, Jiang Y, et al. Levels of components of the urokinase-type plasminogen activator system are related to chronic obstructive pulmonary disease parenchymal destruction and airway remodelling. J Int Med Res 2012; 40: 976–985. [DOI] [PubMed] [Google Scholar]